BAD and KATP channels regulate neuron excitability and epileptiform activity
Figures
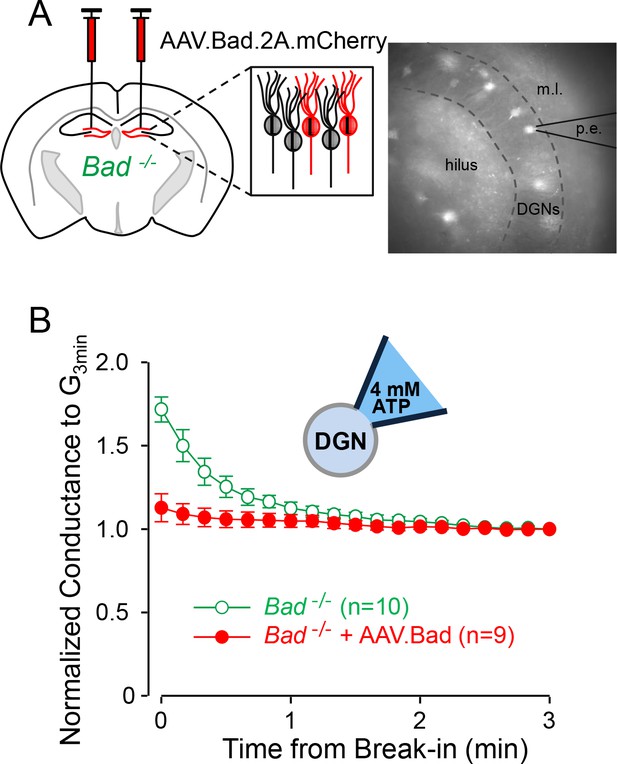
Effect of BAD on KATP channels is cell-autonomous.
(A) Left: schematic representation of intracranial injection of AAV.Bad.2A.mCherry in the hippocampus of Bad−/− mice to reconstitute BAD expression in mCherry labeled cells. Right: epifluorescence picture of a BAD-reconstituted Bad−/− DGN being recorded with a patch electrode (p.e.) in the whole-cell mode. Regions observed in picture: hilus, dentate granule cell layer (DGNs) and molecular layer (m.l.). (B) ‘Washdown’ of an initially high KATP conductance, with high ATP (4 mM) in the patch electrode, seen in Bad−/− but not BAD-reconstituted Bad−/− cells. The time course of slope conductance measured during whole-cell recording was normalized to the value 3 min after break-in. Data are presented as mean ± SEM.
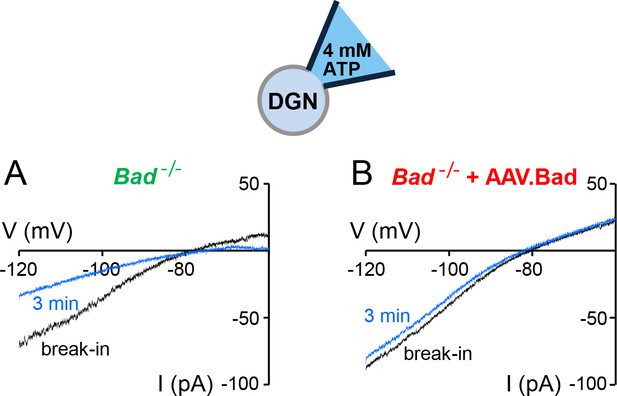
‘Washdown’ of KATP channel conductance in Bad−/− or BAD-reconstituted DGNs.
Representative whole-cell current traces with high ATP (4 mM) in the patch electrode in a Bad−/− (A) or a BAD-reconstituted DGN (B). As the high ATP concentration washed into the cell, the conductance decreased in the Bad−/− cell (A, compare initial black trace to blue trace after 3 min of recording) but remained of comparable magnitude when BAD was reconstituted via AAV.

Total activatable whole-cell KATP conductance is mostly unaffected by overexpressing BAD in Bad−/− DGNs.
(A,B) Representative whole-cell current traces with low ATP (0.3 mM) in the patch electrode (p. (e).) in a Bad−/− (A) or a BAD-reconstituted (B) DGN at the moment of break-in (black traces) or after current had maximally increased (blue traces) due to disinhibition of KATP channels. (C) Typical whole-cell KATP conductance time course with low ATP in the patch electrode in a Bad−/− DGN. Slope conductance gradually increased upon breaking into the cell as KATP channels became uninhibited. This KATP conductance ‘run-up’ was reversed by application of 200 nM glibenclamide (arrow), a KATP channel inhibitor. (D) Maximum activatable KATP conductance (mean ±SEM) determined from the difference between the slope conductance at the time of break-in and after conductance run-up (Gmax), with low ATP (0.3 mM) in the patch electrode, for Bad−/− and BAD-reconstituted Bad−/− as indicated. **p<0.01; ***p<0.001; two-tailed Student’s t-test.
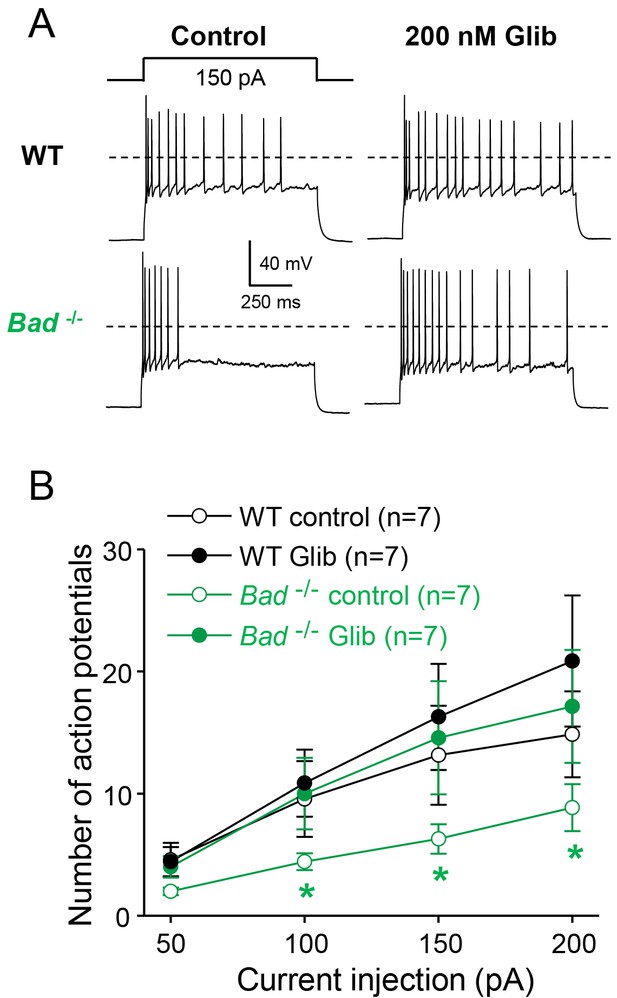
Dentate granule neurons lacking BAD are less excitable.
(A) Bad−/− DGNs fire fewer action potentials that wild-type (WT), and 200 nM glibenclamide reverses Bad knockout effect on DGN excitability. Representative perforated-patch voltage recordings in response to a 1 s, 150 pA current pulse for WT and Bad−/− DGNs, in the presence or absence of 200 nM glibenclamide, as indicated. Dotted lines indicate 0 mV. (B) Glibenclamide application increases firing of Bad−/− DGNs but not that of WT. Number of action potentials (mean ± SEM) plotted against magnitude of current injection pulse. *p<0.05; 1-way ANOVA.
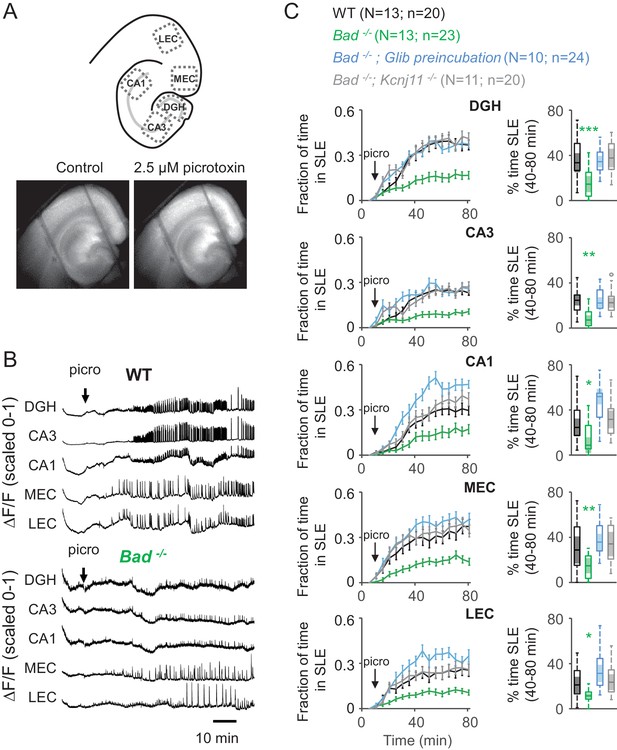
Deletion of BAD reduces picrotoxin-elicited epileptiform activity.
(A) Top: Schematic representation of the analyzed regions of interest in EC-HC acute slices; dentate gyrus/hilus (DGH), CA3, CA1, medial entorhinal cortex (MEC) and lateral entorhinal cortex (LEC). Bottom: representative epifluorescence images of an EC-HC slice expressing GCaMP6f in ACSF (Control; left) or in the presence of 2.5 µM picrotoxin as a SLE is occurring (right). (B) Picrotoxin application triggers SLEs. Representative ΔF/F traces (normalized from 0 to 1) in wild-type (WT; top) or Bad−/− (bottom) slices for the indicated regions of interest. The arrows indicate extracellular application of 2.5 µM picrotoxin in the extracellular bath. (C) Genetic deletion of BAD reduced the time EC-HC slices spent in SLEs, and this effect was mediated by KATP channels. Fraction of time spent in SLEs (mean ± SEM) plotted versus time (left column) or percent time in SLEs calculated from the 40–80 min range when seizure-like activity plateaus (right column) for each region of interest analyzed, and genotypes and conditions specified. *p<0.05; **p<0.005; ***p<0.0005; 1-way ANOVA.

Picrotoxin-triggered changes in GCaMP6f fluorescence intensity correspond to stereotypical epileptiform field potentials.
(A) Schematic representation of extracellular field potential electrode placement in CA1 or lateral entorhinal cortex (LEC). (B) Changes in GCaMP6f fluorescence faithfully represent picrotoxin-triggered field potentials. Representative field potential traces (upper) or ΔF/F traces (lower) simultaneously recorded in CA1 or LEC in the presence of 2.5 µM picrotoxin.
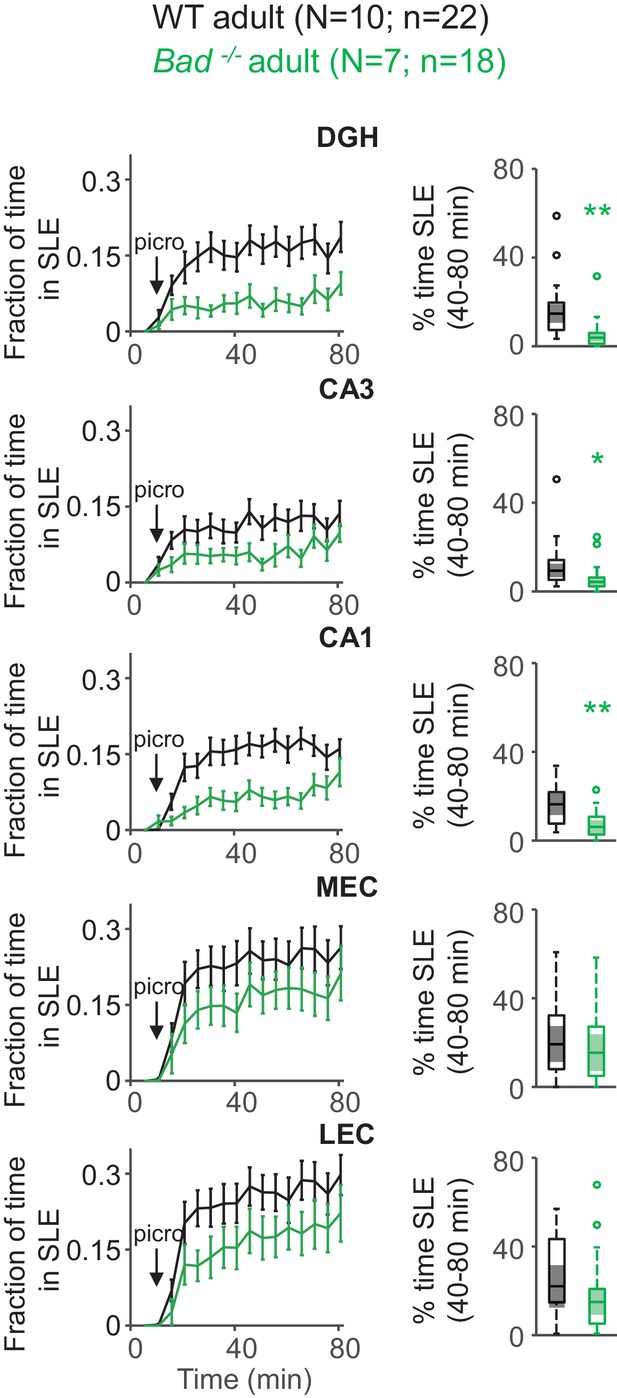
Deletion of BAD decreases picrotoxin-triggered seizure-like activity in adult EC-HC slices.
Genetic deletion of BAD reduced the time hippocampal regions spent in SLEs in adult slices. Left column: fraction of time spent in SLEs (mean ± SEM) plotted versus time. The arrows indicate extracellular application of 50 µM picrotoxin in the extracellular bath. Right column: percent time in SLEs calculated from the 40–80 min range when seizure-like activity plateaus for each region of interest analyzed and genotypes specified. *p<0.05; **p<0.005; two-tailed Student’s t-test.

Picrotoxin-elicited epileptiform activity in adult EC-HP slices.
Picrotoxin application triggers SLEs in adult EC-HP slices. Representative ΔF/F traces (normalized from 0 to 1) in adult wild-type (A) or adult Bad−/− (B) slices for the indicated regions of interest. The arrows indicate extracellular application of 50 µM picrotoxin in the extracellular bath.
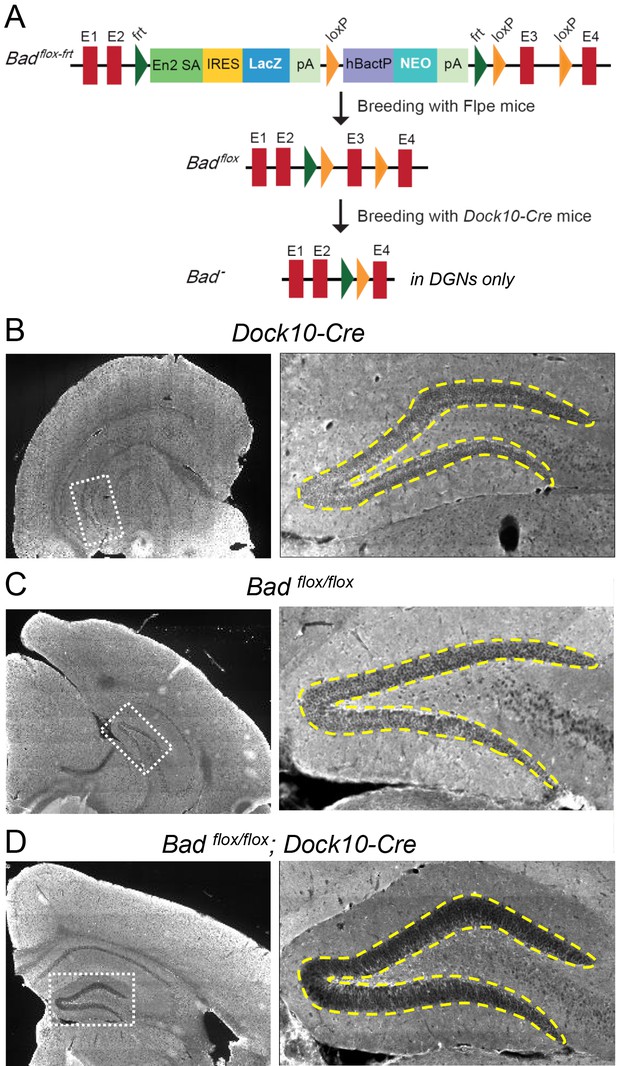
Targeted knockout of Bad expression in the dentate gyrus but not in the rest of the hippocampus.
(A) Schematic illustration of the targeting construct and strategy to generate the conditional Badflox allele. Abbreviations: En2 SA: En2 splice acceptor; IRES: Internal ribosomal entry site; pA: polyadenylation sequence; Frt: Flipase (Flpe) recombinase target; loxP: Cre recombinase target; hBactP: human beta actin promoter; NEO: neomycin resistance gene. (B, C, D) Immunostaining for BAD in brain coronal sections from mice of the specified genotypes. Notice lack of BAD expression only in DGNs from Badflox/flox; Dock10-Cre brain. In the right column dashed line area demarcates the DGN cell body layer. Also notice the lower fluorescence intensity in the entire molecular layer of the dentate gyrus (containing the dendrites of the DGNs) in Badflox/flox; Dock10-Cre compared to the parental genotypes.
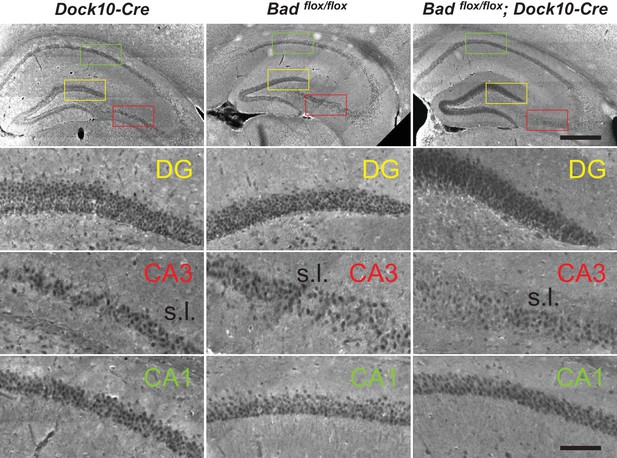
Targeted knockout of Bad expression in the dentate gyrus: coronal sections.
Immunostaining for BAD in brain coronal sections from mice of the specified genotypes. BAD expression is only absent in DGNs from Badflox/flox; Dock10-Cre brain. Notice decreased BAD staining in stratum lucidum (s.l.) of Badflox/flox; Dock10-Cre CA3; this region contains the mossy fiber axons of the DGNs. Color squares in the top row indicate the expanded areas in the other rows as indicated. Scale bars: 500 μm (top) and 100 μm (bottom).
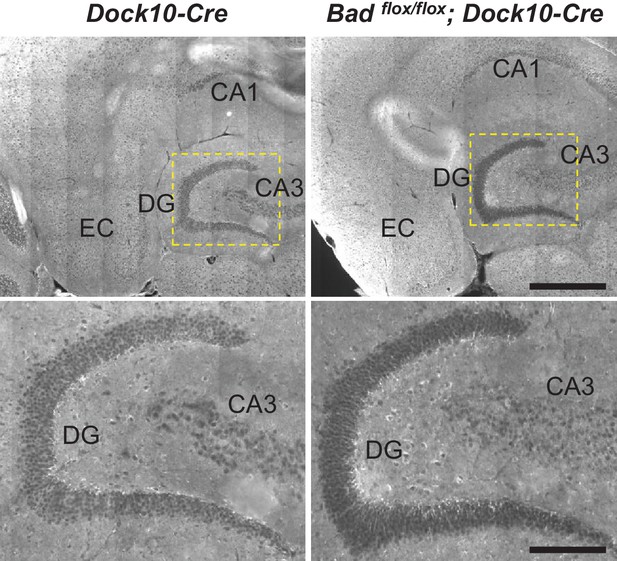
Targeted knockout of Bad expression in the dentate gyrus: horizontal sections.
Immunostaining for BAD in brain horizontal sections from mice of the specified genotypes. BAD expression is only absent in DGNs from Badflox/flox; Dock10-Cre brain. Notice appreciable BAD expression in EC of both genotypes. Dashed yellow squares in top row indicate the expanded areas in bottom row. Scale bars: 500 μm (top) and 175 μm (bottom).
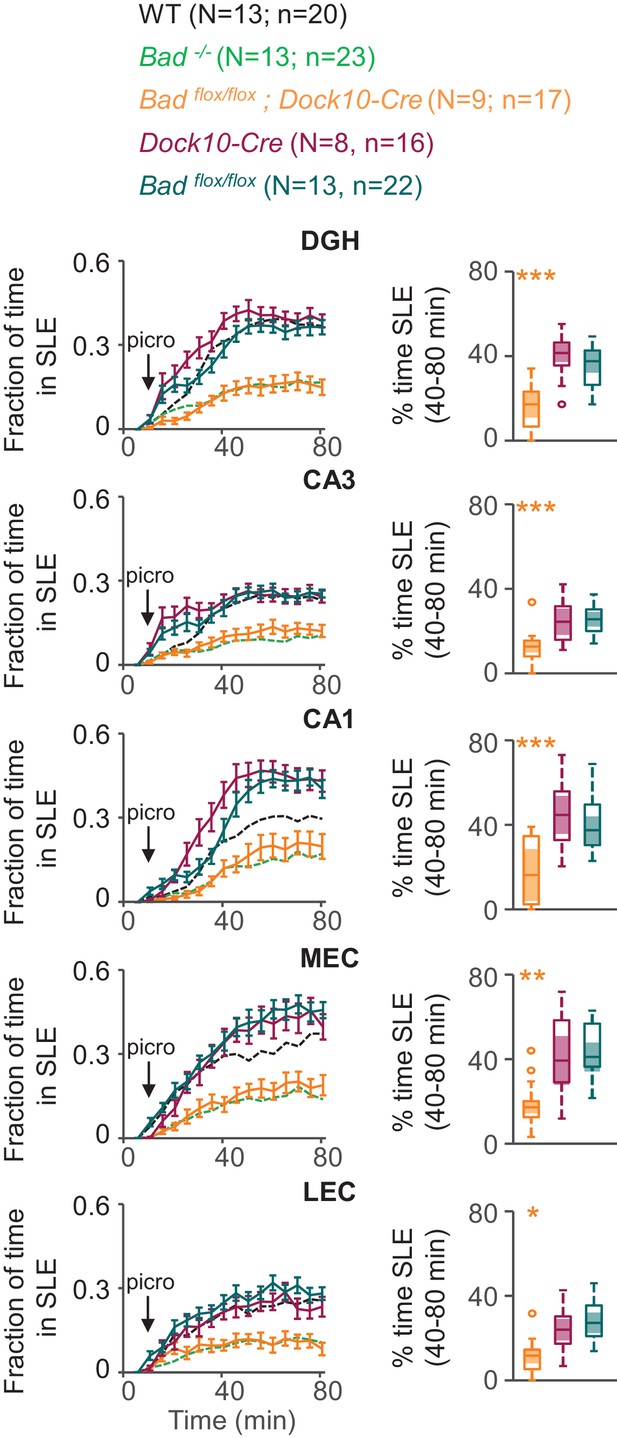
DGN-specific BAD ablation is sufficient to reduce seizure-like activity.
Genetic deletion of Bad in DGNs alone (Badflox/flox; Dock10-Cre, orange) reduced the time hippocampal regions spent in SLEs in adult slices. Left column: fraction of time spent in SLEs (mean ± SEM) for Badflox/flox; Dock10-Cre (orange), Dock10-Cre (dark magenta) or Badflox/flox (teal) plotted versus time. For comparison, WT (black) and Bad−/− (green) data are reproduced from Figure 3 as dashed lines without error bars. Right column: percent time in SLEs calculated from the 40–80 min range when seizure-like activity plateaus for each region of interest analyzed. *p<0.05; **p<0.005; ***p<0.0005; 1-way ANOVA.
Videos
Seizure-like events in the presence of 2.5 μM picrotoxin.
Fluorescence intensity as imaged in Figure 3 showing epileptiform activity in an acute EC-HP slice in the presence of 2.5 μM picrotoxin. Video was created using ImageJ 1.51 s (Fiji).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Mus musculus) | Bad | this paper | HEPD0750_4_D04 | for generation of Badflox/flox mice |
| Strain, strain background (Mus musculus) | male or female C57BL/6 mice | Charles River Laboratories | ||
| Genetic reagent (Mus musculus) | male or female homozygous Bad−/− mice | doi:10.1038/nm1717 | ||
| Genetic reagent (Mus musculus) | male or female homozygous Bad−/−; Kcnj11−/− mice | doi:10.1016/j.neuron. 2012.03.032 | ||
| Genetic reagent (Mus musculus) | female heterozygous Dock10-Cre+/− mice | doi:10.1038/nn.3614 | ||
| Genetic reagent (Mus musculus) | male or female homozygous Badflox/flox mice | this paper | ||
| Genetic reagent (Mus musculus) | male or female heterozygous Badflox/flox; Dock10-Cre+/− mice | this paper | ||
| Strain, strain background (Adeno-associated virus) | AAV2/8.CAG.BAD.2A.mCherry | BAD-2A-mCherry plasmid packed into AAV8 at Viral Core Boston Children’s Hospital | ||
| Strain, strain background (Adeno-associated virus) | AAV2/9.CAG.GCaMP6f | Penn Vector Core | Cat. no. AV-9-PV3081 | |
| Antibody | anti-Bad (rabbit monoclonal) | Abcam | ab32445; RRID:AB_725614 | (1:100) |
| Antibody | anti-Rabbit IgG (goat polyclonal), CF 594 | Sigma | SAB4600107 | (1:1000) |
| Other | VECTASHIELD mounting medium with DAPI | Vector Laboratories | H-1200; RRID:AB_2336790 | |
| Software | MATLAB 2016b | Mathworks | RRID:SCR_001622 | |
| Software | Fiji/ImageJ 1.51 s | NIH | RRID:SCR_002285 | |
| Software | Origin 9.1 | OriginLab | ||
| Software | Patchmaster v2x43 | HEKA | ||
| Software | TILLvisION 4.0.7.2 | Thermo Fisher Scientific |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.32721.015





