Micropattern differentiation of mouse pluripotent stem cells recapitulates embryo regionalized cell fate patterning
Figures
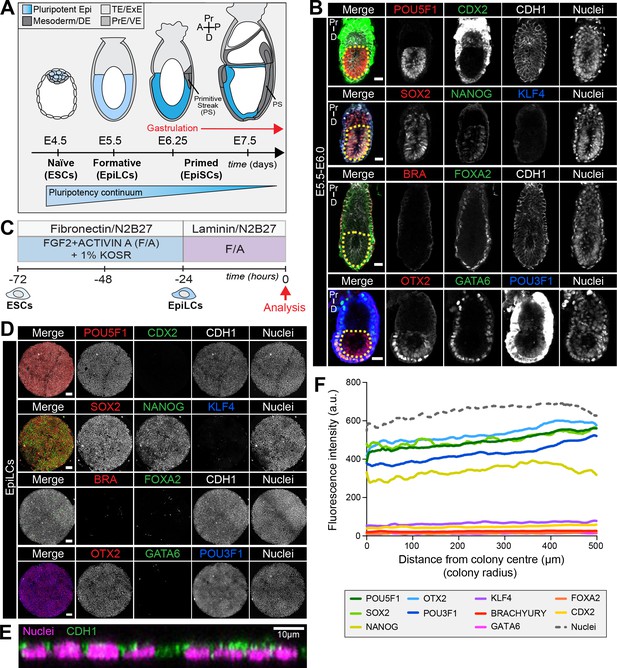
EpiLCs represent a pluripotent state correlating to the pre-streak epiblast of the embryo.
(A) Development of the mouse pluripotent epiblast (Epi) from embryonic day (E) 4.5 to 7.5 and correlating in vitro pluripotent states. ESCs, embryonic stem cells; EpiLCs, epiblast-like cells; EpiSCs, epiblast stem cells; TE/ExE, trophectoderm/extraembryonic ectoderm; PrE/VE, primitive endoderm/visceral endoderm; DE, definitive endoderm; A, anterior; P, posterior; Pr, proximal; D, distal. (B) Sagittal sections of immunostained E5.5-E6.0 embryos. Yellow dashed line demarcates Epi. Scale bars, 25 μm. Non-nuclear anti-BRACHYURY/CDX2/POU3F1 VE fluorescence represents non-specific binding. (C) ESCs were converted to EpiLCs on Fibronectin in N2B27 with FGF2 and ACTIVIN A (F/A) and knockout serum replacement (KOSR) for 48 hr. EpiLCs were plated onto Laminin-coated micropatterns overnight and analyzed the following day (0 hr). (D) Maximum intensity projections of immunostained 1000 μm diameter EpiLC micropatterned colonies. Scale bars, 100 μm. (E) Confocal image showing a z-axis (side profile) region of an immunostained EpiLC micropatterned colony. (F) Quantification of immunostaining voxel fluorescence intensity from center (0) to edge (500). Data represents average voxel intensity across multiple colonies. Dashed line represents average fluorescence of Hoechst nuclear stain. n = 6 NANOG/KLF4/SOX2/nuclei; n = 14 GATA6/OTX2/POU3F1; n = 14 BRACHYURY/FOXA2. BRA, BRACHYURY.
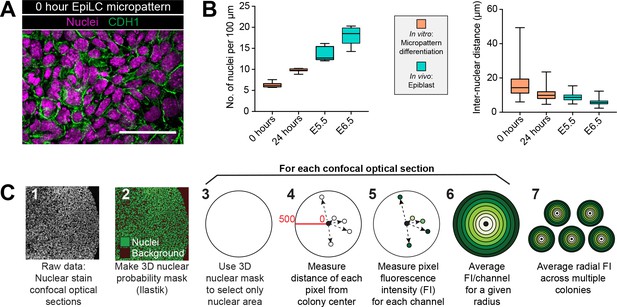
Micropatterned EpiLC colonies begin as an epithelial monolayer that increases density over time.
(A) Confocal optical section of a region of an immunostained EpiLC micropatterned colony at 0 hr. Scale bar, 50 μm. (B) Quantification of nuclei/100 μm and distance between nuclei (in μm) in vitro after 0 hr and 24 hr of micropattern differentiation and in vivo at embryonic day (E) 5.5 and 6.5. For nuclei/100 μm, the number of nuclei across the width of a micropatterned colony at 10 different positions was quantified. In vivo quantification was carried out on sections of 5 different embryos at E5.5 and 6.5. For quantification of inter-nuclear distance, 0 and 24 hr of micropattern differentiation: n = 150, E5.5: n = 128 (5 embryos), E6.5: n = 189. Quantification details can be found in the Materials and methods section. (C) Schematic diagram of quantification of radial fluorescence intensity of immunostaining on micropatterns.
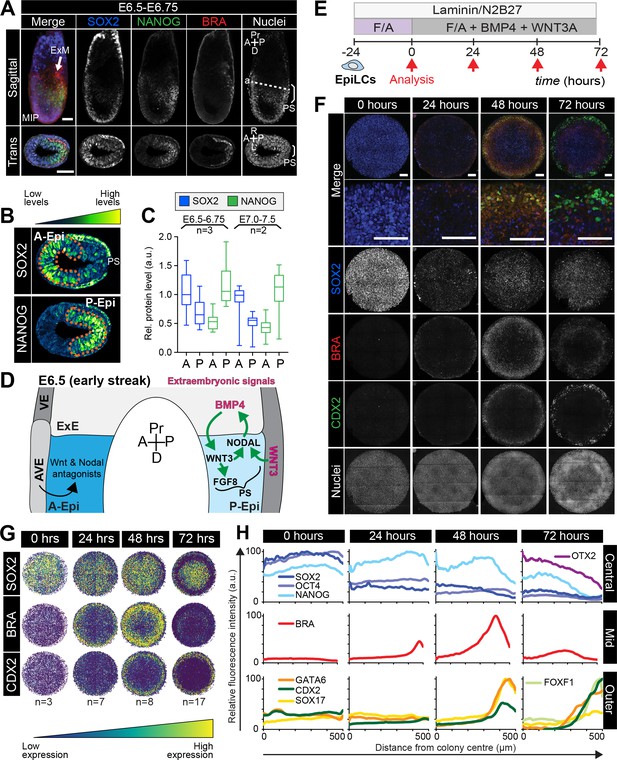
Micropatterned EpiLCs undergo spatially organized differentiation.
(A) Maximum intensity projection (MIP), sagittal and transverse sections of an embryonic day (E) 6.5 mouse embryo. Dashed line marks transverse plane. Non-nuclear anti-BRACHYURY/CDX2/SOX2 VE fluorescence likely represents non-specific binding. ExM, extraembryonic mesoderm; PS, primitive streak; A, anterior; P, posterior; Pr, proximal; D, distal. Scale bars, 50 μm. (B) Lookup table (LUT) of SOX2 marking anterior Epi (A-Epi) and NANOG marking posterior Epi (P-Epi). Orange dashed lines delineate regions of interest. (C) Quantification (5 sections/embryo/ stage) of SOX2 and NANOG in manually selected (panel B) anterior (A) and posterior (P) Epi of E6.5-E6.75 and E7.0-E7.5 embryos, normalized to Hoechst fluorescence. Data depicts mean fluorescence intensity ± S.D. N, number of embryos. No NANOG was observed in the A-Epi hence ~0.5 a.u. equates to background signal. (D) BMP, Wnt, Nodal, FGF signaling initiates gastrulation at the P-Epi - extraembryonic ectoderm (ExE) boundary. BMP4 produced by ExE stimulates Wnt3 expression within proximal Epi. WNT3 produced by Epi and visceral endoderm (VE) triggers Nodal and Fgf8 expression. NODAL promotes Bmp4 expression in the ExE. The anterior VE (AVE) expresses Wnt and Nodal pathway antagonists, restricting signaling activity to P-Epi. (E) EpiLCs were plated onto Laminin-coated micropatterns overnight (−24 hr) in N2B27 with F/A. The following day medium was changed to F/A, BMP4, WNT3A for 72 hr. Colonies were analyzed at 24 hr intervals. (F) MIPs of immunostained 1000 μm diameter colonies. All subsequent data represents 1000 μm diameter colonies. Upper two panels represent a merge of the markers shown below. Second panel shows high magnification of colony edge. Scale bars, 100 μm. BRA, BRACHYURY. (G) Depiction of average positional marker expression across multiple colonies. Each dot represents a single cell. (H) Quantification of voxel fluorescence intensity from colony center (0) to edge (500). Data represents average voxel intensity relative to maximum voxel intensity across time course/marker. For 0,24,48,72 hr respectively, POU5F1/NANOG n = 5,3,3,3, SOX2 n = 15,7,21,20, BRACHYURY n = 11,9,10,12, GATA6/SOX17/CDX2 n = 3,5,6,5. Markers grouped by spatial distribution within colonies. OTX2 and FOXF1 only analyzed at 72 hr.
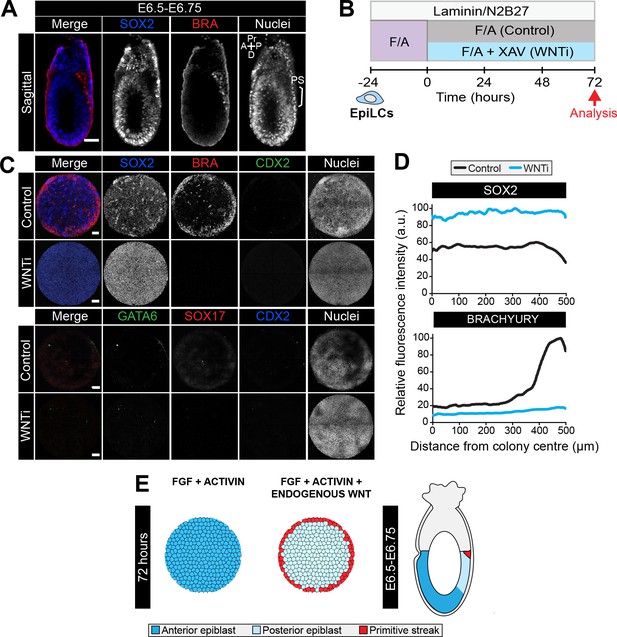
FGF, ACTIVIN and endogenous WNT induce a primitive streak state.
(A). Confocal sagittal optical section of an early streak embryo. Non-nuclear anti-BRACHYURY VE fluorescence likely represents non-specific binding. PS, primitive streak; A, anterior; P, posterior; Pr, proximal; D, distal. Scale bars, 50 μm. (B) EpiLCs were generated as in Figure 1C. EpiLCs were plated overnight onto Laminin-coated micropatterned surfaces (−24 hr) in N2B27 medium with 12 ng/ml FGF2 and 20 ng/ml ACTIVIN A (F/A). Cells were cultured for a further 72 hr in either N2B27 with F/A or F/A with 10 μM of XAV939 (Wnt signaling inhibitor, WNTi). (C) Representative confocal maximum intensity projections of immunostained 1000 μm diameter colonies after 72 hr in the conditions in panel D. Scale bars, 100 μm. (D) Quantification of SOX2 and BRACHYURY immunostaining voxel fluorescence intensity, in arbitrary units (a.u.), from colony center (0) to edge (500). Data represents average voxel intensity across multiple colonies. Control: n = 32, WNTi: n = 7. (E) Schematic diagram depicting cell fates generated after 72 hr of in vitro micropattern differentiation with FGF2 and ACTIVIN A (+XAV) or FGF2, ACTIVIN A and endogenous Wnt signaling and corresponding in vivo cell types.
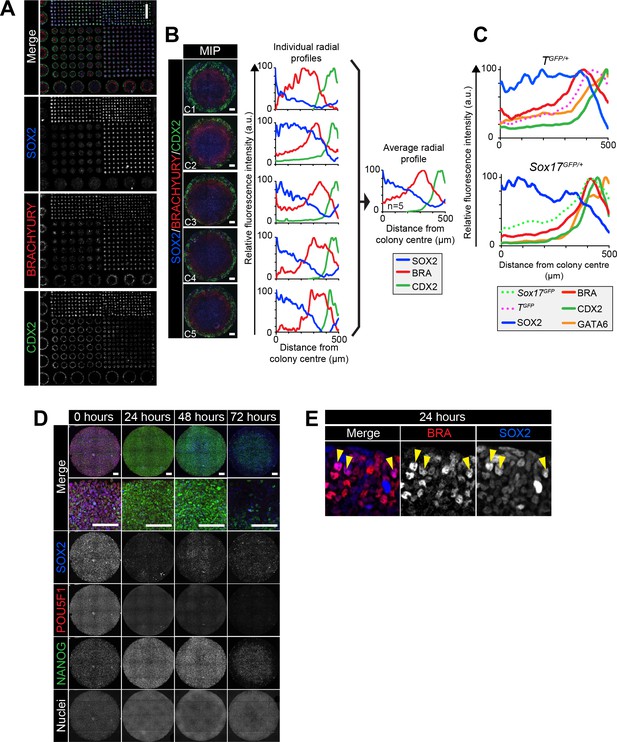
Robust micropattern differentiation of EpiLCs.
(A) Confocal maximum intensity projection (MIP) showing micropatterned colonies of 1000, 500, 250, 140 and 80 μm diameter. Scale bar, 1000 μm. (B) Left panels show confocal MIP images of 5 independent 1000 μm diameter colonies (C1-5). Middle panels show quantification of SOX2, BRACHYURY and CDX2 immunostaining voxel fluorescence intensity, in arbitrary units (a.u.), from colony center (0) to edge (500). Data relative to maximum voxel intensity for each marker. Right panel shows average radial profile of these five colonies. (C) While much of the work in this study utilized E14 ESCs, comparable patterning was observed with other mouse ESC lines. Quantification of immunostaining voxel fluorescence intensity of differentiated TGFP/+ and Sox17GFP/+ cell lines, in arbitrary units (a.u.), from colony center (0) to edge (500). Data represents average voxel intensity across multiple colonies relative to maximum voxel intensity for each marker. For TGFP/+ cells, GATA6, SOX2: n = 10, BRACHYURY, CDX2, TGFP: n = 11. For Sox17GFP/+ cells, SOX2: n = 5, BRACHYURY, CDX2, Sox17GFP: n = 6, GATA6: n = 5. (D) Confocal MIPs of immunostained colonies differentiated as in Figure 2E. Scale bars, 100 μm. (E) High magnification confocal image of the colony edge after 24 hr in conditions defined in Figure 2E. Yellow arrowheads mark cells coexpressing BRACHYURY and SOX2.
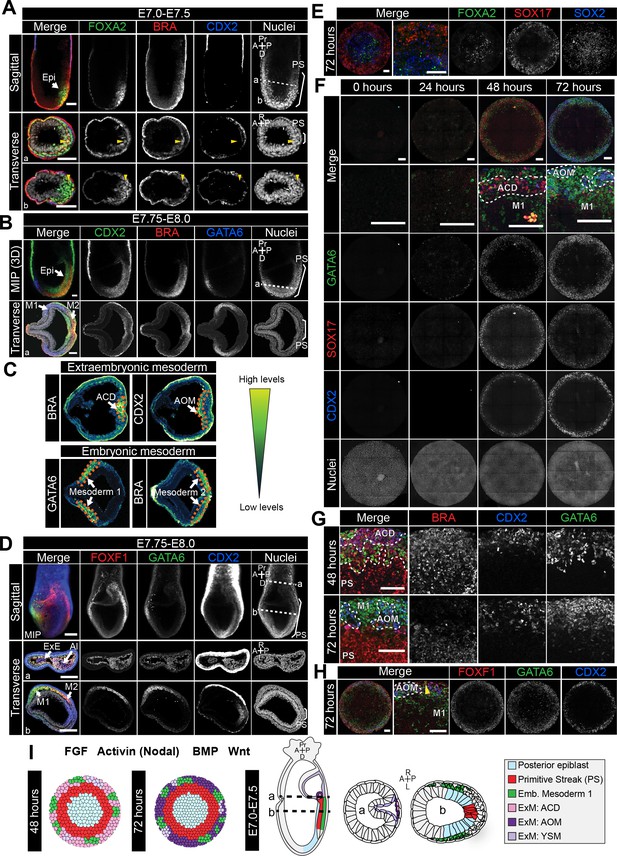
Assignment of cell identities to micropattern-differentiated EpiLC populations.
(A,B,D) Confocal maximum intensity projections (MIP), sagittal optical sections and transverse cryosections of immunostained gastrulating embryos. Dashed lines mark transverse plane. Epi, epiblast; PS, primitive streak; M1, Mesoderm1; M2, Mesoderm2; ACD, allantois core domain; AOM, allantois outer mesenchyme; ExE, extraembryonic ectoderm; Al, allantois; ExM, extraembryonic mesoderm; A, anterior; P, posterior; Pr, proximal; D, distal; R, right; L, left. Scale bars, 50 μm. (A) Yellow arrowheads mark BRACHYURY/FOXA2-coexpressing cells within the anterior PS. (C) LUT of immunostaining of BRACHYURY marking extraembryonic mesoderm allantois core domain (ACD) and CDX2 expressed highly in allantois outer mesenchyme (AOM) (upper panels) as well as GATA6 marking anteriorly migrated embryonic mesoderm (Mesoderm 1) and BRACHYURY marking embryonic mesoderm close to the PS (Mesoderm 2) (lower panels). Orange dashed lines delineate regions of interest. (E,F,H) MIPs of immunostained micropatterns. High magnification shows region at the colony edge. Yellow arrowhead H marks GATA6/FOXF1 cell. Scale bars, 100 μm. (G) High magnification of colony edge. Outer domain represents a mixture of populations often organized in clusters, highlighted by dashed lines. At 48 hr the ACD population coexpressed BRACHYURY and CDX2, M1 expressed GATA6. By 72 hr, outer cells expressed CDX2 (AOM) or GATA6 (M1). BRACHYURY marked PS cells. (I) Schematic diagram summarizing the cell identities observed at 48 and 72 hr of in vitro differentiation, under conditions described in B and corresponding in vivo fates. Dashed lines mark transverse plane. ExM, extraembryonic mesoderm.
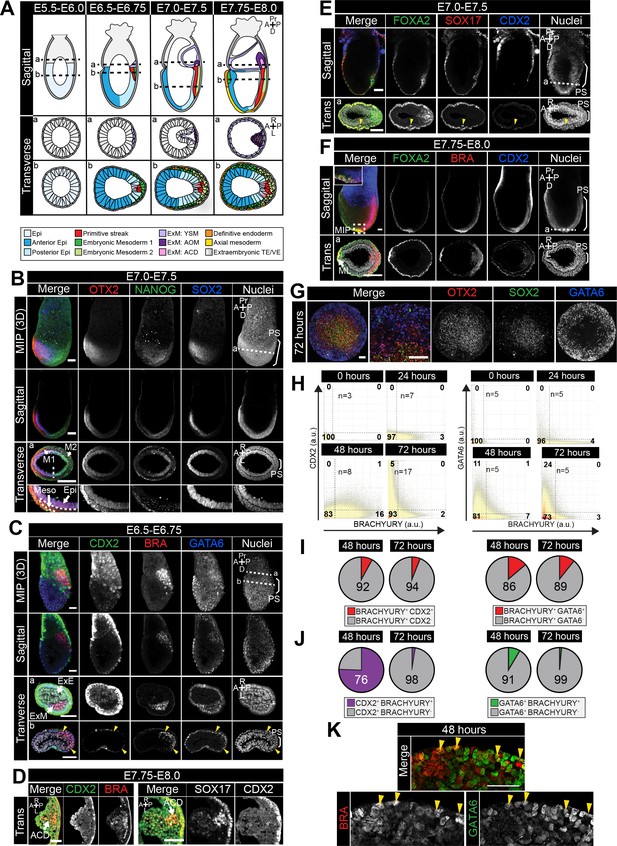
Populations arising during in vivo and in vitro differentiation.
(A) Schematic diagram showing cell types arising during mouse gastrulation. Upper panels show sagittal sections. Lower panels show transverse sections. Dotted lines demarcate approximate position of transverse sections. A, anterior; P, posterior; Pr, proximal; D, distal; R, right; L, left. Cell fates color-coded as in legend. Epi, epiblast; ExM, extraembryonic mesoderm; YSM, yolk sac mesoderm; AOM, allantois outer mesenchyme; ACD, allantois core domain; TE, trophectoderm; VE, visceral endoderm. (B–F) Confocal maximum intensity projections (MIP), sagittal optical sections and transverse cryosections of gastrulating mouse embryos. Dashed lines mark transverse plane. Non-nuclear anti- BRACHYURY/CDX2 VE fluorescence represents non-specific binding. PS, primitive streak; M1, Mesoderm 1; M2, Mesoderm 2; Meso, mesoderm; Epi, epiblast; ExE, extraembryonic ectoderm; ExM, extraembryonic mesoderm; ACD, allantois core domain; ML, midline, A, anterior; P, posterior; Pr, proximal; D, distal. Scale bars, 50 μm. (D) Images of transverse cryosections through posterior extraembryonic region of the allantois of the embryo. (E) Yellow arrowhead marks SOX17/FOXA2 -coexpressing definitive endoderm cell. (G) MIP of immunostained micropatterned colony. High magnification views shows a region at the colony edge. Scale bars, 100 μm. (H) Quantification of marker coexpression by voxel. Each dot indicates the fluorescence intensity of a single voxel, in arbitrary units (a.u.). Color represents voxel density within the plot. Gates were defined based on the 0 hr time point where BRACHYURY, CDX2 and GATA6 were not expressed. Numbers within each quadrant represent the % of voxels within gate, rounded to the nearest whole number. N, number of colonies. (I,J) Pie charts illustrating % of BRACHYURY-positive cells that coexpress CDX2 or GATA6 (I) and % of CDX2 or GATA6-positive cells coexpressing BRACHYURY (J). Percentages shown within the largest fraction. (K) High magnification confocal image of a region of the outer colony edge after 48 hr of differentiation under the conditions described in Figure 2E. Yellow arrowheads highlight GATA6/BRACHYURY-coexpressing cells. Scale bars, 50 μm.
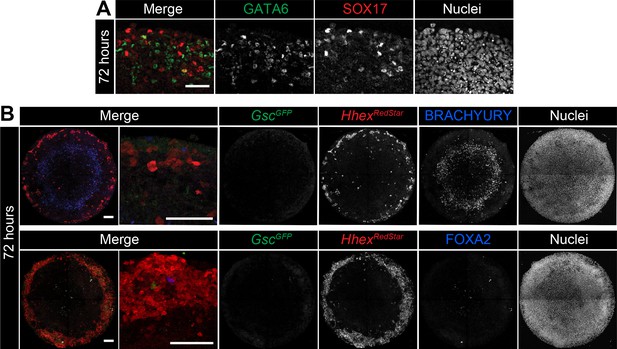
Micropatterns in the presence of BMP express SOX17 and Hhex but not Gsc.
EpiLCs were differentiated as in Figure 2E, with FGF2, ACTIVIN A, BMP4, WNT3A for 72 hr. (A) High magnification confocal optical section of a region of the colony edge after 72 hr of differentiation with FGF2, ACTIVIN A, WNT3A, BMP4. (B) Confocal MIPs of differentiated and immunostained GscGFP/+;HhexRedStar/+ dual reporter cells. Scale bars, 100 μm.
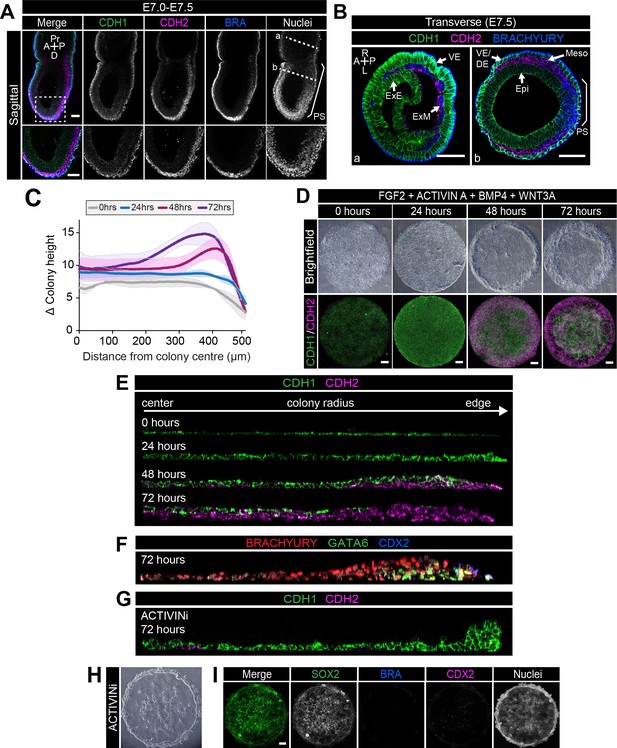
EMT is associated with micropatterned EpiLC differentiation.
Data from colonies differentiated as in Figure 2E. (A,B) Sagittal (A) and transverse sections (B) of late streak embryo. Dashed box marks high magnification region in lower panel. Dashed lines mark transverse planes in B. Non-nuclear anti-BRACHYURY VE fluorescence represents non-specific binding. A, anterior; P, posterior; Pr, proximal; D, distal; L, left; R, right; VE/DE, visceral endoderm/definitive endoderm; ExE, extraembryonic ectoderm; ExM, extraembryonic mesoderm; Epi, epiblast; Meso, mesoderm. Scale bars, 50 μm. (C) Quantification of colony height from colony center (0) to edge (500) across multiple colonies, three independent experiments, 0 hr: n = 11, 24 hr: n = 15, 48 hr: n = 17, 72 hr: n = 18. (D) Time-course showing brightfield images (upper panels) and MIPs of comparable immunostained colonies (lower panels). Scale bars, 100 μm. (E–G) Images of z-axis profile from colony center (left) to edge (right). (G–I) EpiLCs were plated onto micropatterns overnight with F/A. The following day medium was changed to F/A, BMP4, WNT3A (E,F) or medium blocking Activin/Nodal signaling - FGF2, BMP4, WNT3A, SB431542 (ACTIVINi, (G–I). (H) brightfield image of ACTIVINi colony. (I) MIPs of immunostained ACTIVINi colonies at 72 hr differentiation. Scale bars, 100 μm. BRA, BRACHYURY.
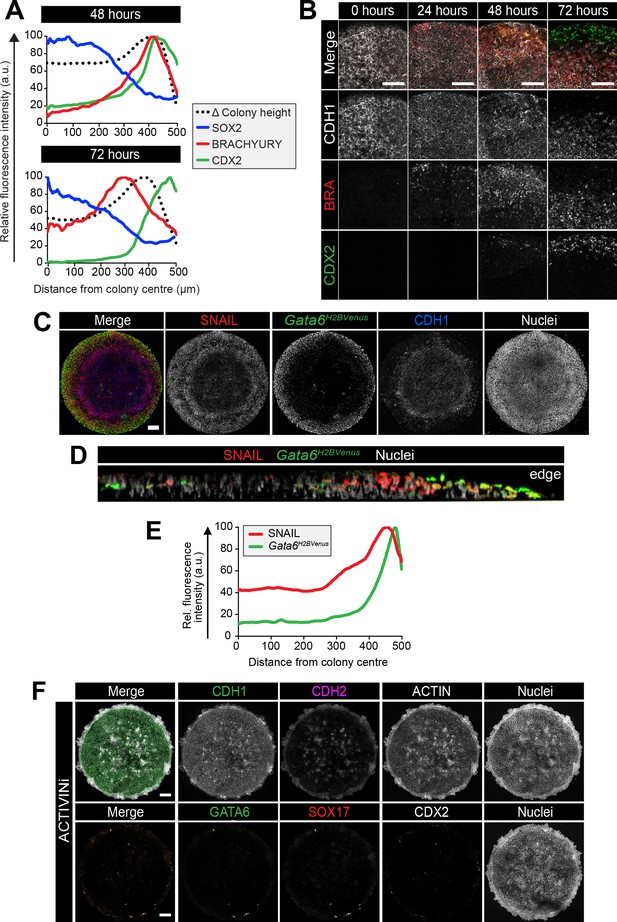
Cells undergo an epithelial to mesenchymal transition during gastrulation and in vitro differentiation.
(A) Quantification of immunostaining voxel fluorescence intensity (in arbitrary units, a.u.) of SOX2, BRACHYURY and CDX2 from colony center (0) to edge (500) as well as relative colony height. Data represents average voxel intensity or change in colony height across multiple colonies relative to the maximum value for each marker. (B) Confocal optical sections of the outer colony edge. Scale bars, 100 μm. (C) MIPs of immunostained 72 hr colonies. Scale bars, 100 μm. (D) Images of z-axis profile from colony center (left) to edge (right). (E) Quantification of immunostaining. Voxel fluorescence intensity was measured from colony center (0) to edge (500). Data represents average voxel intensity across multiple colonies (n = 9) relative to maximum voxel intensity for each marker. (F) EpiLCs were plated onto micropatterns overnight in N2B27 with 12 ng/ml FGF2 and 20 ng/ml ACTIVIN A (F/A). The following day medium was changed to 12 ng/ml FGF2, 50 ng/ml BMP4, 200 ng/ml WNT3A and 10 μM SB431542 (ACTIVINi). Confocal maximum intensity projections of immunostained colonies after 72 hr of differentiation. Scale bars, 100 μm. BRA, BRACHYURY.
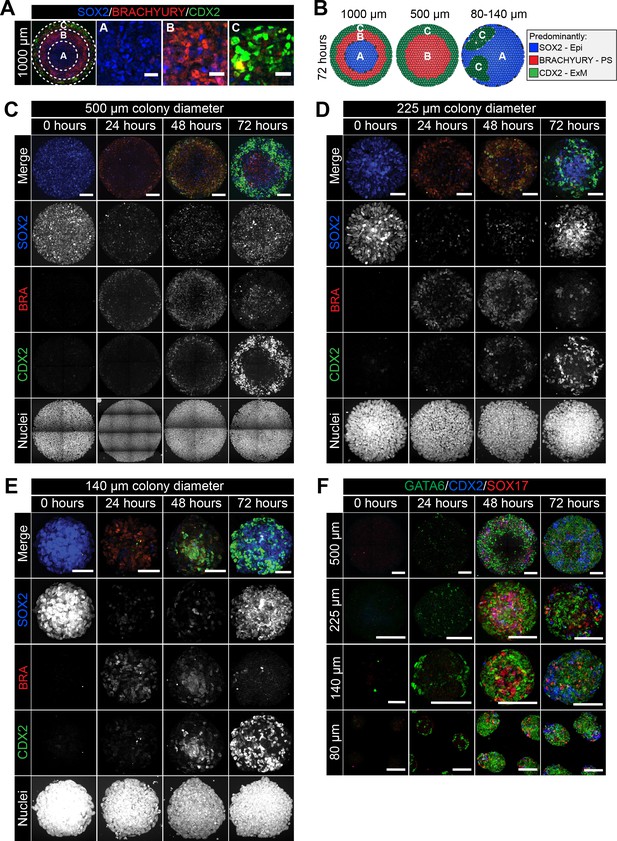
Smaller diameter colonies pattern in the same order of events but lose central populations.
(A) EpiLCs were differentiated with FGF2 and ACTIVIN A (F/A), BMP4 and WNT3A as described in Figure 2E. Confocal optical section of a representative 1000 μm diameter colony after differentiation. Dashed circles define 3 regions of distinct marker expression, shown at higher magnification in adjacent panels. While SOX2 is expressed quite broadly, regions were defined based on the marker that was predominantly expressed. Region A (central) = SOX2 (blue), Region B (intermediate) = BRACHYURY (red), Region C (outer) = CDX2 (green). Scale bars, 25 μm. (B) Schematic diagram showing the changing marker expression in colonies of different diameters. (C–F) Representative confocal maximum intensity projections of colonies at 0, 24, 48 and 72 hr after addition of BMP4 and WNT3A to F/A medium. Images show colonies of 500 μm, 225 μm, 140 μm and 80 μm diameter. Scale bars, 100 μm.
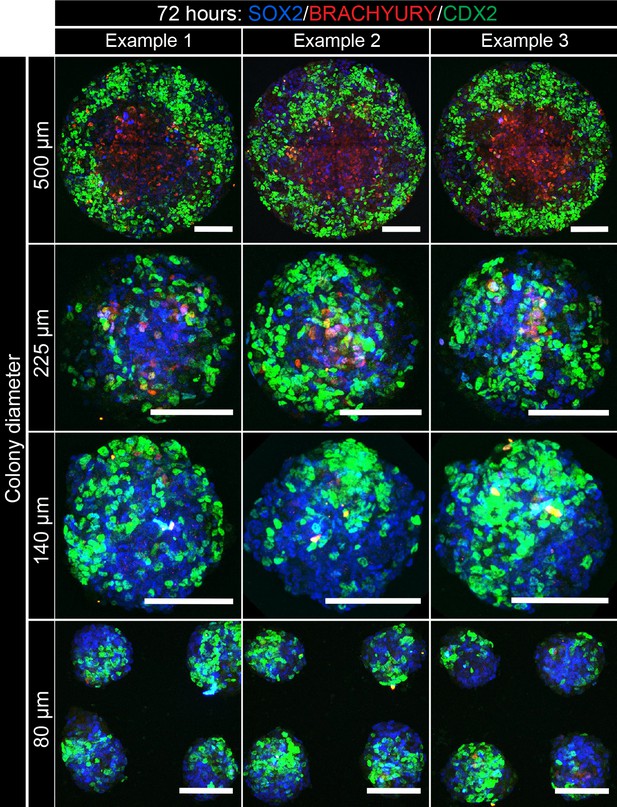
Patterning of cell fates is altered at different colony diameters.
EpiLCs were differentiated as previously described in Figure 2E on different diameter micropatterns. Images show confocal maximum intensity projections of immunostained colonies after 72 hr of differentiation. Three representative examples are shown for each colony diameter. Scale bars, 100 μm.
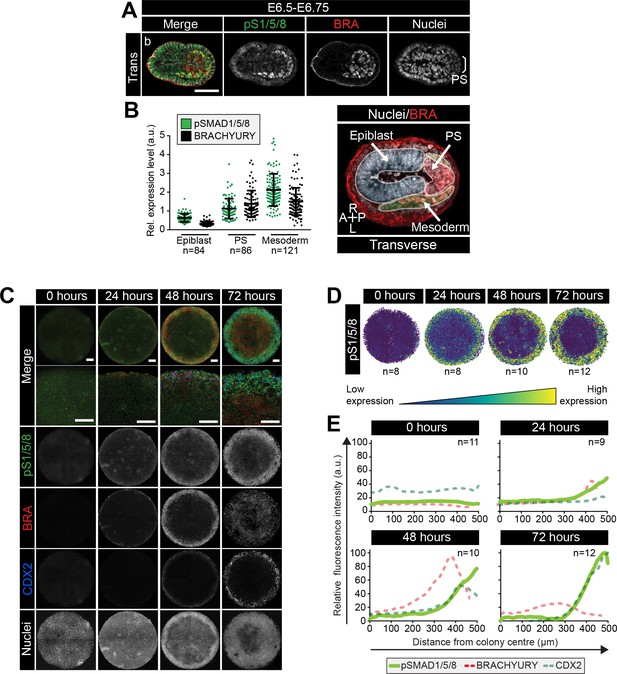
BMP signaling in micropatterns and embryos correlates with embryonic and extraembryonic mesoderm fates.
(A) Transverse cryosection of immunostained embryo in Figure 6—figure supplement 1B. Scale bar, 50 μm. (B) Quantification of pSMAD1/5/8 and BRACHYURY fluorescence intensity in E6.5 embryos. Cells within the epiblast, primitive streak (PS) and mesoderm were manually selected on confocal images of transverse cryosections in ImageJ as shown in right-hand panel. PS = BRACHYURY positive cells at embryo posterior. Mesoderm = cells positioned between VE and Epi. Quantification was carried out on three cryosections per embryo. N, number of cells. Data represents mean fluorescence intensity ± S.D. normalized to Hoechst fluorescence. (C) MIPs of immunostained colonies differentiated as in Figure 2E. Second panel depicts high magnification of colony edge. Scale bars, 100 μm. BRA, BRACHYURY; pS1/5/8, phosphorylated SMAD1/5/8. (D) Depiction of spatial patterning across multiple colonies. Each dot represents a single cell. (E) Quantification of voxel fluorescence intensity of pSMAD1/5/8 from colony center (0 μm) to edge (500 μm). Data represents average voxel intensity across multiple colonies. pSMAD1/5/8 colony numbers (n) in upper right corner. Data relative to maximum voxel intensity across the time course for each marker.
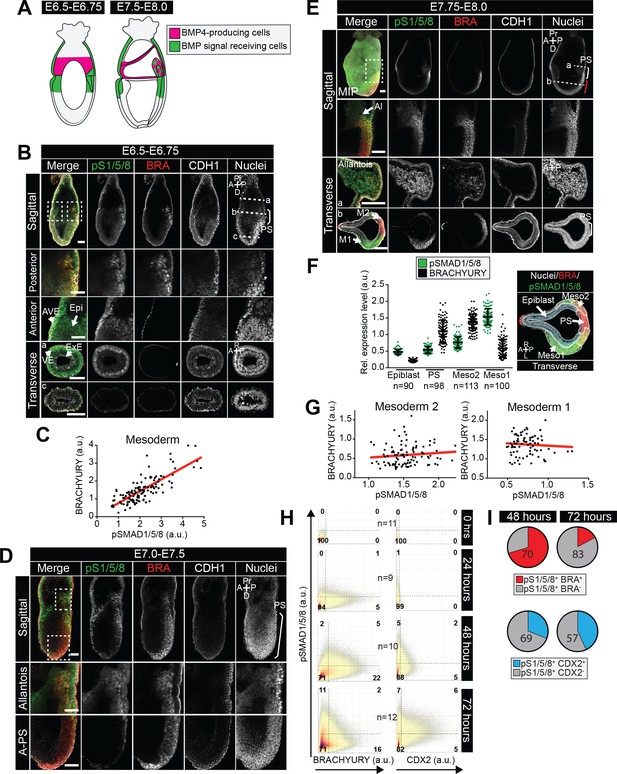
BMP signaling is active in the posterior primitive streak, embryonic and extraembryonic mesoderm.
(A) Schematic diagram depicting the sources of BMP (pink) within gastrulating embryos, described in (Lawson et al., 1999). At the start of gastrulation (E6.5-E6.75), BMP4 is expressed by the extraembryonic ectoderm and later (E7.5-E8.0) in the allantois, amnion and chorion. (B,D,E) Representative confocal images of immunostained gastrulating embryos showing BMP signaling activity based on nuclear localization of pSMAD1/5/8. Images represent maximum intensity projections (MIP), sagittal optical sections and transverse cryosections. Transverse section ‘b’ from panel B is shown in Figure 6A. Non-nuclear anti-BRACHYURY VE fluorescence represents to non-specific binding. AVE, anterior visceral endoderm; Epi, epiblast; ExE, extraembryonic ectoderm; VE, visceral endoderm; A-PS, anterior primitive streak; Al, allantois; M1, mesoderm 1; M2, mesoderm 2; A, anterior; P, posterior; Pr, proximal; D, distal; R, right; L, left; BRA, BRACHYURY; E-CAD, E-CADHERIN; pS1/5/8, pSMAD1/5/8. Scale bars, 50 μm. Dashed lines mark transverse plane. Dashed boxes outline regions in higher magnification in lower panels. In F, white bracket demarcates the posterior primitive streak and red bracket the anterior primitive streak. (C,G) Scatter plots showing the levels of pSMAD1/5/8 and BRACHYURY in arbitrary units (a.u.) within single mesoderm cells of early streak (E6.5-E6.75 - C) and early headfold (E7.75-E8.0 - G) embryos. Each dot represents a single cell. Linear regression curves were fitted to the points (red line). (F) Quantification of pSMAD1/5/8 and BRACHYURY fluorescence intensity in arbitrary units (a.u.) in early headfold (E7.75-E8.0) embryos. Cells within the epiblast, primitive streak (PS) and Mesoderm 1 and 2 (Meso1/2) were manually selected on confocal images of transverse cryosections using ImageJ software as shown in right-hand panel. The PS was identified as BRACHYURY-positive cells at the embryo posterior. Mesoderm cells were separated into two categories, those that had migrated further anteriorly and expressed low BRACHYURY and high pSMAD1/5/8 (Meso1) and those closest to the PS expressing high BRACHYURY (Meso2). Quantification was carried out on three cryosections per embryo. N, number of cells. Data represents mean fluorescence intensity normalized to the fluorescence intensity of Hoechst nuclear stain ± S.D. (H) Quantification of marker coexpression by voxel. Each dot indicates the fluorescence intensity of a single voxel, in arbitrary units (a.u.). Color represents voxel density within the plot. Gates were defined based on the 0 hr time point where BRACHYURY and CDX2 were not expressed and pSMAD1/5/8 was expressed only at low levels. Numbers within each quadrant represent the % of voxels within gate, rounded to the nearest whole number. N, number of colonies. (I) Pie charts illustrating the % of pSMAD1/5/8-positive cells that coexpress BRACHYURY or CDX2. Percentages shown within the largest fraction.
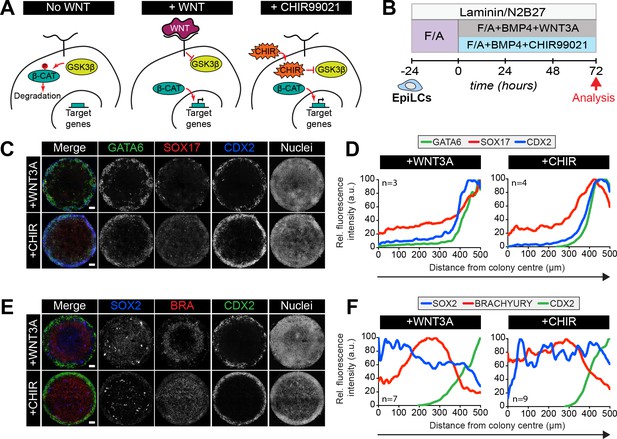
Bypassing the WNT receptor alters spatial patterning.
(A) Simplified schematic of the Wnt pathway. In the absence of WNT receptor binding (left panel), GSKβ phosphorylates β-catenin, targeting it for degradation. β-catenin target genes are inactive. When WNT binds to the receptor (middle panel), factors downstream of the receptor are activated and inhibit GSKβ activity. β-catenin is not phosphorylated and not degraded, but instead translocates to the nucleus and activates target genes. CHIR99021 (CHIR) is a small molecule inhibitor of GSKβ. CHIR directly inactivates GSKβ independent of WNT receptor activation (right panel). As with WNT receptor binding, inactive GSKβ cannot phosphorylate β-catenin hence target genes are activated. (B) Schematic diagram of differentiation conditions. EpiLCs were generated as described in Figure 1C. EpiLCs were plated overnight onto Laminin-coated micropatterns (−24 hr) in N2B27 medium with 12 ng/ml FGF2 and 20 ng/ml ACTIVIN A (F/A). Various conditions were then used for further differentiation, F/A with BMP4 (50 ng/ml) and WNT3A (200 ng/ml) (Control) or F/A, BMP4 and 3 μM CHIR99021, a small molecule inhibitor of GSK3. Colonies were analyzed after 72 hr differentiation. (C,E) Confocal maximum intensity projections of immunostained colonies after 72 hr of differentiation. Scale bars, 100 μm. (D,F) Quantification of immunostaining voxel fluorescence intensity, in arbitrary units (a.u.), from colony center (0) to edge (500). Data represents average voxel intensity across multiple colonies.
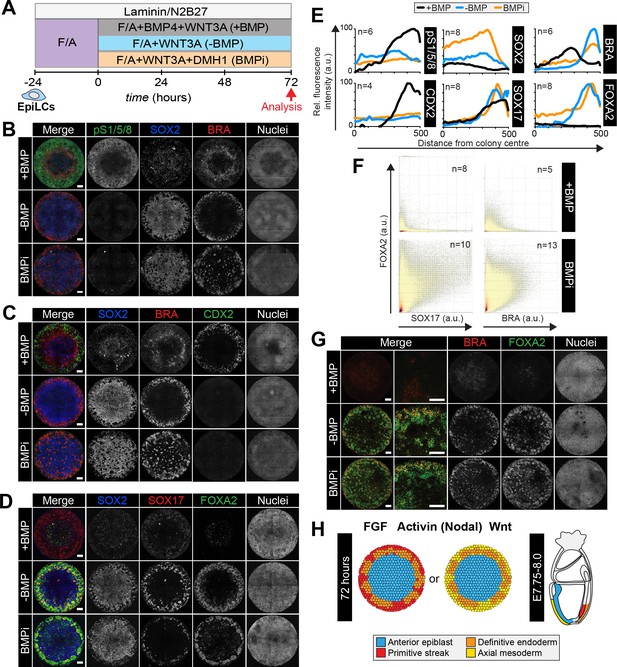
Anterior primitive streak fates are specified in the absence of BMP.
(A) EpiLCs generated as in Figure 1C were plated overnight onto Laminin-coated micropatterns (−24 hr) in N2B27 medium with F/A. Various conditions were used for further differentiation - F/A, BMP4, WNT3A (+BMP), F/A, WNT3A (-BMP) or F/A, WNT3A with DMH1 BMP signaling inhibitor (BMPi). Colonies were analyzed after 72 hr differentiation. (B–D, G). MIPs of immunostained 72 hr colonies. Scale bars, 100 μm. (E) Quantification of immunostaining. Voxel fluorescence intensity was measured from colony center (0) to edge (500). Data represents average voxel intensity across multiple colonies relative to maximum voxel intensity for each marker. (F) Quantification of marker coexpression by voxel. Each dot indicates fluorescence intensity of a single voxel. Color represents voxel density within the plot. Numbers within quadrants show % of voxels within the gate. N, number of colonies. (H) Schematic diagram summarizing the cell fates observed after 72 hr in vitro differentiation under conditions described in A and corresponding in vivo cell types at E7.75-E8.0. The outer domain of the micropattern colony comprises cells that coexpress SOX17 and FOXA2, representing definitive endoderm and cells that coexpress BRACHYURY and FOXA2, representing anterior primitive streak or axial mesoderm cells.
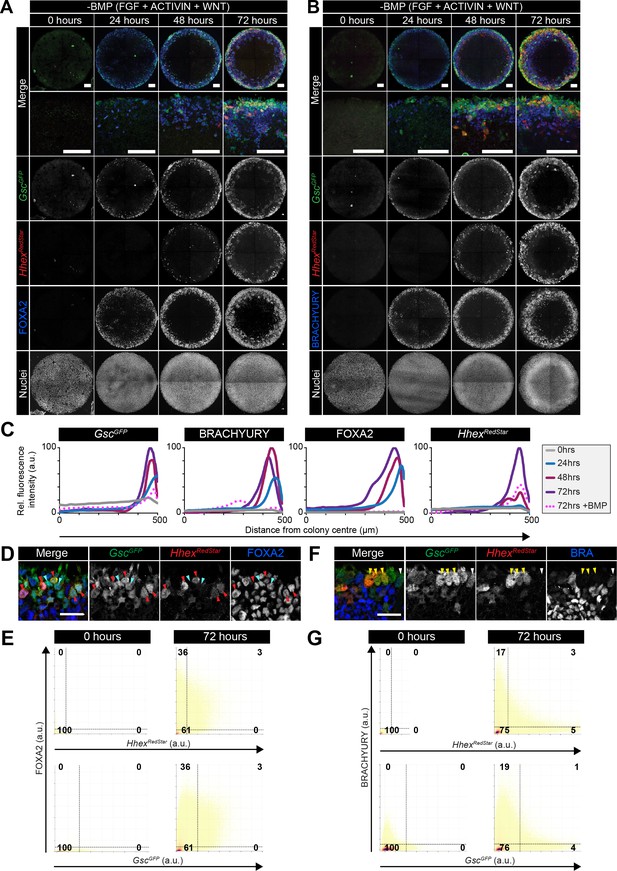
Anterior primitive streak and definitive endoderm populations are formed in the absence of BMP.
GscGFP/+;HhexRedStar/+ dual reporter ESCs were differentiated as in Figure 7A, with FGF2, ACTIVIN A, WNT3A for 72 hr. (A,B) Confocal MIPs of immunostained colonies. Scale bars, 100 μm. (C) Quantification of immunostaining voxel fluorescence intensity in arbitrary units (a.u.), from colony center (0) to edge (500). Data represents average voxel intensity across multiple colonies (n = 10/time point) relative to maximum voxel intensity for each marker. (D,F) High magnification confocal optical sections of a region of the colony edge after 72 hr of differentiation. (D) Red arrowheads mark FOXA2/GscGFP/HhexRedStar -expressing cells. Cyan arrowheads mark FOXA2/GscGFP -expressing cells. (F) Yellow arrowheads mark GscGFP/HhexRedStar –expressing cells. White arrowheads mark GscGFP/BRACHYURY –expressing cells. Scale bars, 25 μm (E,G) Quantification of marker coexpression by voxel. Each dot indicates the fluorescence intensity of a single voxel, in arbitrary units (a.u.). Color represents voxel density within the plot. Gates were defined based on the 0 hr time point when FOXA2, BRACHYURY, GscGFP and HhexRedStar were not expressed. Numbers within each quadrant represent the % of voxels within gate, rounded to the nearest whole number. n = 10 colonies/time point.
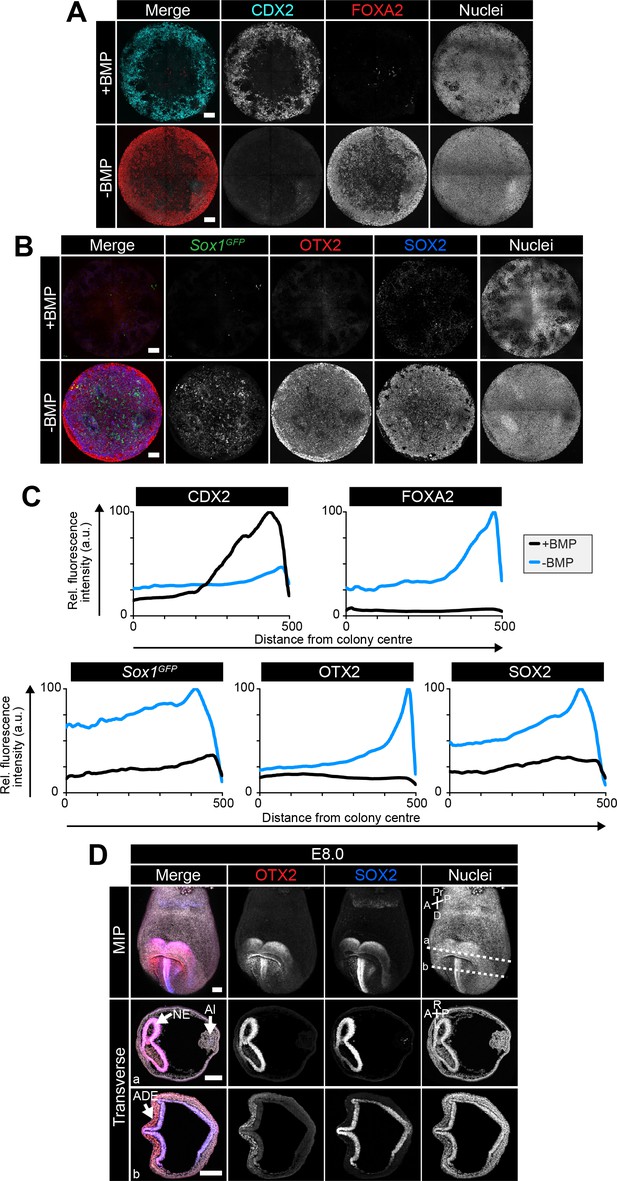
An anterior epiblast/neurectoderm population is formed in the absence of BMP.
Sox1GFP/+ EpiLCs generated as in Figure 1C were plated overnight onto Laminin-coated micropatterns (−24 hr) in N2B27 medium with F/A. Various conditions were used for further differentiation - F/A, BMP4, WNT3A (+BMP) or F/A, WNT3A (-BMP). Colonies were analyzed after 72 hr differentiation. (A,B) MIPs of immunostained 72 hr colonies. Sox1GFP/+ cells were stained with an anti-GFP antibody. Scale bars, 100 μm. (C) Quantification of immunostaining. Voxel fluorescence intensity was measured from colony center (0) to edge (500). Data represents average voxel intensity across multiple colonies relative to maximum voxel intensity for each marker. n = 10. (D) Confocal maximum intensity projections (MIP) and transverse cryosections of an E8.0 mouse embryo. Dashed lines mark transverse plane. Al, allantois; NE, neurectoderm; ADE, anterior definitive endoderm; A, anterior; P, posterior; Pr, proximal; D, distal; L, left; R, right. Scale bars, 100 μm.
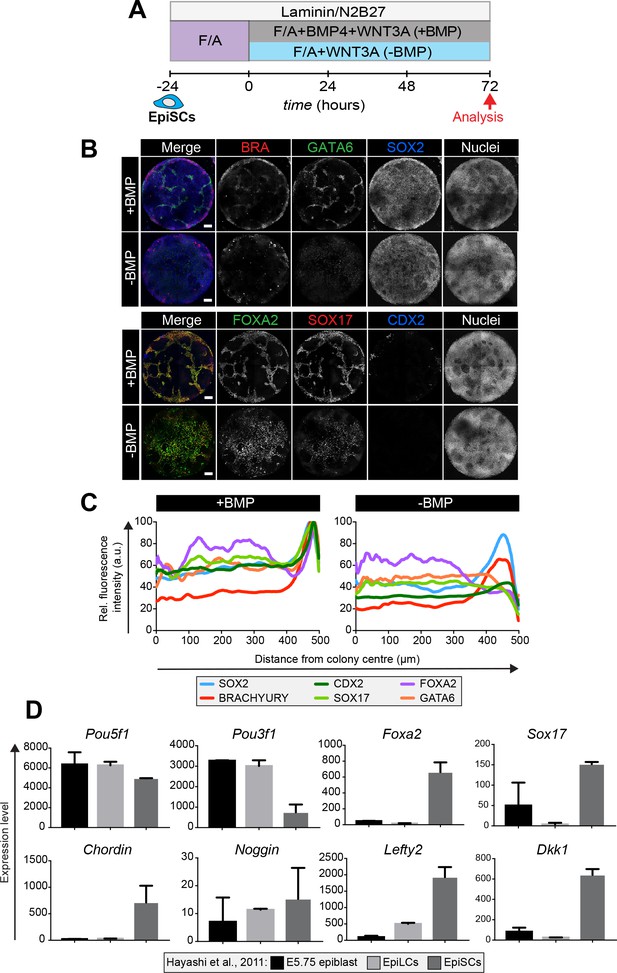
Epiblast stem cells undergo definitive endoderm differentiation in the presence or absence of BMP.
(A) Epiblast stem cells (EpiSCs) of the EpiSC9 line (Najm et al., 2011) were cultured in the presence of 12 ng/ml FGF2 and 20 ng/ml ACTIVIN A (F/A) on fibronectin. EpiSCs were then plated overnight onto Laminin-coated micropatterns (−24 hr) in N2B27 medium with F/A. Various conditions were used for further differentiation - F/A, BMP4, WNT3A (+BMP) or F/A, WNT3A (-BMP). Colonies were analyzed after 72 hr differentiation. (B) MIPs of immunostained 72 hr colonies. Scale bars, 100 μm. (C) Quantification of immunostaining. Voxel fluorescence intensity was measured from colony center (0) to edge (500). Data represents average voxel intensity across multiple colonies (n = 10/condition) and is shown relative to maximum voxel intensity for each marker across both conditions. (D) Graphs showing the expression level of a number of genes from the published microarray dataset of Hayashi et al. from E5.75 in vivo epiblast, EpiLCs and EpiSCs (Hayashi et al., 2011). Data shown is from amplified RNA samples and represents the mean ± S.D for two independent replicates.
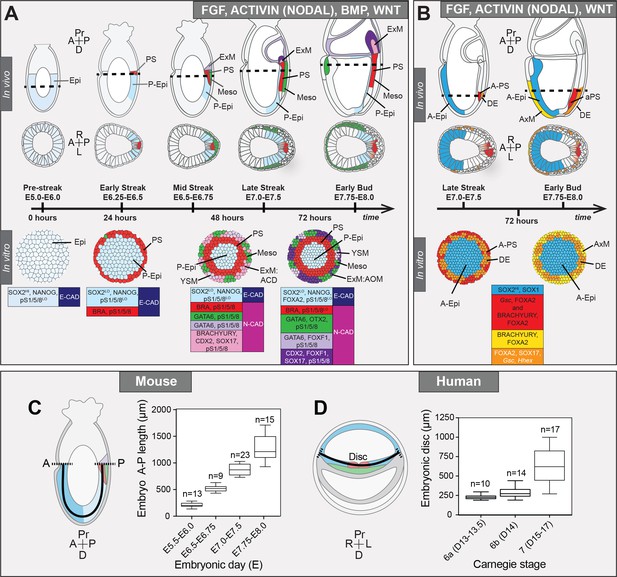
Micropattern differentiation of mouse pluripotent stem cells recapitulates cell fate specification in the posterior or anterior primitive streak.
(A) Summary of embryo gastrulation (upper) and correlation with in vitro micropattern differentiation (lower). With FGF2, ACTIVIN A, BMP4 and WNT3A, mouse PSC differentiation recapitulated differentiation in the proximal posterior of the gastrulating embryo. Epi-like cells (EpiLCs) correlated to the embryonic day (E) 5.5–6.0 pre-streak epiblast (Epi). After 24 hr, cells in the colony center adopted a posterior Epi (P-Epi) identity and a primitive streak (PS)-like population arose at the colony edge, as E6.25-E6.5. After 48 hr, clusters of cell populations emerged at the outer colony edge correlating to embryonic Mesoderm 1 (Meso), and extraembryonic mesoderm (ExM) allantois core domain (ACD) arising at E6.75-E7.0. After 72 hr, cells in the colony center represented the distal P-Epi of E7.0-E7.25 embryos. Meso, ExM and PS populations were maintained. However, ACD cells were replaced by cells with an allantois outer mesenchyme (AOM) identity. Cells were highly confluent and could not be maintained under these conditions after 72 hr. LO, low expression. Dashed lines mark transverse plane shown below. (B) Summary of correlation between in vitro micropattern differentiation with FGF, ACTIVIN and WNT and in vivo gastrulating embryos. Under these conditions, mouse PSCs recapitulated differentiation of distal posterior (left panels) or distal posterior and anterior of embryo. After 72 hr, the central population expressed elevated levels of SOX2 compared to BMP4 conditions, likely representing anterior Epi (A-Epi). Cells coexpressed FOXA2, SOX17, Gsc and Hhex representing definitive endoderm (DE), FOXA2 and BRACHYURY representing anterior PS (A–PS) or AxM and or FOXA2 and Gsc likely representing A-PS. HI, high expression. A, anterior; P, posterior; Pr, proximal; D, distal. Color-coded legends highlight key markers of different cell states at each time point. (C) Box plots showing Epi length along the anterior-posterior (A–P) axis at pre- (E5.5-E6.0) early (E6.5-E6.75), mid- (E7.0-E7.5) and late (E7.75-E8.0) streak stages of mouse embryonic development. The A-P length was measured on sagittal confocal optical sections through the middle of the embryo with ImageJ software, as depicted in the schematic diagram. N, number of embryos. (D) Box plots showing human embryonic disc measurements compiled from human embryo data collections. Abnormal embryo data were excluded. D = embryonic day. Carnegie stage 6a, pre-streak; 6b, early streak; 7, early-mid gastrulation.
Tables
Summary of cell fates arising in the presence of various signaling factors.
Cell fates generated after 72 hr of mouse micropattern differentiation with described cytokine combinations. It should be noted that cells do not detect these signals homogeneously. WNT inhibition (WNTi) refers to XAV treatment; ACTIVIN inhibition (ACTIVINi) refers to the absence of ACTIVIN and SB431542.
| Signaling pathways | Outcome |
|---|---|
| FGF, ACTIVIN, (WNTi) | Epiblast |
| FGF, ACTIVIN, endogenous WNT | Epiblast PS |
| FGF, ACTIVIN, BMP, WNT | Posterior epiblast PS Extraembryonic mesoderm Embryonic mesoderm |
| FGF, ACTIVIN, WNT | Anterior epiblast Definitive endoderm Axial mesoderm or Anterior PS |
| FGF, BMP, WNT (ACTIVINi) | Epiblast |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | Crl:CD1 (ICR) | RRID:IMSR_CRL:22 | CD1 Mus musculus wild-type outbred mouse | |
| Cell line (Mus musculus) | ES-E14 | (Hooper et al., 1987) | RRID:CVCL_C320 | Embryonic stem cell line:Mus musculus |
| Cell line (Mus musculus) | ES-R1 | (Nagy et al., 1993) | RRID:CVCL_2167 | Embryonic stem cell line:Mus musculus |
| Cell line (Mus musculus) | Sox17GFP/+ | (Kim et al., 2007) | Embryonic stem cell line:Mus musculus | |
| Cell line (Mus musculus) | TGFP/+ | (Abe and Naski, 2004) | Embryonic stem cell line: Mus musculus | |
| Cell line (Mus musculus) | 46 C cell line (Sox1GFP) | (Ying et al., 2003) | RRID:CVCL_Y482 | Embryonic stem cell line:Mus musculus |
| Cell line (Mus musculus) | GscGFP/+; HhexRedStar/+ | (Villegas et al., 2013) | Embryonic stem cell line:Mus musculus | |
| Cell line (Mus musculus) | EpiSC9 | (Najm et al., 2011) | Epiblast stem cell line: Mus musculus | |
| Antibody | anti-BRACHYURY | R and D Systems | Cat# AF2085, RRID:AB_2200235 | 1:200 |
| Antibody | anti-CDH1 | Sigma-Aldrich | Cat# U3254, RRID:AB_477600 | 1:500 |
| Antibody | anti-CDH2 | Santa Cruz Biotechnology | Cat# sc-7939, RRID:AB_647794 | 1:300 |
| Antibody | anti-CDX2 | BioGenex | Cat# AM392, RRID:AB_2650531 | 1:200 |
| Antibody | anti-DsRed | Clontech Laboratories, Inc. | Cat# 632496, RRID:AB_10013483 | 1:500 |
| Antibody | anti-FOXA2 | Abcam | Cat# ab108422, RRID:AB_11157157 | 1:500 |
| Antibody | anti-FOXF1 | R and D Systems | Cat# AF4798, RRID:AB_2105588 | 1:200 |
| Antibody | anti-GATA6 | R and D Systems | Cat# AF1700, RRID:AB_2108901 | 1:100 |
| Antibody | anti-GATA6 | Cell Signaling Technology | Cat# 5851, RRID:AB_10705521 | 1:500 |
| Antibody | anti-GFP | Aves Labs | Cat# GFP-1020, RRID:AB_10000240 | 1:500 |
| antibody | anti-KLF4 | R and D Systems | Cat# AF3158, RRID:AB_2130245 | 1:200 |
| Antibody | anti-NANOG | Thermo Fisher Scientific | Cat# 14-5761-80, RRID:AB_763613 | 1:200 |
| Antibody | anti-NANOG | Cosmo Bio Co | Cat# REC-RCAB0002PF, RRID:AB_567471 | 1:500 |
| Antibody | anti-OTX2 | R and D Systems | Cat# AF1979, RRID:AB_2157172 | 1:500 |
| Antibody | anti-POU3F1 | Millipore Sigma | MABN738 | 1:100 |
| Antibody | anti-POU5F1 | Santa Cruz Biotechnology | Cat# sc-5279, RRID:AB_628051 | 1:100 |
| Antibody | anti-pSMAD1/5/8 | a gift from Dr. Ed Laufer, Columbia University, New York, NY | N/A | 1:200 |
| Antibody | anti-SNAIL | R and D Systems | Cat# AF3639, RRID:AB_2191738 | 1:100 |
| Antibody | anti-SOX2 | Thermo Fisher Scientific | Cat# 14-9811-82, RRID:AB_11219471 | 1:200 |
| Antibody | anti-SOX17 | R and D Systems | Cat# AF1924, RRID:AB_355060 | 1:200 |
| Software, algorithm | Ilastik | http://ilastik.org/ | RRID:SCR_015246 | 3-D Nuclear mask generation |
Additional files
-
Supplementary file 1
Summary of expression data for a panel of factors used for cell fate assignments in this study.
Where data was not generated in this study, appropriate literature references are provided – listed in bold lowercase letters, full references provided below the table. Where primary data is included in this study, the reference is given in the following format – (Main text figure (Fig._) – figure supplement (S_): Supplementary file 2). Cell fates are color-coded as in Figure 3—figure supplement 1A and in table legend. At each gastrulation stage, each cell type was classified as 1 = factor expressed, 0.5 = factor expressed heterogeneously or at low levels, 0 = not expressed (white), ?=expression unclear from immunostaining and exact localization is unclear/unknown from published literature. FOXF1 is expressed in the embryonic mesoderm and also reported to be expressed within the PS at E7.0-E7.5 but PS expression is unclear. Gsc and Hhex are both expressed in the AxM although their expression has been reported to be transient in some studies and the exact timing of expression is not clear. To note, some cells of the PS coexpressed BRACHYURY, SOX2 and NANOG, likely a transition state between Epi and PS before SOX2 and NANOG are downregulated. It was unclear whether GATA6 was absent or expressed only at low levels in the ACD and early AOM. Certain markers were expressed in some but not all embryos at a particular stage – likely indicating the onset of expression, for example SOX17 within the ExM at E7.0-E7.5. *** At E7.0-E7.5 both DE and mesoderm cells arise from the anterior PS and are spatially intermixed hence it is difficult to identify DE cells based only on localization. At this time DE therefore refers to presumptive DE cells arising from the anterior PS based on known marker expression.
- https://doi.org/10.7554/eLife.32839.023
-
Supplementary file 2
Representative data of immunostained gastrulating mouse embryos utilized to identify marker signatures of distinct cell states.
Supplemental file comprising representative images of embryo data used in this study to identify in vivo marker signatures for particular cell states. Embryos were collected at different stages of development throughout gastrulation, from embryonic day (E) 6.25-E8.5 and immunostained for trios of marker combinations. File contains images of wholemount embryos and cryosections acquired by confocal microscopy. The expression patterns of the markers determined using this data are summarized in Supplementary file 1.
- https://doi.org/10.7554/eLife.32839.024





