Mechanistic insights into the active site and allosteric communication pathways in human nonmuscle myosin-2C
Figures
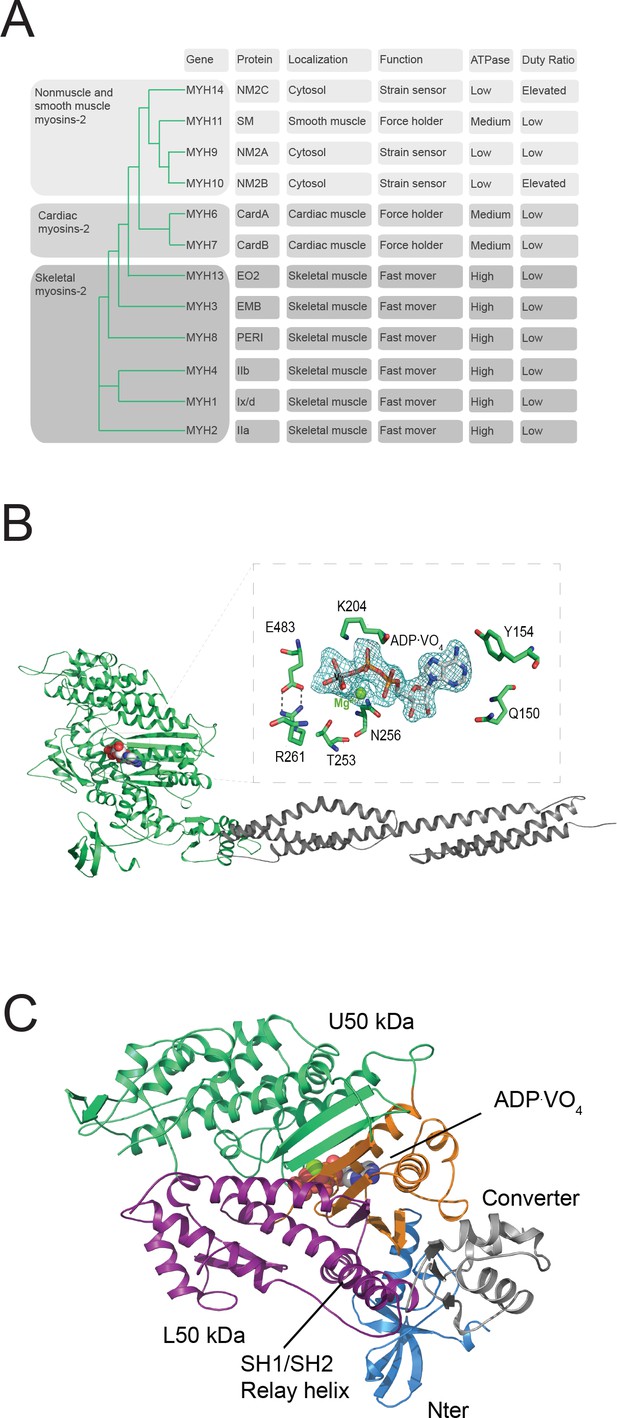
Myosin-2 phylogeny, overall topology, and active site characteristics of human NM2C.
(A) Phylogenetic analysis divides human myosins-2 in the three subfamilies (i) nonmuscle and smooth muscle myosins-2, (ii) cardiac, (iii) and skeletal muscle myosins-2 (Foth et al., 2006). Nonmuscle myosin-2s are essential for the structural integrity of the cytoplasmic architecture during cell shape remodeling and motile events of eukaryotic cells, whereas all other myosins-2 play eminent roles in the contraction of smooth, cardiac and striated muscle cells (Sellers, 2000). Abbreviations used: NM2A: nonmuscle myosin-2A, NM2B: nonmuscle myosin-2B; NM2C: nonmuscle myosin-2C; SM: smooth muscle myosin-2; CardA: α-cardiac myosin-2; CardB: β-cardiac myosin-2; EO2: extraocular myosin-2; EMB: embryonic myosin-2; PERI: perinatal myosin-2; IIb: fast skeletal muscle myosin-2; IIx/d: skeletal muscle myosin-2; IIa: slow skeletal muscle myosin-2. (B) Architecture of the crystallized NM2C construct in the pre-powerstroke state. The myosin motor domain and the α-actinin repeats are shown in cartoon representation in green and grey color. The nucleotide is shown in spheres representation. Inset, Conserved key residues that interact with the nucleotide in the NM2C active site. The Fo-Fc omit map of Mg2+·ADP·VO4 is contoured at 4σ. The salt bridge between switch-1 R261 and switch-2 E483 is highlighted. (C) Subdomain architecture of NM2C. The U50 kDa is shown in green, the L50 kDa in purple, the converter in grey, and the Nter in blue. The region shown in orange corresponds to the active site and the junction of U50 kDa and L50 kDa. The bound nucleotide is shown in spheres representation. The location of the SH1-SH2 helix and the relay helix in the L50 kDa is highlighted.
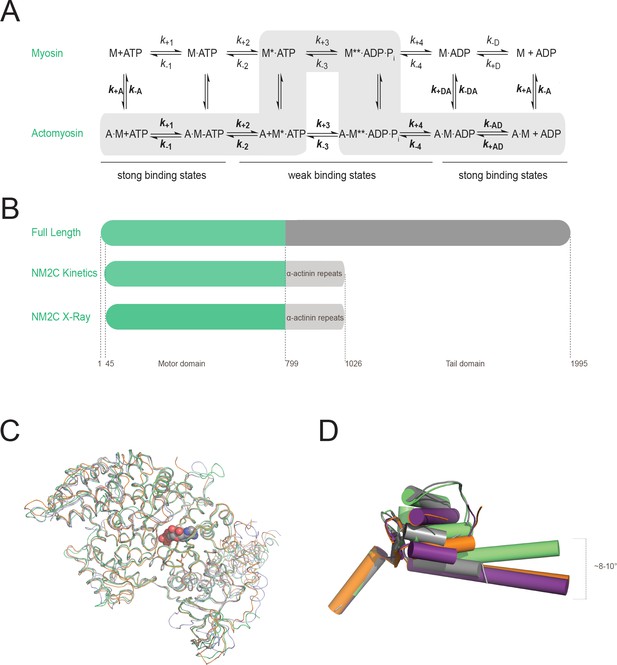
Myosin-2 ATPase cycle, expression constructs, and structural alignment of the NM2C Cα coordinates.
(A) Consensus scheme of the myosin and actomyosin ATPase cycle. The upper part represents the myosin (M) ATPase cycle. The lower part represents the ATPase cycle in the presence of F-actin (A). The asterisk denotes enhanced states of the intrinsic myosin fluorescence, which are attributed to the nucleotide-induced changes in the microenvironment of the conserved relay loop W525. Lowercase k denotes a rate constant. k+D=k+6, k-D = k-6, k+AD = k+6, k-AD = k-6. An uppercase K denotes a dissociation equilibrium constant (K = k-x/k+x) throughout this work. Normal and bold face notation denote the respective kinetic constants in the absence and presence of F-actin. The main pathway of the actomyosin ATPase cycle is highlighted in grey. Strong and weak actin binding states are indicated. (B) Schematic representation of the expression constructs used in this study. Top, Nonmuscle myosin-2C contains a N-terminal motor domain (green) followed by a neck and a tail domain (dark grey). Middle, For kinetic studies of NM2C, R788K, and R788E, the motor domain was directly fused to spectrin repeats 1 and 2 from Dictyosteliumα-actinin (light grey) which serves as an artificial lever arm. The concept of the artificial lever arm has been successfully used in structural and kinetic studies of myosins-2 (Heissler and Manstein, 2011; Furch et al., 1999; Kliche et al., 2001; Münnich et al., 2014). Bottom: For structural studies, the N-terminal 45 amino acids were deleted. The numbering refers to the amino acid sequence of the full-length protein. (C) The NM2C Cα atoms (green) in ribbon representation superimpose with a root mean square deviation (r.m.s.d.) of 0.57 Å to chicken smooth muscle myosin-2 (grey, PDB entry 1BR2), with 0.78 Å to scallop striated muscle myosin-2 (orange, PDB entry 1QVI), and 0.73 Å to Dictyostelium nonmuscle myosin-2 (blue, PDB entry 2XEL), underlining a strong correlation between Cα geometry and overall motor domain fold. The JK-loop and the nucleotide are highlighted in color and spheres representation. (D) Relative orientation of converter and lever in NM2C (green), chicken smooth muscle myosin-2 (PDB entry 1BR2, grey), and scallop striated muscle myosin-2 (PDB entry 1QVI (orange), 1DFL (violet)) motor domain structures in cartoon representation. α-helices are depicted as cylinders.
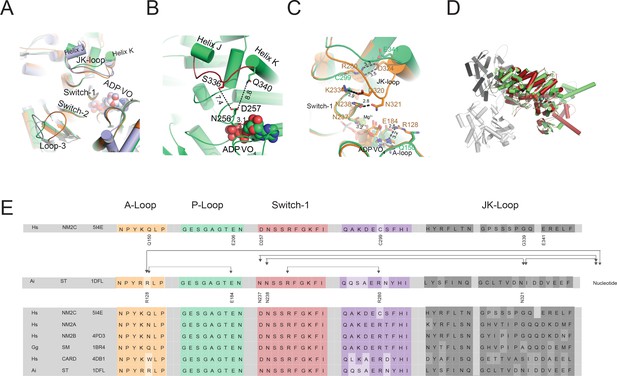
Conformational changes of the JK-loop in the myosin active site.
(A) Top view on the NM2C active site in the pre-powerstroke state (green) superimposed on pre-powerstroke state structures from chicken smooth muscle myosin-2 (grey, PDB entry 1BR4), Dictyostelium nonmuscle myosin-2 (blue, PDB entry 2XEL), and scallop striated muscle myosin-2 (orange, PDB entry 1QVI). The ATP analog ADP⋅VO4 is shown in spheres representation. (B) Conformation of the JK-loop in vicinity to the NM2C active site. The JK-loop flanks the active site and connects helices J and K. The distance between the residue Q340 of the JK-loop in the U50 kDa and the D257 of switch-1 of the active site is ~8.8 Å. The distance between residue S336 of the JK-loop and switch-1 D257 is ~7.4 Å. Switch-1 residue N256 interacts with α-phosphate (3.1 Å) and β-phosphate (3.5 Å) group of ADP⋅VO4 in the active site. NM2C is colored in green/orange, the JK-loop is colored in brick red and ADP⋅VO4 is shown in spheres. (C) Interactions between the JK-loop and the switch-1 region are compared between the NM2C (green) and scallop striated muscle myosin-2 (orange, PDB entry 1QVI). A-loop residue R128 is coordinating the interaction to the ADP adenosine in the active site of striated muscle myosin-2. The distance between the residues is 3.2 Å. R128 further forms a hydrogen bond (2.8 Å) with E184 of the P-loop. JK-loop N321 is in hydrogen bond interaction with switch-1 N238, located at a distance of 4.6 Å to the hydroxyl group of the C2’ of the ADP ribose. The connectivity between switch-1 and the nucleotide is further strengthened by a hydrogen bond between N237 and the ADP ribose. NM2C lacks all interactions described for scallop striated muscle myosin-2 due to the replacement of R128 with Q150 and JK-loop shortening which increases the distance to the adenosine in the active site to 5.8 Å and disrupts constrains between swich-1 and the JK-loop. All residues in the JK-loop region are labeled for scallop striated muscle myosin-2 (PDB entry 1QVI). For NM2C only amino acid substitutions are labeled for legibility. (D) Superimposition of the NM2C pre-powerstroke state structure (green) and the actin-bound near-rigor actoNM2C complex (red) shows that the nucleotide-binding site does not undergo major structural changes. Actin subunits are colored in shades of grey and the nucleotide is shown in spheres representation. (E) Sequence alignment of select structural elements in the myosin motor domain that interact with the JK-loop. Interactions of A-loop R128 are highlighted with brackets for scallop striated muscle myosin-2 (PDB entry 1QVI). All highlighted interactions are absent in NM2C due to the presence of Q150 in the A-loop. Abbreviations used: Hs NM2C: human NM2C (NP_079005.3); Hs NM2A: human nonmuscle myosin-2A (NP_002464.1); NM2B: human nonmuscle myosin-2B (NP_005955.3); Gg SM: chicken smooth muscle myosin-2 (NP_990605.2); Hs CARD: human beta β-cardiac muscle myosin-2 (NP_000248.2); Ai ST: scallop striated muscle myosin-2 (P24733.1). PDB entries are indicated when available.
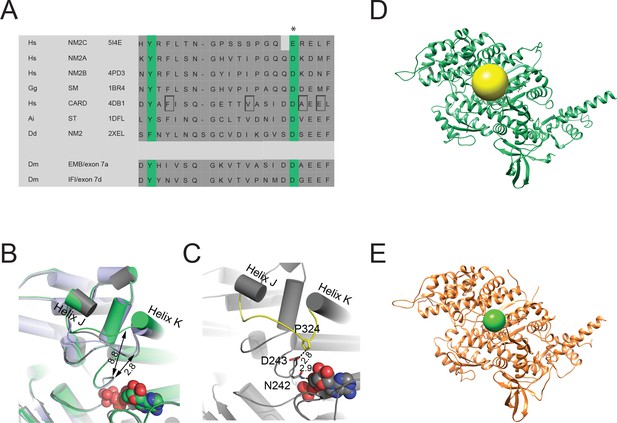
Active site characteristics in myosin-2 motor domains.
(A) Sequence alignment of myosin-2 JK-loops. The asterisk indicates the invariant, negatively charged residues corresponding to NM2C E341. The abbreviations used are as follows: Hs NM2C: human NM2C (NP_079005.3); Hs NM2A: human nonmuscle myosin-2A (NP_002464.1); NM2B: human nonmuscle myosin-2B (NP_005955.3); Gg SM: chicken smooth muscle myosin-2 (NP_990605.2); Hs CARD: human beta β-cardiac muscle myosin-2 (NP_000248.2); Ai ST: scallop striated muscle myosin-2 (P24733.1); Dd NM2: Dictyostelium nonmuscle myosin-2: (XP_637740.1); Dm EMB: Drosophila embryonic body wall muscle myosin-2 (P05661 with spliced exon 7a); Dm IFI: Drosophila indirect flight muscle myosin-2 (P05661 with spliced exon 7d). PDB entries are indicated when available. Cardiomyopathy-associated mutations in β-cardiac myosin-2 on positions F312 (F312C), V320 (V320M), A326 (A326P), and E328 (E328G) are highlighted in the boxed areas. (B) Close-up view of the active site region of pre-powerstroke state structures of NM2C (green), chicken smooth muscle myosin-2 (grey, PDB entry 1BR2), and Dictyostelium nonmuscle myosin-2 (blue, PDB entry 2XEL). The distance between the JK-loops of smooth muscle myosin-2 and Dictyostelium nonmuscle myosin-2 and switch-1 is ~2.8 Å and 8.8 Å for NM2C. The ADP⋅VO4 complex in the active site is shown in spheres. (C) Close-up view of the active site region of pre-powerstroke state chicken smooth muscle myosin-2 (grey, PDB entry 1BR2). The distance between residue P324 of the JK-loop (yellow) in the U50 kDa and D243 of switch-1 is ~2.8 Å. Switch-1 residue N242 interacts with α-phosphate (2.8 Å) and β-phosphate (3.1 Å) group of ADP⋅ALF4 (shown in spheres) in the active site. (D) The volume of the active site, indicated by the spheres, was determined to 3065 Å3 in NM2C by fitting a sphere with a radius (r) of 9.012 Å to the active site with UCSF Chimera (Pettersen et al., 2004). The volume of the sphere was calculated based on its radius using the equation volume = 4/3πr3. (E) The volume of the active site of scallop striated muscle myosin-2 (PDB entry 1QVI) was determined to 697 Å3 based on a radius of r = 5.5 Å as in (D). The structures shown in (D) and (E) are in the pre-powerstroke state.
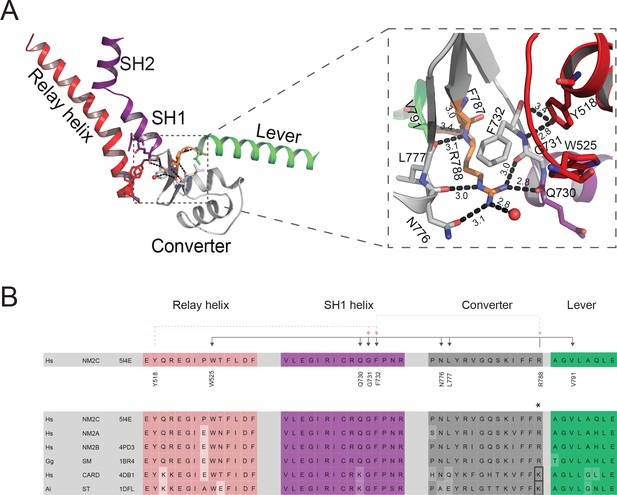
Interdomain connectivity at the converter/Nter/lever junction.
(A) Interaction profile of R788 in the pre-powerstroke state. R788 is shown in orange colored sticks and the converter is colored in white, the relay helix in red, the SH1-SH2 helix in purple, and the lever arm in green colored cartoon representation. The inset shows a close-up view of the complete R788 interaction profile and is rotated 137° respective to the main panel. The guanidinium group of R788 forms hydrogen bonds (2.8 Å) with backbone oxygen atom of Q730 from the SH1 helix and the backbone oxygen atom of G731 (3.0 Å) of the converter. The δ-nitrogen atom of R788 interacts (3.0 Å) with N776 backbone oxygen atom of N776. The R788 guanidinium group interacts (3.1 Å) with the hydroxyl group of N776 of the converter. The backbone nitrogen atom of R788 interacts (3.1 Å) with the carbonyl group of L777 of the converter. The backbone carbonyl group of R788 interacts (3.4 Å) with the backbone nitrogen of V791 of the lever as well as a water molecule (3.0 Å). The hydroxyl group from relay helix Y518 interacts with the backbone nitrogen atom of G731 (2.8 Å) and backbone oxygen atom of F732 (3.4 Å). F732 forms hydrophobic interactions with the methylene groups of R788 with the latter positioned in van der Waals distance to relay loop W525. All the amino acids involved in interactions with R788 are shown as sticks and water molecules as spheres. (B) Sequence alignment of selected regions from relay helix, SH1 helix, converter and lever arm shows the high sequence conservation within the myosin-2 motor domain. The asterisk indicates the invariant, positively charged residue corresponding to NM2C R788. The interactions of NM2C R788 with structural elements of the L50 kDa, the converter, and the lever are highlighted. Abbreviations used: Hs NM2C: human NM2C (NP_079005.3); Hs NM2A: human nonmuscle myosin-2A (NP_002464.1); NM2B: human nonmuscle myosin-2B (NP_005955.3); Gg SM: chicken smooth muscle myosin-2 (NP_990605.2); Hs CARD: human beta β-cardiac muscle myosin-2 (NP_000248.2); Ai ST: scallop striated muscle myosin-2 (P24733.1). PDB entries are indicated when available. Lysine residues that replace R788 in cardiac and striated muscle myosins-2 highlighted in the boxed area.
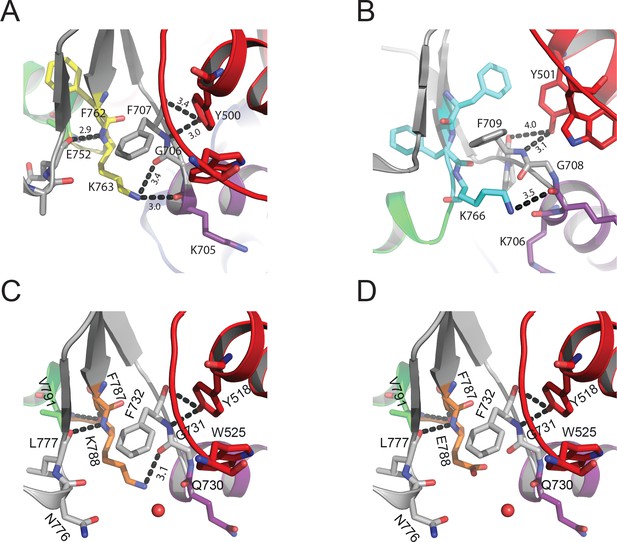
Interdomain connectivity at the converter/Nter/lever junction in muscle myosins-2.
In contrast to NM2C (Figure 3), the substitution of R788 with K763 in scallop striated muscle myosin-2 (PDB entry IQVI) (A) and K766 in human β-cardiac muscle myosin-2 (PDB entry 4DB1) (B) reduces the number of side chain and main chain interactions at the converter/Nter/lever junction. (C) In the conservative mutant R788K, the side chain: side chain and side chain: main chain interactions are reduced compared to NM2C (Figure 3). A further reduction in side chain: side chain and side chain: main chain interactions is achieved in the charge reversal mutant R788E (D). Coloring is according to Figure 3.
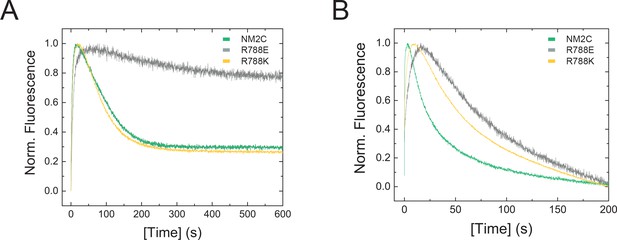
Transient kinetic features of NM2C and R788E.
(A) Interaction between NM2C/R788K/R788E with ATP under single-turnover conditions. Binding 0.375 µM d-mantATP to 0.5 µM myosin results in a transient fluorescence increase that is followed by a short plateau (hydrolysis) and a slow decrease in mantADP fluorescence that is associated with its release. All three phases are reduced in R788E (grey) compared to NM2C (green) and R788K (yellow). The very slow decrease of the fluorescence signal in R788E indicates that either the ATP hydrolysis rate or a subsequent release rate of the hydrolysis products are severely decreased when compared to NM2C and R788K. (B) Interaction between 0.25 µM pyrene-actoNM2C/R788K/R788E with 0.15 µM ATP under single-turnover conditions. Color coding is according to (A).
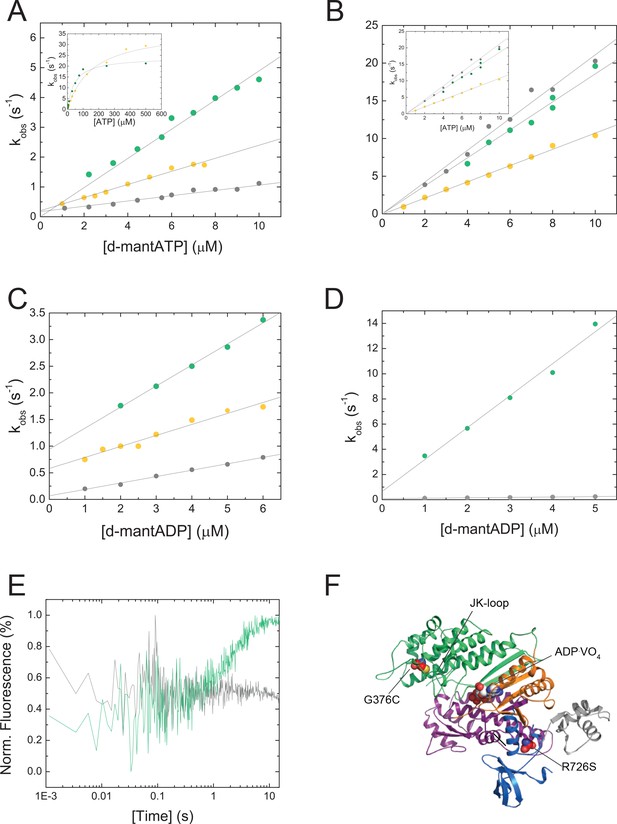
Nucleotide binding characteristics of NM2C, R788K, and R788E and disease causing NM2C mutations.
(A) Dependence of the observed rate constants (kobs) upon d-mantATP binding to 0.25 µM NM2C/R788K/R788E on the nucleotide concentration. Linear fits to the data result in second-order binding rate constants of K1k+2 = 0.48±0.01 µM−1s−1 for NM2C (green), a two-fold reduced ATP binding constant K1k+2 = 0.22±0.01 µM−1s−1 for R788K (yellow), and a five-fold reduced binding rate constant of K1k+2 = 0.09±0.005 µM−1s−1 for R788E (grey) compared to NM2C. Inset, The rate constants (kobs) obtained from the binding reaction of ATP to NM2C/R788K dependent hyperbolically on the ATP concentration. The rate of ATP hydrolysis (k+3+k-3) and the second-order binding rate constants K1k+2 were determined from a fit to the data (NM2C: k+3+k-3=24.32 ± 0.89 s−1; K1k+2 = 0.39±0.01 µM−1s−1; R788K: k+3+k-3=37.31 ± 0.55 s−1; K1k+2 = 0.18±0.01 µM−1s−1). The respective parameters are not experimentally accessible for R788E. (B) The apparent second-order ATP binding rate constants, determined by the ATP-induced dissociation of the pyrene-labeled actoNM2C (green) or actoR788E (grey) complexes are with K1k+2 = 1.86±0.03 µM−1s−1 and K1k+2 = 2.11±0.05 µM−1s−1 similar. The respective constant is reduced to K1k+2 = 1.03±0.03 µM−1s−1 for actoR788K (yellow). Inset, The maximum isomerization rate constants k+2 = 643.06±22.6 s−1 (NM2C), k+2 = 767.43±21.27 s−1 (R788K), and k+2 = 579.27±12.81 s−1 was determined from a hyperbolic fit to the respective data. (C) The R788K (yellow) and the R788E (grey) mutation decrease the second-order ADP-binding rate constant two- to threefold to k+D = 0.21±0.02 µM−1s−1 and k+D = 0.12±0.01 µM−1s−1 when compared to k+D = 0.39±0.01 µM−1s−1 for NM2C (green). (D) The ADP-binding rate constant k+AD = 0.03±0.001 µM−1s−1, as calculated from the slope of the kobs versus [d-mantADP] plot, is 85-fold decreased for R788E (grey) when compared to k+AD = 2.54±0.18 µM−1s−1 for NM2C (green). (E) Time-dependent change in the intrinsic tryptophan signal upon mixing 0.5 mM ATP with 0.25 µM NM2C (grey) or R778E (grey) in the presence of 5 µM ADP. ATP binding increases the fluorescence signal in NM2C but not R788E under identical conditions in a stopped-flow spectrophotometer. (F) Missense mutations G376C and R726S are associated with autosomal dominant hearing impairment (DFNA4) and their location in NM2C is shown in spheres representation. G376C is in proximity to the JK-loop, R726S is in the SH1-SH2 helix. NM2C subdomains are color coded according to Figure 1C and the nucleotide is shown in spheres representation.
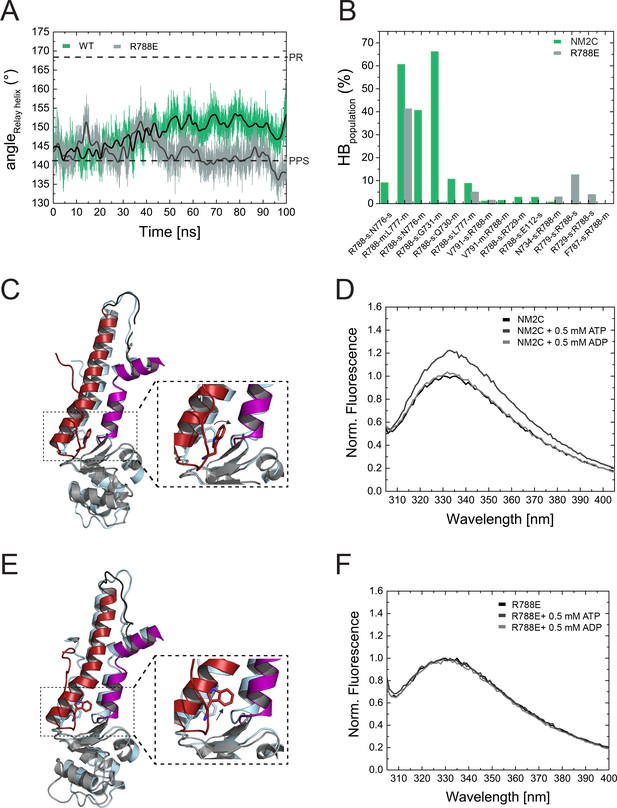
Structural importance of R788 at the converter/Nter/lever junction.
(A) Relay helix angle as a function of MD simulation time, as monitored by the angle between Cα atoms of residues S489, M510, and E521 along the trajectories. The relay helix straightens in NM2C with a steady increase in the angle of the relay helix from approximately 145° to 150°, while the angle does not change significantly in R788E and fluctuates around 145° throughout the 100 ns time course of the simulation. Values for the relay helix angle observed in crystal structures of pre-power stroke (PPS) and post-rigor (PR) are indicated by dotted lines. (B) Population of hydrogen bonds (HB) between R788 (NM2C) or E788 (R788E) and surrounding structural elements over the simulation time of 100 ns. The abbreviations s and m indicate side chain and main chain. (C) Dynamics and conformational changes in NM2C during MD simulations. Snapshots from the start (0 ns simulation time) and end conformations (100 ns simulation time) are shown in light cyan and colored cartoon representation, respectively. The relay helix is shown in red, the SH1-SH2 helix in purple and the converter in grey. Relay loop W525 is shown in red in stick representation. The insets show a close-up view of the conformational changes of W525 along the simulation trajectory. (D) Tryptophan fluorescence emission spectrum of 4 µM NM2C in the absence of nucleotide or the presence of 0.5 mM ATP or 0.5 mM ADP. (E) Dynamics and conformational changes in R788E during MD simulations. Snapshots from the start (0 ns simulation time) and end conformations (100 ns simulation time) are shown in light cyan and colored cartoon representation, respectively. The relay helix is shown in red, the SH1-SH2 helix in purple and the converter in grey. Relay loop W525 is shown in red in stick representation. The insets show a close-up view of the conformational changes of W525 along the simulation trajectory. (F) Tryptophan fluorescence emission spectrum of 4 µM R788E in the absence of nucleotide or presence of 0.5 mM ATP or 0.5 mM ADP.
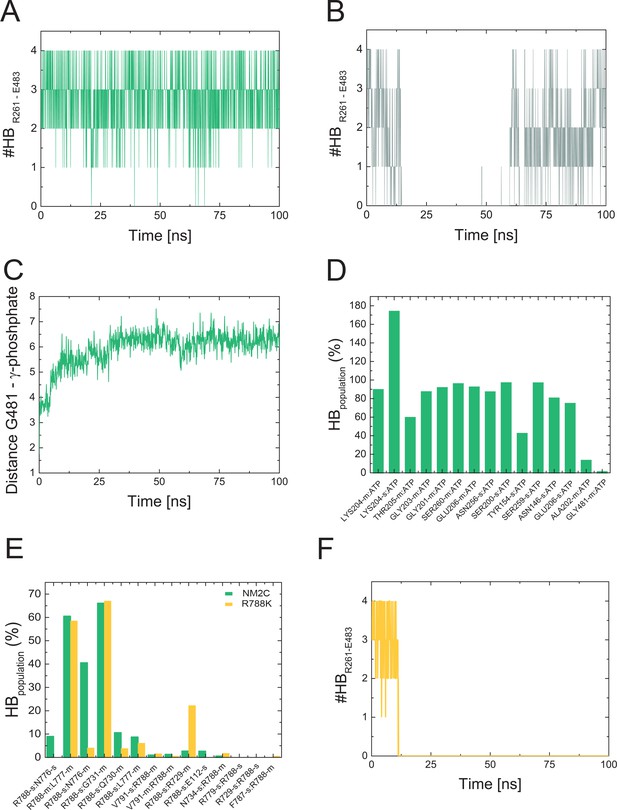
Stability of the salt bridge between switch-1 and switch-2 in MD simulations of NM2C, R788K, and R788E.
(A) Monitored number of hydrogen bond (#HB) interactions between R261 (switch-1) and E483 (switch-2) along the simulation time of NM2C. (B) Monitored number of hydrogen bond (#HB) interactions between R261 (switch-1) and E483 (switch-2) along the simulation time of R788E (B). Hydrogen bonds were detected with cutoff values for the donor-acceptor distance and angle of 3.5 Å and 30°. Note the intermediate breaking of the salt-bridge in R788E (B). (C) Distance between main chain of G481 of switch-2 with the γ-phosphate of ATP during the 100 ns simulation time for NM2C. (D) Population of hydrogen bonds (HB) between structural elements of the active site of NM2C and ATP along the 100 ns simulation. The abbreviations s and m indicate side chain and main chain, respectively. (E) Population of hydrogen bonds (HB) between R788 (NM2C) or K788 (R788K) and surrounding structural elements along the simulation time. The abbreviations s and m indicate side chain and main chain, respectively. (F) Monitored number of hydrogen bond (#HB) interactions between R261 (switch-1) and E483 (switch-2) along the simulation trajectory of R788K. Simulations for NM2C, R788K, and R788E were performed in the presence of ATP in the active site.
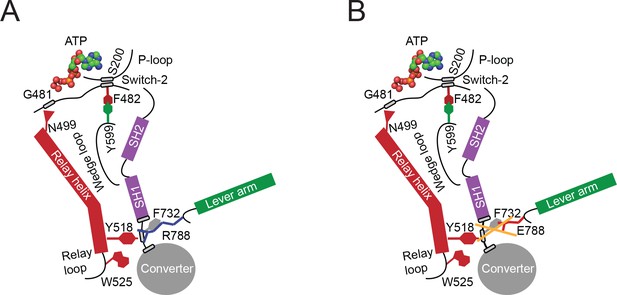
Proposed allosteric communication pathway from the converter to the NM2C active site.
(A) Residue R788 connects the converter to the SH1 interface through main chain and side chain interactions in NM2C. This interface further interacts with Y518 of the relay helix and W525. The interactions are propagated to the active site and vice versa through the relay helix and through an interaction of relay helix residue N499 with the main chain of G481 from switch-2. The latter directly interacts with the nucleotide. Y599 of the wedge loop, that itself contacts the relay helix, establishes an interaction with switch-2 F482. This residue is in contact with S200 of the P-loop by main chain interactions and directly interacts with the nucleotide in the active site. Schematic not drawn to scale. (B) The interface between R788E and both, the SH1 and the relay helix, is perturbed and the allosteric communication from the active site to the converter compromised. The experimental observation that R788E does not change its intrinsic fluorescence upon nucleotide binding which is caused by a lacking conformational change of W525 indicates that the communication pathway is interrupted in the relay helix. The position of W525 in the relay loop at the distal end of the relay helix indicates that the pathway is interrupted before or at Y518, which is supported by the observation that the relay loop does not change its position during the time course of the MD simulation. As a consequence, Y518 cannot establish an interface with E788 and F732 of the converter and SH1 helix. Further, E788 cannot establish interactions with N776 and V791 and completely disrupts the structural integrity of the interface of converter, SH1-SH2 helix, relay helix and the lever arm and uncouples nucleotide-induced changes in the active site from the converter rotation. Only key residues involved are shown in the proposed mechanism. Small grey boxes indicate main chain interactions. Schematic not drawn to scale.
Tables
Crystallographic data collection and refinement statistics for human NM2C.
https://doi.org/10.7554/eLife.32742.004| Parameter | Hs NM2C-ADP⋅VO4 |
|---|---|
| X-ray data reduction statistics | |
| Space group | P22121 |
| Unit cell dimensions | |
| a, b, c | 81.12 Å, 125.61 Å, 153.96 Å |
| α,β,γ | 90°, 90°, 90° |
| Resolution | 97.33 Å – 2.25 Å |
| Last shell | 2.35 Å – 2.25 Å |
| Total measurements | 1116790 |
| No. of unique reflections | 75357 |
| Last shell | 9069 |
| Wavelength | 0.91841 Å |
| Rmerge | 0.12 |
| Last shell | 0.23 |
| I/σ(I) | 13.6 |
| Last shell | 2.63 |
| Completeness | 0.998 |
| Last shell | 1.00 |
| Multiplicity | 14.82 |
| Last shell | 14.97 |
| Refinement statistics | |
| Resolution | 34.77 Å–2.25 Å |
| Last shell | 2.31 Å–2.25 Å |
| No. of reflections (working set) | 71459 |
| No. of reflections (test set) | 3786 |
| R-factor | 0.2210 |
| Last shell | 0.2910 |
| R-free | 0.2410 |
| Last shell | 0.2920 |
| No. of non-hydrogen atoms | |
| Macromolecules | 7697 |
| Ligands | 33 |
| solvent | 666 |
| Average B-factor | |
| Protein | 76.06 Å2 |
| Ligands | 35.59 Å2 |
| Solvent | 63.37 Å2 |
| RMS deviations from ideal values | |
| Bond lengths | 0.01 Å |
| Bond angles | 1.16° |
| Ramachandran favored | 96.22% |
| Ramachandran allowed | 3.78% |
| Ramachandran outliers | 0 |
Structure function relationships in the myosin-2 motor domain.
Interactions between residues in the active site involved in nucleotide binding and release kinetics based on this work and previous biochemical and structural studies on myosin motor domains (Miller et al., 2007; Risal et al., 2004; Swank et al., 2006; Grammer et al., 1993; Szilagyi et al., 1979). It is of note that myosin is a highly allosteric enzyme and nucleotide binding and release kinetic involve numerous interactions and subtle structural rearrangements of residues from different motor subdomains. Kinetic parameters from monomeric myosin motor domain constructs that are associated with a structural interaction are listed for direct comparison. An emerging trend from this analysis is that the myosin-2 kinetic cycle does not have a selectivity of ATP versus ADP. The presence of F-actin results in different allosteric communication pathways in myosins-2 and establishes ATP/ADP binding selectivity. Overall, nucleotide-binding rates are decreased for the group of nonmuscle and smooth muscle myosins-2 compared to myosins-2 from cardiac and skeletal muscle. The lacking salt bridge interactions between JK-loop, U50 kDa and switch-1 in nonmuscle myosins-2 results in decreased second-order binding rate constants for ATP (K1k+2) and ADP (k+D) (Figure 1—figure supplement 1A). Either a salt bridge interaction or hydrophobic interactions between the A-loop and the P-loop of muscle and cardiac myosins-2 at the active site favor fast nucleotide binding kinetics and does not or only marginally discriminate between ADP and ATP. The lack of a salt bridge interactions can have different effects dependent on the coordinating residue in the A-loop: An asparagine in the A-loop of nonmuscle myosins-2A and −2B favors ADP over ATP binding to actomyosin. A glutamine in the NM2C A-loop, which has a longer side chain than asparagine, abolishes ATP/ADP sensitivity in NM2C and the closely related smooth muscle myosin-2. The number of salt bridge interactions between P-loop, switch-1, and the Nter correlates with the thermodynamic and kinetic coupling, and the actin-activated ADP release rates in all myosins-2. Abbreviations used: NM2A: human nonmuscle myosin-2A; human NM2B: nonmuscle myosin-2B (PDB entry 4PD3); NM2C: human nonmuscle myosin-2C; SM: chicken smooth muscle myosin-2 (PDB entry 1BR2); CARD: human β-cardiac myosin-2 (PDB entry 4DB1); Oc ST: rabbit striated muscle myosin-2 (PDB entry 1DFL).
| Myosin | Residue | Residue | Residue | Residue | Parameter | ||
|---|---|---|---|---|---|---|---|
| JK-loop | U50 kDa | JK-loop | Switch-1 | K1k+2 | K+D | ||
| Hs NM2A (Kovács et al., 2003) | D315 | R272 | G312 | D230 | 0.56 | 0.55 | |
| Hs NM2B (Wang et al., 2003) | D322 | R279 | G319 | D237 | 0.65 | 0.81 | |
| Hs NM2C | E341 | C299 | G339 | D257 | 0.48 | 0.39 | |
| Gg SM (Cremo and Geeves, 1998) | D328 | R285 | A325 | D243 | 2.1 | 1.8 | |
| Hs CARD (Deacon et al., 2012) | D325 | R281 | S322 | D239 | 1.5 | 1.5 | |
| Oc ST (Ritchie et al., 1993; Kurzawa-Goertz et al., 1998) | D323 | R280 | N321 | N238 | 3.9 | 1.7 | |
| Residue | Residue | Parameter | |||||
| A-loop | P-loop | k+AD | K1k+2 | k+D | K1k+2 | k+AD/ K1k+2 | |
| Hs NM2A (Kovács et al., 2003) | N126 | E182 | 2.72 | 0.14 | 0.55 | 0.56 | 19.4 |
| Hs NM2B (Wang et al., 2003) | N130 | E186 | 2.41 | 0.24 | 0.81 | 0.65 | 10 |
| Hs NM2C | Q150 | E206 | 2.54 | 1.86 | 0.39 | 0.48 | 1.5 |
| Gg SM (Cremo and Geeves, 1998) | Q129 | E185 | 3.6 | 2 | 1.8 | 2.1 | 4.4 |
| Hs CARD (Deacon et al., 2012) | W130 | V186 | 4.4 | 1.1 | 1.5 | 1.5 | 1.6 |
| Oc ST (Ritchie et al., 1993; Kurzawa-Goertz et al., 1998) | R128 | E184 | 1.6 | 2.5 | 1.7 | 3.9 | 1.6 |
| Residue | Residue | Residue | Parameter | ||||
| P-loop | Switch-1 | Nter | KAD/KD | k-AD/k-D | k-AD | ||
| Hs NM2A (Kovács et al., 2003) | E175 | K228 | H676 | 0.7 | 2.8 | 1.72 | |
| Hs NM2B (Wang et al., 2003) | E179 | K235 | H683 | 0.2 | 0.7 | 0.35 | |
| Hs NM2C | E199 | K255 | H700 | 0.11 | 1 | 0.65 | |
| Gg SM (Cremo and Geeves, 1998) | E178 | K241 | H689 | 4.2 | 12 | 15 | |
| Hs CARD (Deacon et al., 2012) | E179 | R237 | E677 | 42 | 103.3 | 150 | |
| Oc ST (Ritchie et al., 1993; Kurzawa-Goertz et al., 1998) | E177 | R236 | E675 | 49 | 250 | 500 |
Steady-state and transient state kinetic parameters of NM2C, R788K, and R788E.
Numbering of the kinetic constants refers to Figure 1—figure supplement 1A. Measurements of transient kinetic parameters that rely on a change in intrinsic tryptophan fluorescence are not experimentally accessible for R788E. Normal and bold face notation denote the respective kinetic constants in the absence and presence of F-actin.
| Parameter | Signal or calculation | NM2C | R788K | R788E |
|---|---|---|---|---|
| Steady-state ATPase | ||||
| kcat (s−1) | NADH assay* | 0.37 ± 0.02 | 0.26 ± 0.02 | 0.2 ± 0.01 |
| Kapp (µM) | NADH assay* | 129.4 ± 17.2 | 110.5 ± 15.89 | 45.9 ± 13 |
| kcat/Kapp (µM−1s−1) | NADH assay | ~0.003 | ~0.002 | ~0.004 |
| ATP interaction | ||||
| K1k+2 (µM−1s−1) | Tryptophan | 0.39 ± 0.01 | 0.18 ± 0.01 | Not accessible¶ |
| K1k+2 (µM-1s−1) | d-mantATP | 0.48 ± 0.01 | 0.22 ± 0.01 | 0.09 ± 0.005 |
| 1/K0.5 (µM) | Tryptophan | 46.23 ± 5.22 | 158.03 ± 9 | Not accessible¶ |
| k3 + k-3 (s−1) | Tryptophan | 24.2 ± 0.86 | 37.31 ± 0.55 | Not accessible¶ |
| K1k+2 (µM−1s−1) | Pyrene-actin | 1.86 ± 0.03 | 1.03 ± 0.03 | 2.11 ± 0.05 |
| K1k+2 (µM−1s−1) | d-mantATP | 1.64 ± 0.04 | 0.73 ± 0.02 | 1.37 ± 0.04 |
| 1/K1 (µM) | Pyrene-actin | ~318 | ~826 | ~380 |
| k + 2 (s−1) | Pyrene-actin | 643.06 ± 22.6 | 767.43 ± 21.27 | 579.27 ± 12.81 |
| ADP interaction | ||||
| k+D (µM−1s−1) | d-mantADP | 0.39 ± 0.01 | 0.21 ± 0.02 | 0.12 ± 0.01 |
| k-D (s−1) | d-mantADP† | 0.94 ± 0.07 | 0.58 ± 0.05 | 0.06 ± 0.01 |
| k-D (s−1) | d-mantADP‡ | 0.39 ± 0.01 | 0.51 ± 0.01 | 0.05 ± 0.001 |
| k-D (s−1) | Tryptophan‡ | 0.47 ± 0.02 | 0.24 ± 0.001 | Not accessible¶ |
| KD (µM) | k-D/k+D | ~1–2.4 | ~1.1–2.8 | ~0.5 |
| k+AD (µM−1s−1) | d-mantADP | 2.54 ± 0.18 | not accessible** | 0.03 ± 0.001 |
| k-AD (s−1) | d-mantADPb | 0.65 ± 0.06 | not accessible** | 0.09 ± 0.03 |
| k-AD (s−1) | Light scattering‡ | 0.39 ± 0.01 | 0.42 ± 0.002 | 0.15 ± 0.01 |
| k-AD (s−1) | d-mantADP‡ | 0.68 ± 0.01 | 0.59 ± 0.01 | 0.19 ± 0.01 |
| k-AD (s−1) | Pyrene-actin‡ | 0.48 ± 0.01 | 0.46 ± 0.01 | 0.14 ± 0.01 |
| KAD (µM) | k-AD/k+AD | ~0.29 | Not accessible | ~2.68 |
| Actin interaction | ||||
| k+A (µM−1s−1) | Light scattering§,# | 2.49 ± 0.07 | 1.65 ± 0.03 | 0.66 ± 0.03 |
| k-A (s−1) | Pyrene-actin§,# | ~0.15 | ~0.04–0.07 | ~0.031 |
| KA (nM) | k-A/k+A | ~60 | ~24–42 | ~47 |
| k+DA (µM−1s−1) | Light scattering§ | 0.53 ± 0.01 | 2.53 ± 0.05 | 0.21 ± 0.01 |
| k-DA (s−1) | Pyrene-actin§ | ~0.004 | ~0.067–0.11 | ~0.002 |
| KDA (nM) | k-DA/k+DA | ~8 | ~26–43 | ~9 |
-
*In 50 mM NaCl, 2 mM MgATP, T = 25°C.
†From y-intercept.
-
‡From chasing experiment.
§Measured in salt free stopped-flow buffer.
-
#Samples were treated with apyrase prior to the assay to ensure rigor conditions.
¶The lack of a change in intrinsic fluorescence upon nucleotide binding to R788E precludes the direct measurement of this kinetic parameter.
-
**Small amplitudes prevent the direct measurement of this parameter.
Comparative analysis of kinetic signatures of monomeric myosin-2 motor domain constructs.
Abbreviations used: NM2A: human nonmuscle myosin-2A, NM2B: human nonmuscle myosin-2B (PDB entry 4PD3); NM2C: human nonmuscle myosin-2C; SM: chicken smooth muscle myosin-2 (PDB entry 1BR2); CARD: human/chicken β-cardiac myosin-2 (PDB entry 4DB1); ST: rabbit/scallop striated muscle myosin-2 (PDB entry 1DFL).
| Myosin | kcat/Kapp | k+AD/K1k+2 | KAD/KD | k-AD/ k-D | KDA/KA | Duty ratio |
|---|---|---|---|---|---|---|
| Hs NM2C | 0.003 | 1.5 | 0.11 | 1 | 0.13 | ~0.3‡ |
| Hs R788K | 0.002 | n.d. | n.d. | 1 | 1 | ~0.3‡ |
| Hs R788E | 0.004 | 0.26 | 5 | 3 | 0.19 | ~1$ |
| Hs NM2A (Kovács et al., 2003) | 0.002 | 19.4 | 0.7 | 2.8 | 2 | 0.1 |
| Hs NM2B (Wang et al., 2003) | 0.002 | 10 | 0.2 | 0.7 | 0.4 | 0.37 |
| Gg SM (Marston and Taylor, 1980; Cremo and Geeves, 1998) | 0.07 | 4.4 | 4.2 | 12 | 6.9 | <0.05 |
| Hs/Gg CARD (Marston and Taylor, 1980; Deacon et al., 2012) | 0.07† | 1.6 | 42 | 103.3 | 23.9 | <0.02 |
| Oc/Ai ST (Ritchie et al., 1993; Kurzawa-Goertz et al., 1998; Wagner, 1981; Harris and Warshaw, 1993) | 1.6* | 1.6* | 49 | 250* | 30* | <0.04 |
-
*Rabbit striated muscle myosin-2.
†Chicken cardiac muscle myosin-2.
-
‡Calculated at 190 µM F-actin and saturating [ATP].
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Spodoptera frugiperda) | Sf9 | Thermo Fisher Scientific | Thermo Fisher Scientific 11496015 | Maintained in Sf-900 III SFM |
| Recombinant DNA reagent | pFastBac1-NMHC- 2C0-2R-His8 | PMID: 21478157 | N/A | Progenitors: sequence optimized 1–799 human nonmuscle myosin-2C (GenBank accession number NP_079005) directly fused to spectrin repeats 1 and 2 from Dictyostelium discoideum alpha-actinin; vector pFastBac1 |
| Recombinant DNA reagent | pFastBac1-NMHC- 2C0-2R-Flag | this paper | NM2C (kinetic studies) | Progenitor: pFastBac1-NMHC-2C0-2R-His8 |
| Recombinant DNA reagent | pFastBac1-NMHC (45-799) −2C0-2R-His8 | this paper | NM2C (crystal structure) | Progenitor: pFastBac1-NMHC-2C0-2R-His8 |
| Recombinant DNA reagent | pFastBac1-NMHC (R788K) −2C0-2R-Flag | this paper | R788K (kinetic studies) | Progenitor: pFastBac1-NMHC-2C0-2R-Flag |
| Recombinant DNA reagent | pFastBac1-NMHC (R788E) −2C0-2R-Flag | this paper | R788E (kinetic studies) | Progenitor: pFastBac1-NMHC-2C0-2R-Flag |
Additional files
-
Supplementary file 1
Dihedral angles of switch-1 and lever arm residues in crystal structures of NM2C and smooth muscle myosin-2.
- https://doi.org/10.7554/eLife.32742.017
-
Transparent reporting form
- https://doi.org/10.7554/eLife.32742.018





