Mfn2 ubiquitination by PINK1/parkin gates the p97-dependent release of ER from mitochondria to drive mitophagy
Figures
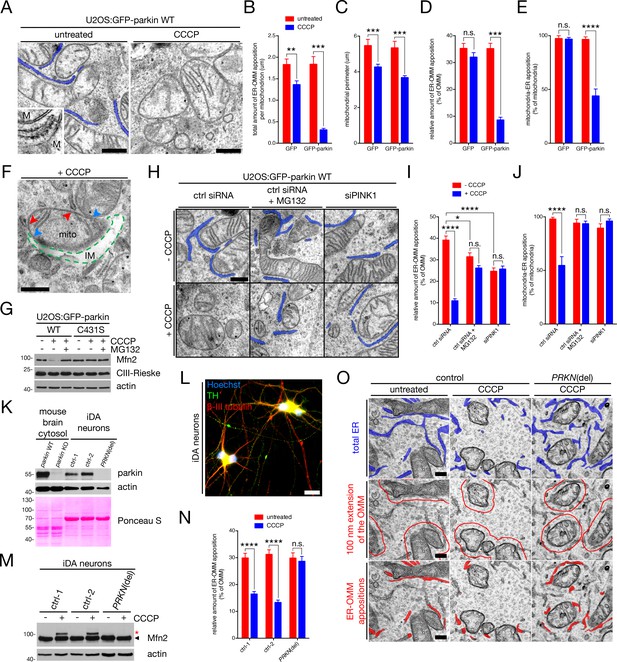
Ultrastructural analysis of ER-mitochondria contact during mitophagy in U2OS cells and dopaminergic neurons.
(A) Representative TEM images of mitochondria (‘M’) in contact with ER (pseudocoloured blue) in untreated and CCCP-treated U2OS:GFP-parkin cells. Scale bars, 500 nm. (B–E) Quantification of TEM from (A) in U2OS:GFP and GFP-parkin WT cells, left untreated (red bars) or treated with 20 μM CCCP for four hours (blue bars). Total apposition length (B), mitochondrial size (C), and the percent of OMM per mitochondrion (D) and mitochondria per field (E) in contact with the ER was quantified. Bars represent mean ± SEM, n = 82 to 152 mitochondria in 15 to 19 fields per condition. n.s., not significant; **, p<0.01; ***, p<0.001; ****, p<0.0001. (F) TEM image of an isolation membrane (‘IM’, broken green line) wrapping a mitochondrion (‘mito’). Blue arrowheads indicate the boundaries of OMM rupture, while red arrowheads indicate ER tubules in contact with the intact portion of the OMM. Scale bar, 500 nm. (G) Immunoblot analysis of whole-cell lysates from U2OS:GFP-parkin WT and C431S cells treated with 20 μM CCCP for four hours with or without 10 μM MG132. In the case of MG132 treatment, cells were first pre-incubated with 10 μM MG132 for 30 min prior to addition of CCCP. (H) Representative TEM images of mitochondria in contact with ER (pseudocoloured blue) in U2OS:GFP-parkin WT cells transfected with the indicated siRNA, and treated with 20 μM CCCP (‘+CCCP’) for four hours, in the presence or absence of 10 μM MG132 as in (G). Scale bar, 500 nm. (I,J) Quantification of TEM from (H) in cells treated with (blue bars) or without (red bars) 20 μM CCCP for four hours. The percent of OMM per mitochondrion (I) and mitochondria per field (J) in contact with the ER were quantified. Bars represent mean ± SEM, n = 101 to 203 mitochondria in 14 to 16 fields per condition. n.s., not significant; *, p<0.05; ***, p<0.001; ****, p<0.0001. (K) Immunoblot analysis of parkin levels in mouse brain cytosol from parkin+/+ and parkin-/- mice, along with whole-cell lysates from iDA neurons derived from iPSCs isolated from control (ctrl) individuals and a PRKN patient ("PRKN(del)"). (L) A representative wide-field image showing that iDA neurons express TH (green) and β-III tubulin (red) (Hoechst, blue). Scale bar, 20 microns. (M) Immunoblot analysis of whole-cell lysates from iDA neurons treated with 20 μM CCCP for one hour. The arrowhead indicates the unmodified Mfn2 band, while the red asterisk indicates ubiquitinated Mfn2. (N) Quantification of the percent of the OMM opposed to the ER in iDA neurons treated with 20 μM CCCP for one hour. Bars represent mean ± SEM, n = 80 to 131 mitochondria per condition. n.s., not significant; ****, p<0.0001. (O) Representative TEM images of mitochondria in contact with ER in iDA neurons. In the top row, total ER is pseudocoloured blue. In the second row, the red line denotes an area within 100 nm of the OMM. In the bottom row, ER tubules within the 100 nm area are pseudocoloured red. Scale bars, 200 nm.
-
Figure 1—source data 1
Numerical source data for Figure 1B–1D, E, I, J and N and Figure 1—figure supplement 1B to D.
- https://doi.org/10.7554/eLife.32866.004

Mitophagy reduces ER-OMM contacts of all intermembrane distances.
(A) Representative TEM image of an untreated U2OS:GFP-parkin WT cell highlighting ER-OMM distances of less than 100, 50 and 25 nm. Intermembrane distances (d) are indicated. Scale bars, 500 and 100 nm. (B) Quantification of the relative amount of OMM in contact with the ER (top) and fraction of mitochondria in contact with the ER per field of view (bottom) for each of the three ER-OMM distance categories from (A), for cells left untreated or treated with 20 μM CCCP for four hours. Bars represent mean ± SEM, n = 83 to 150 mitochondria in 17 to 19 fields per condition. ****, p<0.0001. (C,D) Distribution of ER-OMM contact in CCCP-treated and untreated U2OS:GFP-parkin WT cells, displayed as the percentage of OMM corresponding to each intermembrane distance (C) or as a percentage of all ER-OMM contacts (D).
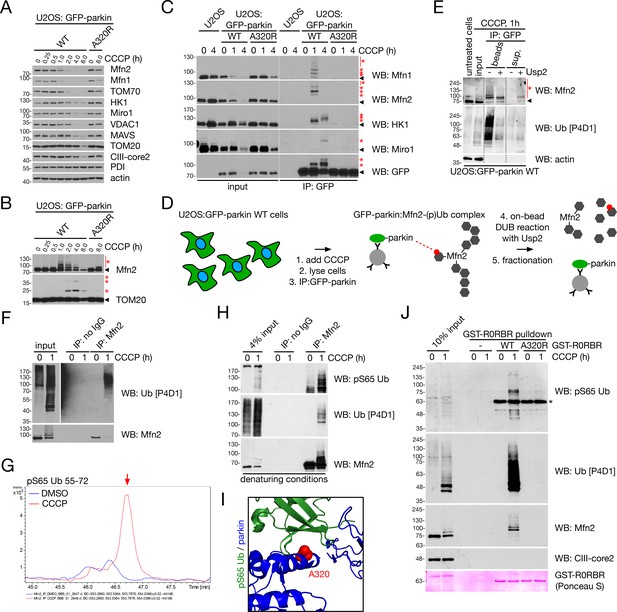
Mfn2 is rapidly phosphoubiquitinated upon induction of mitophagy.
(A) Immunoblot analysis of protein turnover in glucose-maintained U2OS:GFP-parkin WT and A320R cells treated with 20 μM CCCP for the indicated time. (B) Higher exposures of Mfn2 and TOM20 immunoblots from (A). Red asterisks indicate ubiquitinated forms of Mfn2 and TOM20. (C) Co-immunoprecipitation of parkin substrates with GFP-parkin WT or A320R in U2OS cells treated with 20 μM CCCP for the indicated time, using an anti-GFP antibody. Immunoprecipitates were separated, along with 4% input, by SDS-PAGE and immunoblotted for the indicated protein. The arrowhead indicates the unmodified form of the protein, while the red asterisks denote ubiquitinated forms. (D) Workflow for the on-bead deubiquitination of Mfn2. U2OS:GFP-parkin WT cells were treated for one hour with 20 μM CCCP, and GFP-parkin was immunoprecipitated as in (C). Immunoprecipitates were then treated with Usp2 deubiquitinase and the beads were re-isolated by centrifugation. (E) Immunoblot detection of Mfn2 after on-bead deubiquitination, as described in (D). Immunoprecipitates were either incubated at 37°C in the absence or presence of Usp2 catalytic domain for 30 min. Samples were then centrifuged to separate beads and supernatant (‘sup.’), which were denatured in sample buffer prior to separation by SDS-PAGE. Arrowheads indicate unmodified forms of Mfn2, while the red asterisks denote ubiquitinated forms. (F) Immunoprecipitation of Mfn2 for LC/MS analysis. Immunoprecipitates were separated, along with 4% input, by SDS-PAGE and immunoblotted for Ub. (G) Extracted ion chromatogram for the pS65 Ub peptide (TLSDYNIQKEpSTLHLVLR, a.a. 55–72) from Mfn2 immunoprecipitates from DMSO- (blue line) and CCCP- (red line) treated U2OS:GFP-parkin WT cells, immunoprecipitated as in (F). The red arrow indicates the peak corresponding to the peptide. (H) Immunoprecipitation of Mfn2 under denaturing conditions. Cells were lysed in buffer containing 1% SDS (see Materials and methods). Immunoprecipitates were separated, along with 4% input, by SDS-PAGE and immunoblotted for Ub and pS65 Ub. (I) Crystal structure of parkin complexed with pUb (PDB ID 5N2W, Kumar et al., 2017). The A320 residue at the pUb/parkin interface is highlighted in red, with parkin coloured blue and ubiquitin in green. (J) GST-R0RBR pulldown of pUb from U2OS:GFP-parkin WT cells. Pulldowns were performed with WT or A320R GST-R0RBR, with no GST-R0RBR (‘-’) as a further negative control. Pulldowns were separated, along with 10% input, by SDS-PAGE and immunoblotted for the indicated protein. The asterisk represents a cross-reaction between the pS65 antibody and the GST-R0RBR module.
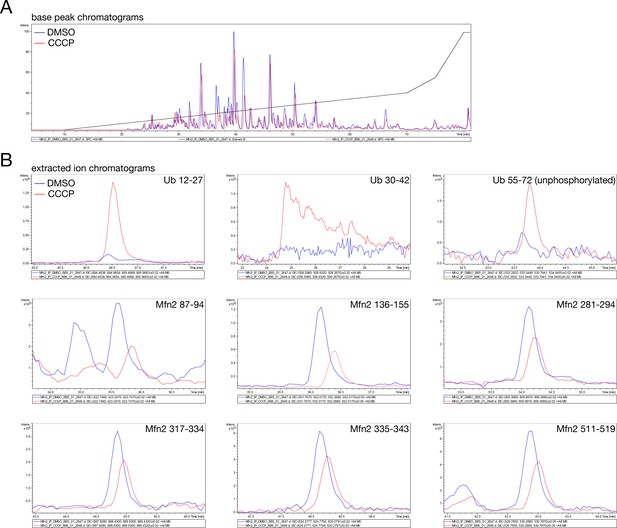
LC/MS of immunoprecipitated Mfn2.
(A) Base peak chromatograms indicating equal loading of both DMSO- and CCCP-treated samples from Figure 2F and G. (B) Extracted ion chromatograms of the indicated Ub and Mfn2 peptides from both DMSO- (blue line) and CCCP- (red line) treated samples.
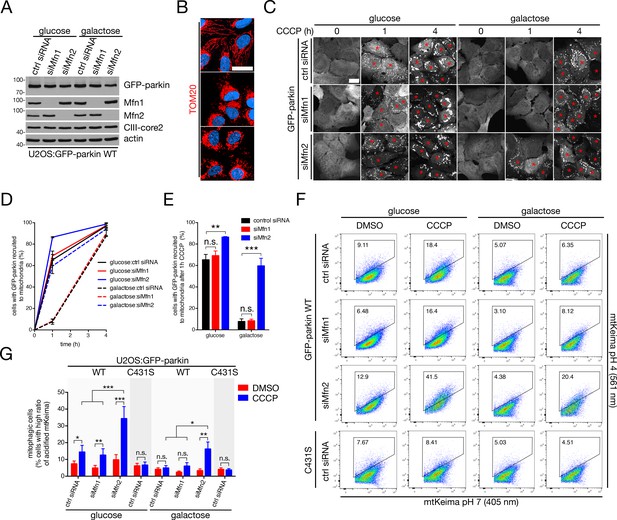
Mfn2 antagonizes mitophagy.
(A) Immunoblot analysis of whole-cell lysates from cells cultured in glucose or galactose transfected with control siRNA or siRNA targeting Mfn1 (‘siMfn1’) or Mfn2 (‘siMfn2’). (B) Mitochondrial morphology in glucose-maintained cells transfected with the indicated siRNA, as revealed by confocal imaging of TOM20 (red) staining (Hoechst, blue). Scale bar, 30 microns. (C) Representative confocal images of GFP-parkin recruitment to mitochondria as a function of time in U2OS:GFP-parkin cells treated with 20 μM CCCP. Red asterisks indicate cells in which GFP-parkin has fully translocated to mitochondria. Scale bar, 20 microns. (D) Quantification of parkin recruitment in cells from (C). Data points represent mean ± SEM, n = 3 replicates cells per condition, with >100 cells counted per condition for each replicate. (E) Parkin recruitment at one hour CCCP in cells from (C) arranged as a histogram. Bars represent mean ± SEM. n.s., not significant; **, p<0.01; ***, p<0.001. (F) U2OS:mtKeima cells were transfected with the indicated siRNA and GFP-parkin WT or C431S, and were treated with 20 μM CCCP (or DMSO) for four hours. mtKeima fluorescence in GFP-positive cells was measured using flow cytometry by excitation at 405 nm (neutral pH) and 561 nm (acidified). The data are represented as scatter plots of fluorescence emission from excitation at both wavelengths. The gated area encloses cells undergoing mitophagy (high acidified:neutral Keima ratio), and the percentage of cells within this gate is indicated in the top-left corner of each plot. (G) Quantification of the percent of cells undergoing mitophagy in cells from (F) treated with DMSO (red bars) or CCCP (blue bars) for four hours. Bars represent mean ± SEM, n = 2 experiments. n.s., not significant; *, p<0.05; **, p<0.01; ***, p<0.001.
-
Figure 3—source data 1
Numerical source data for Figure 3D, E and G, Figure 3—figure supplement 1B–D, Figures 2B, 3C, D, H, I, 4F, H and 5C.
- https://doi.org/10.7554/eLife.32866.013

Mfn2 is a mitochondrion-ER tether.
(A) Representative TEM images of U2OS:GFP-parkin cells transfected with the indicated siRNA. ER tubules are pseudocoloured blue. Scale bar, 500 nm. (B–D) Quantification of mitochondrial length (B), relative percentage of OMM in contact with the ER (C) and percentage of mitochondria in contact with ER per field of view (D) in cells from (A). Bars represent mean ± SEM, n = 66 to 70 mitochondria in 5 to 7 fields per condition. n.s., not significant; *, p<0.05; ***, p<0.001; ****, p<0.0001.
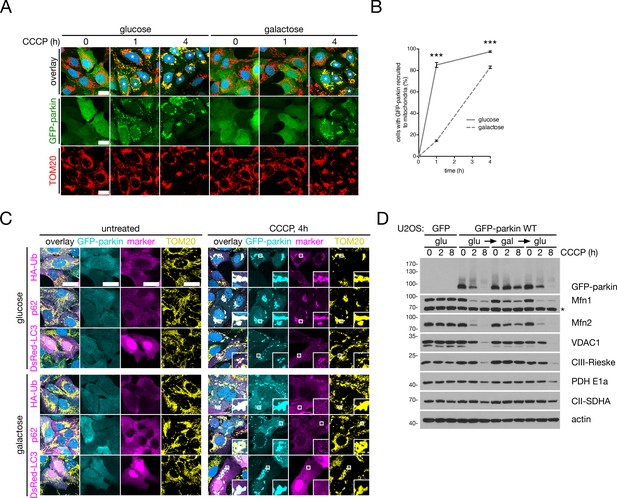
Mitochondrial respiration impedes mitophagy.
(A) Representative confocal images of U2OS:GFP-parkin (green) cells, grown on either glucose or galactose, treated with 20 μM CCCP for the indicated times. Cells were then fixed and stained for TOM20 (red) (Hoechst, blue). Cells marked with asterisks display parkin fully translocated to mitochondria. Scale bars, 20 microns. (B) Quantification of parkin recruitment to mitochondria in cells treated in (A). Data points represent mean ± SEM, n = 3 replicates cells per condition, with >100 cells counted per condition for each replicate. ***, p<0.001. (C) Representative confocal images of U2OS:GFP-parkin (cyan) cells expressing the indicated construct, treated with 20 μM CCCP for four hours and then fixed and stained for TOM20 (yellow) and the indicated tag (magenta) (Hoechst, blue). In the case of p62 (middle panels), an antibody against endogenous p62 was used. Scale bars, 30 microns. (D) Immunoblot analysis of whole-cell lysates from U2OS:GFP and GFP-parkin cells – grown either on glucose (‘glu’), converted to galactose (‘glu>gal’) or back to glucose (‘glu>gal>glu’) – treated with 20 μM CCCP for the indicated times. The asterisk indicates a non-specific band.

Parkin recruitment kinetics in cells lacking both Mfns and other mitochondria-ER tethering factors.
(A) Immunoblot analysis of whole-cell lysates from cells cultured in glucose or galactose transfected with control siRNA or siRNA targeting Mfn1 (‘siMfn1’), Mfn2 (‘siMfn2’) or both mitofusins (‘siMfn1 +2’). (B) Representative confocal images of GFP-parkin recruitment to mitochondria as a function of time in U2OS:GFP-parkin cells treated with 20 μM CCCP. Red asterisks indicate cells in which GFP-parkin has fully translocated to mitochondria. Scale bar, 20 microns. (C) Quantification of parkin recruitment in cells from (B). Data points represent mean ± SEM, n = 3 replicates cells per condition, with >100 cells counted per condition for each replicate. (D) Parkin recruitment at one hour CCCP in cells from (B) arranged as a histogram. Bars represent mean ± SEM. n.s., not significant; *, p<0.05; **, p<0.01; ***, p<0.001. (E) Immunoblot analysis of whole-cell lysates from glucose-maintained U2OS:GFP-parkin cells transfected with the indicated siRNA targeting tethering-promoting proteins. (F) Mitochondrial morphology in cells from (E), as revealed by confocal imaging of TOM20 (red) staining (Hoechst, blue). Scale bar, 20 microns. (G) Representative confocal images of GFP-parkin recruitment to mitochondria as a function of time in U2OS:GFP-parkin cells treated with 20 μM CCCP. Red asterisks indicate cells in which GFP-parkin has fully translocated to mitochondria. Scale bar, 20 microns. (H) Quantification of parkin recruitment in cells from (G). Data points represent mean ± SEM, n = 3 replicates cells per condition, with >100 cells counted per condition for each replicate. (I) Parkin recruitment at one hour CCCP in cells from (G) arranged as a histogram. Bars represent mean ± SEM. n.s., not significant; **, p<0.01; ***, p<0.001.
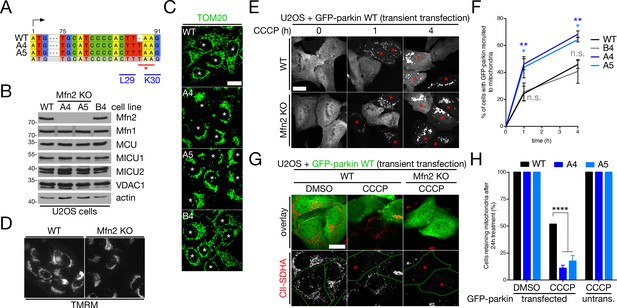
Analysis of mitophagy in Mfn2 KO U2OS cells.
(A) Genomic sequence of human Mfn2 (exon 3) that was mutated in U2OS cells using CRISPR/Cas9. The arrow indicates the codon corresponding to methionine-1; leucine-29 (‘L29’), lysine-30 (‘K30’) and the introduced stop codon (‘*’) are also indicated. (B) Immunoblot analysis of whole-cell lysates from Mfn2 KO clones (A4 and A5). (C) Mitochondrial morphology in Mfn2 KO cells, as revealed by confocal imaging of TOM20 (green) staining. The asterisks indicate nuclei. Scale bar, 20 microns. (D) Representative wide-field images of mitochondrial polarization in live WT and Mfn2 KO (clone A4) cells as indicated by TMRM staining. (E) Representative confocal images of GFP-parkin recruitment to mitochondria as a function of time in WT or Mfn2 KO (clone A4) U2OS cells, transfected with GFP-parkin and treated with 20 μM CCCP. Red asterisks indicate cells in which GFP-parkin has fully translocated to mitochondria. Scale bar, 20 microns. (F) Quantification of parkin recruitment in cells from (E). Data points represent mean ± SEM, n = 3 replicates per condition, with >100 cells counted per condition for each replicate. n.s., not significant; *, p<0.05; **, p<0.01. Significance (or lack thereof) relative to WT cells is colour-coded according to genotype. (G) Representative images of glucose-cultured WT and Mfn KO cells, transfected with GFP-parkin (green) and treated with 20 μM CCCP for 24 hr, analyzed for their mitochondrial content (represented by SDHA, red). Green lines delineate the boarders of parkin-expressing cells, and red asterisks indicate cells devoid of SDHA signal. ‘Untransfected’ refers to cells in the experiment lacking parkin expression. Scale bar, 20 microns. (H) Quantification of complete mitochondrial turnover in cells from (G). Bars represent mean ± SEM, n = 3 replicates cells per condition, with 38 to 63 cells counted per condition for each replicate. ****, p<0.0001.
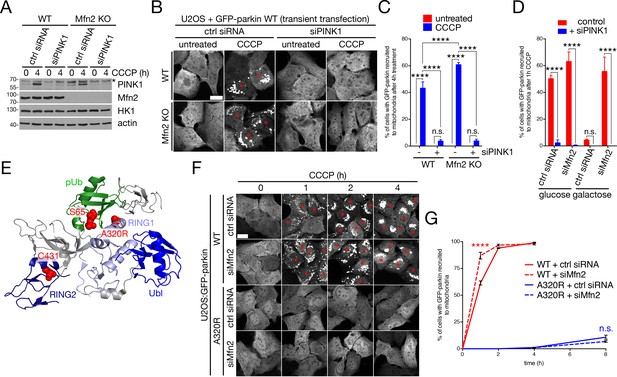
Parkin recruitment in Mfn2-depleted cells requires PINK1 and phosphoubiquitin binding.
(A) Immunoblot analysis of PINK1 depletion in WT and Mfn2 KO (clone A4) U2OS cells treated with 20 μM CCCP for four hours. The arrowhead indicates the PINK1 band, while the asterisk indicates a non-specific band. (B) U2OS cells from (A) were transfected with GFP-parkin and treated with 20 μM CCCP for four hours prior to fixation. Red asterisks mark cells in which parkin has been recruited to mitochondria. Scale bar, 10 microns. (C) Quantification of parkin-expressing cells from (A), left untreated (red bars) or treated with 20 μM CCCP for four hours (blue bars). Bars represent mean ± SEM, n = 3 replicates cells per condition, with >100 GFP-positive cells counted per condition for each replicate. n.s., not significant; ****, p<0.0001. (D) Quantification of parkin recruitment in U2OS:GFP-parkin cells, grown on glucose or galactose, treated with 20 μM CCCP for one hour prior to fixation. Cells were transfected with control siRNA (‘ctrl siRNA’) or siMfn2, and either additional ctrl siRNA (red bars) or siPINK1 (blue bars). Bars represent mean ± SEM, n = 3 replicates cells per condition, with >100 cells counted per condition for each replicate. n.s., not significant; ****, p<0.0001. (E) Crystal structure of parkin complexed with pUb (PDB ID 5N2W, Kumar et al., 2017). Sites of Ub phosphorylation (S65 in Ub), pUb binding (A320 in parkin) and catalysis (C431 in parkin) are highlighted in red, with relevant domains of parkin coloured different shades of blue, and ubiquitin in green. (F) Representative confocal images of U2OS cells stably expressing WT or A320R mutant parkin. Cells were treated with 20 μM CCCP for the indicated time prior to fixation. Red asterisks indicate cells in which GFP-parkin has been recruited to mitochondria. Scale bar, 20 microns. (G) Quantification of parkin recruitment in cells from (F). Data points represent mean ± SEM, n = 3 replicates cells per condition, with >100 cells counted per condition for each replicate. n.s., not significant; ****, p<0.0001.
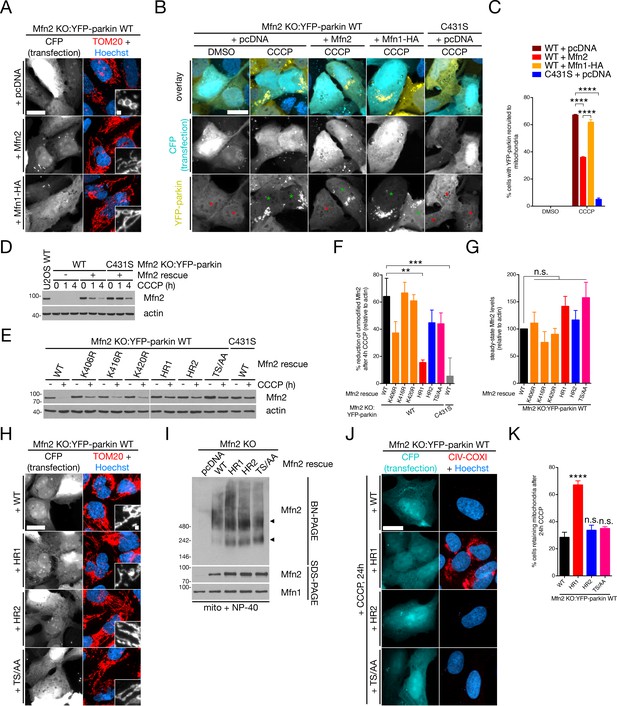
Parkin ubiquitinates Mfn2 in the HR1 domain to derepress mitophagy.
(A) Mfn2 KO:YFP-parkin WT cells were transfected with the indicated plasmid and CFP in a 3:1 ratio, then fixed and immunostained for TOM20 (red) and counterstained with Hoechst 33342 (blue). Scale bars, 20 and 1 microns. (B) Mfn2 KO:YFP-parkin WT and C431S cells, transfected as in (A), were treated with 20 μM CCCP for four hours prior to fixation, then scored for YFP-parkin recruitment. Green and red asterisks indicated CFP-positive cells with mitochondrial and cytosolic YFP-parkin, respectively. Scale bar, 20 microns. (C) Quantification of recruitment in (B). Bars represent mean ± SEM, n = 3 replicates cells per condition, with >50 cells counted per condition for each replicate. ****, p<0.0001. (D) Immunoblot analysis of Mfn2 KO:YFP-parkin cells (WT and C431S) transfected with Mfn2 and treated with 20 μM CCCP for the indicated time. An untreated U2OS cell lysate is included as a control for endogenous Mfn2 levels. (E) Representative immunoblot analysis of Mfn2 KO:YFP-parkin WT cells transfected with the indicated Mfn2 mutant and treated with 20 μM CCCP for four hours. (F) Quantification of Mfn2 modification in immunoblot analyses from (E), given as the percent of Mfn2 reduction after CCCP relative to actin. Bars represent mean ± SEM, n = 4 replicates. **, p<0.01; ***, p<0.001. (G) Quantification of steady-state (‘- CCCP’) levels of Mfn2 in immunoblot analyses from (E), relative to actin. Bars represent mean ± SEM, n = 4 replicates. n.s., not significant. (H) Mfn2 KO:YFP-parkin WT cells were transfected with the indicated plasmid and CFP in a 3:1 ratio, then fixed and immunostained for TOM20 (red) and counterstained with Hoechst 33342 (blue). Scale bars, 20 and 1 microns. (I) Immunoblot analysis of BN- and SDS-PAGE gels of solubilized mitochondria from cells from (H). Arrows indicate two Mfn2-containing complexes in the native condition. (J) Representative wide-field images of Mfn2 KO:YFP-parkin WT cells transfected with the indicated Mfn2 construct. Cells were treated with 20 μM CCCP for 24 hr prior to fixation, then stained with CIV-COX1 (red) and Hoechst (blue). Scale bar, 20 microns. (K) Quantification of mitophagy in (J). Bars represent mean ± SEM, n = 4 replicates per condition, with >50 cells counted per condition for each replicate. ****, p<0.0001; n.s., not significant.
-
Figure 4—source data 1
Numerical source data for Figure 4C, F, G and K.
- https://doi.org/10.7554/eLife.32866.016
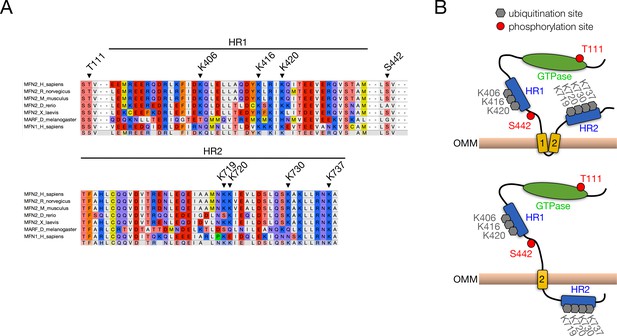
Location and conservation of ubiquitination and phosphorylation sites in Mfn2.
(A) Sequence alignment of sites of Mfn2 modification across species. Ubiquitinated lysines and phosphorylated serines and threonines are indicated by arrowheads. Residue numbering is according to the human sequence. HR, heptad repeat domain. (B) Diagram of Mfn2 post-translational modification by parkin-mediated ubiquitination (Sarraf et al., 2013) and PINK1-mediated phosphorylation (Chen and Dorn, 2013) for both double- (top) and single- (bottom) pass topologies. Phosphosites are denoted in red, while sites of ubiquitination are marked in grey. HR, heptad repeat domain; OMM, outer mitochondrial membrane.
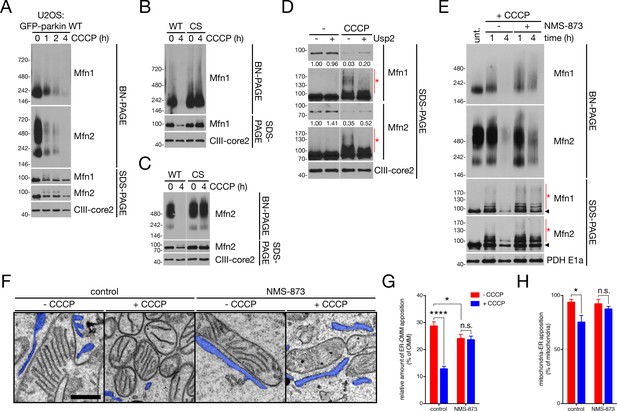
p97 governs ER-OMM contact via the extraction of Mfn2 complexes.
(A) Immunoblot analysis of NP-40-solubilized mitochondria, isolated from U2OS:GFP-parkin WT cells treated with 20 μM CCCP for the indicated time, separated by blue native- (BN-) and SDS-PAGE. (B, C) Immunoblot analysis of Mfn1- (B) and Mfn2- (C) containing complexes in NP-40-solubilized mitochondria, isolated from U2OS:GFP-parkin WT and C431S cells treated with 20 μM CCCP for four hours, separated by BN- and SDS-PAGE. (D) Mitochondria isolated from U2OS:GFP-parkin WT cells treated with 20 μM CCCP for one hour were, after solubilization in NP-40, incubated with 1 μM Usp2 for 30 min at 37°C prior to separation by SDS-PAGE. Red asterisks indicate ubiquitinated species of Mfn1 and Mfn2. Densitometry calculations for the Mfn1 and Mfn2 bands (shorter exposure) relative to CIII-core2 are shown under the respective immunoblots. (E) Immunoblot analysis of NP-40-solubilized mitochondria, isolated from U2OS:GFP-parkin WT cells treated with 20 μM CCCP in the presence or absence of 25 μM NMS-873 for the indicated time, separated by blue native- (BN-) and SDS-PAGE. Red asterisks indicate ubiquinated Mfn species visible by SDS-PAGE, while the arrowhead denotes the unmodified band. (F) Representative TEM images of mitochondria in contact with ER (pseudocoloured blue) in U2OS:GFP-parkin cells treated with 20 μM CCCP (‘+CCCP’) for four hours in the presence or absence of 25 μM NMS-873. Scale bar, 500 nm. (G,H) Quantification of TEM from (F) in cells treated with (blue bars) or without (red bars) 20 μM CCCP for four hours. The percent of OMM per mitochondrion (G) and mitochondria per field (H) in contact with the ER was quantified. Bars represent mean ± SEM, n = 99 to 187 mitochondria in 12 to 14 fields per condition. n.s., not significant; *, p<0.05; ***, p<0.001; ****, p<0.0001.
-
Figure 5—source data 1
Numerical source data for Figure 5G and H.
- https://doi.org/10.7554/eLife.32866.018
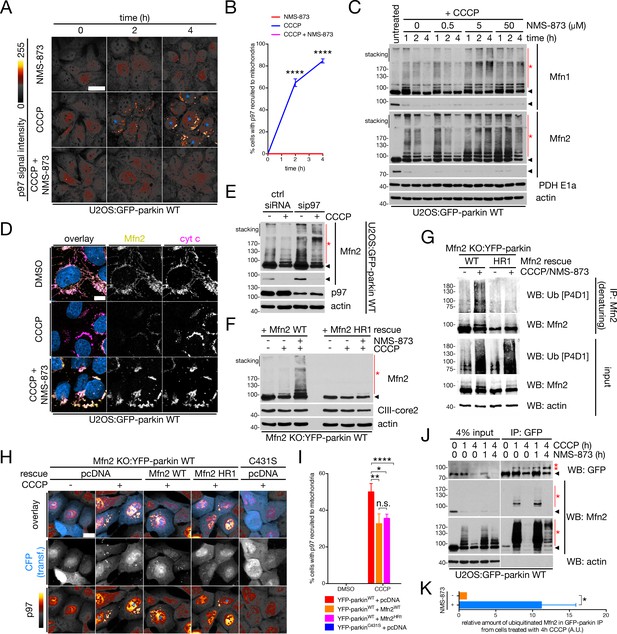
Degradation of ubiquitinated Mfn2 involves p97 recruitment and activity.
(A) Representative confocal images of p97 recruitment to mitochondria in cells treated with 20 μM CCCP and/or 25 μM NMS-873 for the indicated time. Blue asterisks denote cells with mitochondrial p97, and p97 signal intensity is represented as a heat map. Scale bar, 20 microns. (B) Quantification of cells with p97 translocation to mitochondria in cells treated with either 25 μM NMS-873 (red line), 20 μM CCCP (blue line) or both simultaneously (magenta line). Bars represent mean ± SEM, n = 3 replicates per condition, with >100 cells counted per condition for each replicate. ****, p<0.0001. (C) Immunoblot analysis of whole-cell lysates from U2OS:GFP-parkin cells treated with 20 μM CCCP and the specified concentration of NMS-873 for the indicated time, separated by SDS-PAGE. For each Mfn, longer (upper panel) and shorter (lower panel) exposures are shown. Red asterisks indicate ubiquitinated Mfn species, while the arrowheads denote the unmodified band. (D) U2OS:GFP-parkin cells were treated with 20 μM CCCP in the presence or absence of 25 μM NMS-873 for four hours, then fixed and immunostained for Mfn2 (yellow) and cytochrome c (magenta). Scale bar, 10 microns. (E) Immunoblot analysis of Mfn2 ubiquitination in U2OS:GFP-parkin WT cells transfected with siRNA targeting p97 (sip97) or control (ctrl siRNA) and treated with 20 μM CCCP for two hours. Arrowheads indicate the unmodified Mfn2 band (two exposures), while the red asterisk denotes ubiquitinated Mfn2. (F) Immunoblot analysis of exogenous Mfn2 in Mfn2 KO:YFP-parkin WT cells reconstituted with the indicated Mfn2 construct. Cells were treated with 25 μM NMS-873 and/or 20 μM CCCP for four hours prior to lysis. The arrowhead indicates the unmodified Mfn2 band and the red asterisk denotes ubiquitinated Mfn2 conjugates. (G) Immunoprecipitation of Mfn2 under denaturing conditions from Mfn2 KO:YFP-parkin WT cells reconstituted with the indicated Mfn2 construct. Cells were lysed in buffer containing 1% SDS (see Materials and Methods). Immunoprecipitates were separated by SDS-PAGE and immunoblotted for Ub. (H) Representative widefield images of p97 translocation to mitochondria (pseudocoloured as in [A]) in Mfn2 KO:YFP-parkin WT or C431S cells, reconstituted with the indicated plasmid, treated with 20 μM CCCP (or DMSO) for four hours. CFP (blue) is included as a marker of Mfn2 transfection, and blue asterisks indicate cells where p97 has translocated to mitochondria. Scale bar, 20 microns. (I) Quantification of mitochondrial recruitment of p97 in Mfn2 KO:YFP-parkin cells from (H). Bars represent mean ± SEM, n = 3 replicates per condition, with >50 cells counted per condition for each replicate. *, p<0.05; **, p<0.01; ****, p<0.0001. (J) Co-immunoprecipitation of mitofusins with GFP-parkin in U2OS:GFP-parkin WT cells treated with 20 μM CCCP in the presence or absence of 25 μM NMS-873 for the indicated time, using an anti-GFP antibody. Immunoprecipitates were separated, along with 4% input, by SDS-PAGE and immunoblotted for the indicated protein. The arrowhead indicates the unmodified form of the protein, while the asterisks denote ubiquitinated forms. (K) Quantification of the relative amount of ubiquitinated Mfn2 co-immunoprecipitated with GFP-parkin in cells from (J). Bars represent mean ± SEM, n = 3 replicates. *, p<0.05.
-
Figure 6—source data 1
Numerical source data for Figure 6I and K and Figure 6—figure supplement 1C.
- https://doi.org/10.7554/eLife.32866.021
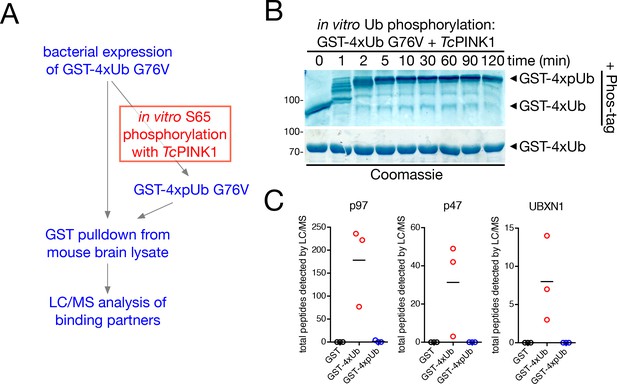
Analysis of pUb interactors from mouse brain.
(A) Workflow of protein purification, phosphorylation, pull-down and LC/MS. GST-4xUb G76V, which cannot be cleaved by the cellular Ub processing machinery, was phosphorylated on S65 by Tribolium castaneum PINK1 (TcPINK1) to form GST-4xpUb G76V. Both GST-4xUb and GST−4xpUb were incubated with mouse brain lysate, and binding partners were analyzed by LC/MS. See Materials and Methods for more detail. (B) Ub phosphorylation was determined by separation by SDS-PAGE over Phos-tag gel, which slows the migration of phosphorylated proteins. (C) Quantification of the number of peptides corresponding to p97-related factors identified by LC/MS in GST (black dots), GST-4xUb (red dots) and GST-4xpUb (blue dots) pull-downs from mouse brain lysate. Bars represent the mean, n = 3 independent experiments. See Supplementary file 1 for complete lists of identified interactors.
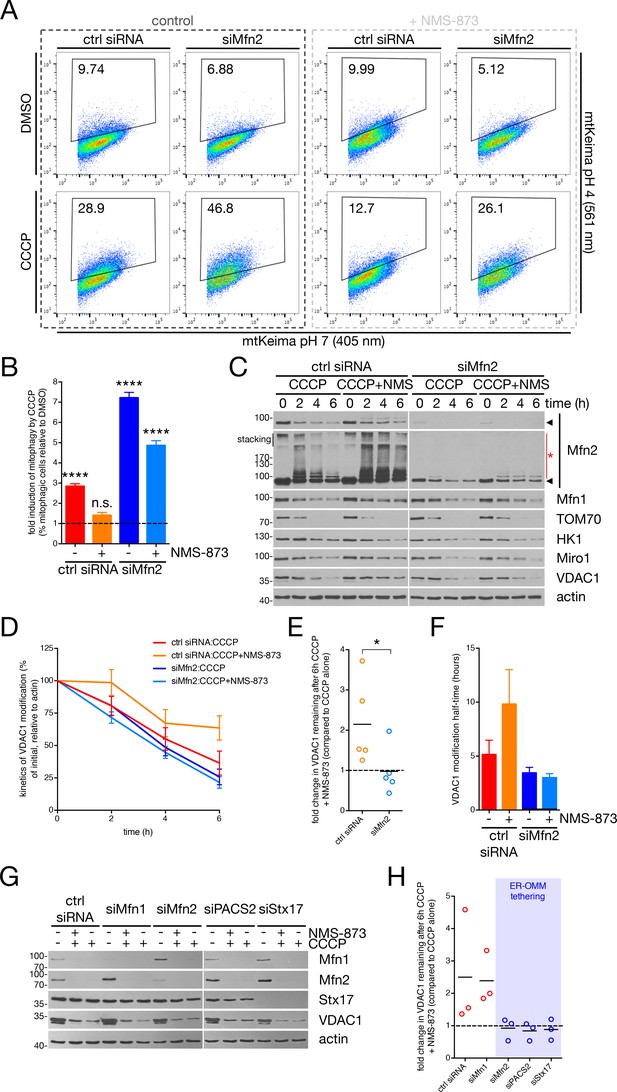
p97 and Mfn2 effect mitophagy through parkin substrate availability.
(A) U2OS:mtKeima cells were transfected with the indicated siRNA and GFP-parkin WT, and were treated with 20 μM CCCP (or DMSO) for five hours in the presence (dark grey box) or absence (light grey box) of 25 μM NMS-873. mtKeima fluorescence in GFP-positive cells was measured using flow cytometry by excitation at 405 nm (neutral pH) and 561 nm (acidified). The data are represented as scatter plots of fluorescence emission from excitation at both wavelengths. The gated area encloses cells undergoing mitophagy and the percentage of cells within this gate is indicated in the top-left corner of each plot. (B) Quantification of the percent of cells undergoing mitophagy in cells from (A), expressed as a ratio of CCCP-treated cells to those treated with DMSO. Bars represent mean ± SEM, n = 2 experiments. n.s., not significant; ****, p<0.0001. (C) Immunoblot analysis of U2OS:GFP-parkin cells, transfected with siRNA targeting Mfn2 (siMfn2) or control (ctrl siRNA), treated with 20 μM CCCP in the presence or absence of 25 μM NMS-873 over a period of six hours. (D) Immunoblot quantification of VDAC1 levels (relative to actin) from cells from (C). Bars represent mean ± SEM, n = 5 experiments. (E) The 6 hr time-point data from (D) is represented as a fold change in VDAC1 remaining when NMS-873 is added. Data points are represented on the graph, n = 5 experiments. *, p<0.05. (F) Quantification of the half-time (t1/2) of VDAC1 modification in cells from (C) over 6 hr. Half-times were obtained from decay curves generated with the time-points in (C). Bars represent mean ± SEM, n = 5 experiments. (G) Immunoblot analysis of U2OS:GFP-parkin cells, transfected with the indicated siRNA, treated with 20 μM CCCP in the presence or absence of 25 μM NMS-873 for six hours. (H) Immunoblot quantification of VDAC1 levels (relative to actin) in cells from (G), represented as a fold change in VDAC1 remaining when NMS-873 is added. Data points are represented on the graph, n = 3 experiments. Factors promoting ER-OMM contact are contained within the blue box.
-
Figure 7—source data 1
Numerical source data for Figure 7B, D–F and H.
- https://doi.org/10.7554/eLife.32866.023
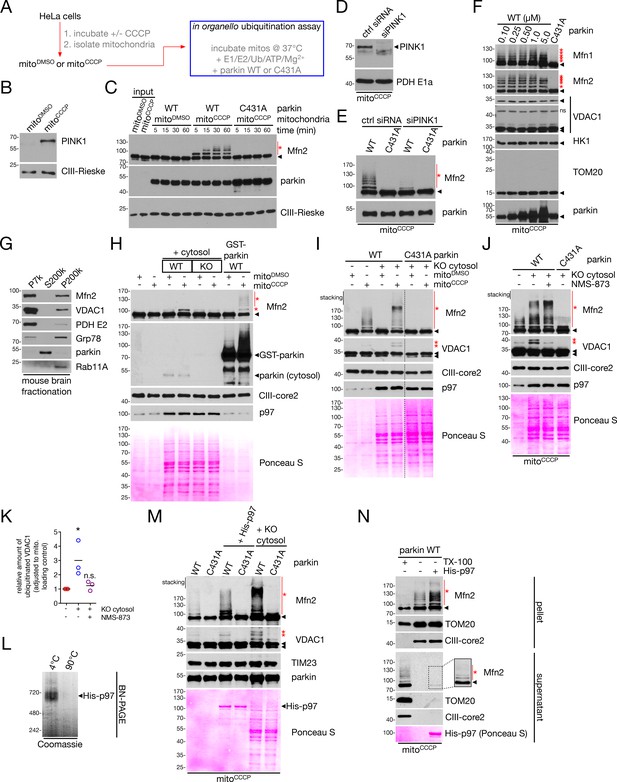
In organello ubiquitination of Mfn2 and VDAC1.
(A) Workflow for the in organello ubiquitination assay, where HeLa cells are depolarized with 20 μM CCCP for four hours and mitochondria are isolated (‘mitoCCCP’, with control ‘mitoDMSO’). These are combined with ubiquitination assay components (blue box) and incubated at 37°C (see Materials and Methods for full details). (B) Immunoblot analysis of PINK1 levels in mitochondria isolated from depolarized (‘mitoCCCP’) or control (‘mitoDMSO’) cells. (C) In organello ubiquitination assays, using depolarized or control mitochondria and 100 nM of the indicated parkin construct, were incubated at 37°C for the indicated time, and reactions were quenched with SDS-PAGE sample buffer. Mfn2 ubiquitination was analyzed by immunoblot. Ubiquitinated species are indicated by red asterisks, while unmodified bands are denoted by arrowheads. (D) Immunoblot analysis of PINK1 levels in mitochondria isolated from depolarized cells transfected with control siRNA (ctrl siRNA) or siRNA targeting PINK1 (siPINK1). (E) Mitochondria from (D) were used for 30 min in organello ubiquitination assays using 100 nM WT or C431A parkin, and Mfn2 ubiquitination was analyzed by immunoblot. Ubiquitinated species are indicated by red asterisks, while unmodified bands are denoted by arrowheads. (F) Depolarized mitochondria were used for 30 min in organello ubiquitination assays with the indicated concentration of WT parkin, or 100 nM parkin C431A as a negative control. Ubiquitinated species are indicated by red asterisks, while unmodified bands are denoted by arrowheads. (G) Immunoblot analysis of mouse brain fractionation. Mouse brain homogenate was separated into heavy membrane (P7k), cytosolic (S200k) and light membrane (P200k) fractions. Distribution of mitochondrial (Mfn2, VDAC1, PDH E2), ER (Grp78), soluble (parkin) and endosomal (Rab11A) markers are shown. (H) CCCP-uncoupled (‘mitoCCCP’) or control (‘mitoDMSO’) mitochondria were incubated for 60 min with 2 mg/ml cytosol from WT mouse brain (‘WT cytosol’) or from the brain of parkin-/- mice (‘KO cytosol’). As a positive control, mitochondria were incubated with 100 nM uncleaved GST-parkinWT (without cytosol). Ubiquitinated species are indicated by red asterisks, while unmodified bands are denoted by arrowheads. (I) CCCP-uncoupled (‘mitoCCCP’) or control (‘mitoDMSO’) mitochondria were incubated for 60 min with 100 nM parkin WT or C431A and in the presence or absence of 2 mg/ml cytosol from parkin-/- mouse brain (‘KO cytosol’). Mfn2 and VDAC1 ubiquitination were assayed by immunoblot. Ubiquitinated species are indicated by red asterisks, while unmodified bands are denoted by arrowheads. (J) In organello ubiquitination reactions were performed with parkin-/- mouse brain (‘KO cytosol’) in the presence of absence of 25 μM NMS-873. Reactions were incubated on ice for 30 min prior to a 60 min 37°C incubation. In the immunoblot analysis, ubiquitinated species are indicated by red asterisks, while unmodified bands are denoted by arrowheads. (K) Quantification of the level of ubiquitinated VDAC1 as compared to control, relative to mitochondrial loading control (TIM23 or CIII-core2). Data points are represented on the graph, n = 3 experiments. *, p<0.05; n.s., not significant. (L) Recombinant, hexameric His-p97 runs as a ~ 700 kDa complex as assayed by BN-PAGE. Prior to separation on the gel, samples were incubated at the indicated temperature for 10 min. (M) Immunoblot analysis of 60 min in organello ubiquitination assays using depolarized mitochondria, 100 nM parkin, 200 nM His-p97 hexamer, and 2 mg/ml parkin KO brain cytosol. Ubiquitinated species are indicated by red asterisks, while unmodified bands are denoted by arrowheads. Recombinant His-p97 is additionally indicated on the Ponceau. (N) In organello retrotranslocation of Mfn2. In organello ubiquitination reactions with or without recombinant p97 were centrifuged at 10,000 g to separate mitochondria (pellet) from soluble factors (supernatant). As a control, reactions were lysed in 1% TX-100 prior to centrifugation. The inset on the supernatant Mfn2 blot shows ubiquitination (red asterisks) of the protein at higher exposure.
-
Figure 8—source data 1
Numerical source data for Figure 8K.
- https://doi.org/10.7554/eLife.32866.026
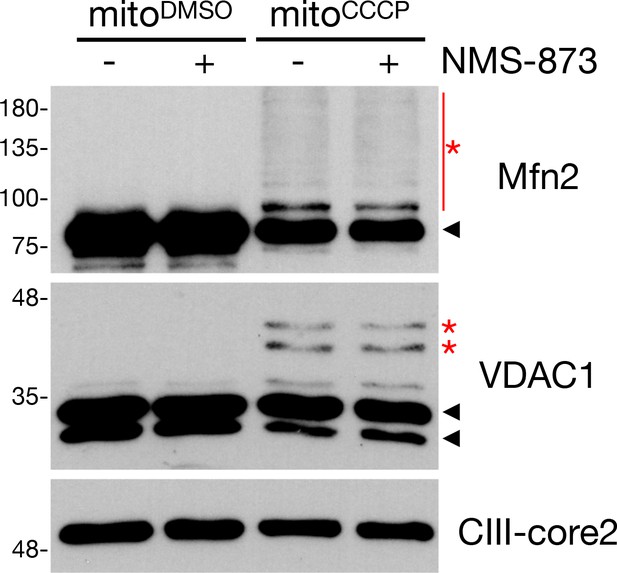
Effect of NMS-873 on cytosol-free ubiquitination.
In organello ubiquitination reactions were performed in the presence or absence of 25 μM NMS-873. Reactions were incubated on ice for 30 min prior to a 60 min 37°C incubation. In the immunoblot analysis, ubiquitinated species are indicated by red asterisks, while unmodified bands are denoted by arrowheads.
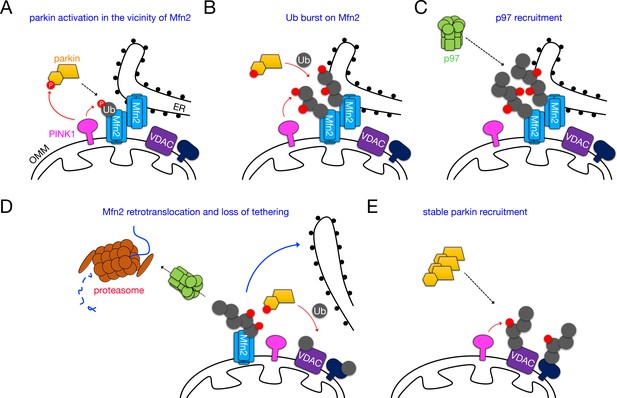
Dismantling of Mfn2 interorganellar bridges by PINK1, parkin and p97 during mitophagy.
(A) PINK1-phosphorylated Ub on Mfn2 initially recruits parkin to Mfn2 complexes, where it is phosphorylated and activated by PINK1. (B) Parkin and PINK1 cooperate to catalyze a pUb burst on Mfn2. (C) Ubiquitinated Mfn2 HMW complexes are recognized by p97, which translocates to mitochondria. (D) Ubiquitinated Mfn2 is retrotranslocated from the OMM and degraded by the proteasome. (E) VDACs and possibly other substrates become available to the parkin/PINK1 system, and their phosphoubiquitination stabilizes parkin on mitochondria to drive mitophagy.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| cell line (Homo sapiens) | U2OS | PMID 24446486 | ||
| cell line (Hs) | U2OS:GFP | PMID 24446486 | ||
| cell line (Hs) | U2OS:GFP-parkin WT | PMID 24446486 | ||
| cell line (Hs) | U2OS:GFP-parkin A320R | PMID 28276439 | ||
| cell line (Hs) | Mfn2 KO | this paper | see Plasmids and transfection | |
| cell line (Hs) | Mfn2 KO:YFP-parkin WT | this paper | see Plasmids and transfection | |
| cell line (Hs) | Mfn2 KO:YFP-parkin C431S | this paper | see Plasmids and transfection | |
| cell line (Hs) | HeLa | PMID 24446486 | ||
| cell line (Hs) | control-1 | NIH | NCRM-1 | |
| cell line (Hs) | control-2 | PMID 27641647 | ||
| cell line (Hs) | PRKN(del) | PMID 20885945 | ||
| transfected construct (Hs) | HA-Ub | PMID 25216678 | ||
| transfected construct (Hs) | DsRed2-LC3 | PMID 18596167 | ||
| transfected construct (Hs) | Mfn1-HA | PMID 15878861 | ||
| transfected construct (Hs) | Mfn2 WT | PMID 15878861 | ||
| transfected construct (Hs) | Mfn2 K406R | this paper | see Plasmids and transfection | |
| transfected construct (Hs) | Mfn2 K416R | this paper | see Plasmids and transfection | |
| transfected construct (Hs) | Mfn2 K420R | this paper | see Plasmids and transfection | |
| transfected construct (Hs) | Mfn2 HR1 | this paper | see Plasmids and transfection | |
| transfected construct (Hs) | Mfn2 HR2 | this paper | see Plasmids and transfection | |
| transfected construct (Hs) | Mfn2 TS/AA | this paper | see Plasmids and transfection | |
| transfected construct (Hs) | GFP-parkin WT | PMID 24446486 | ||
| biological sample (Mus musculus) | parkin WT brain cytosol | this paper | see In organello ubiquitination assays | |
| biological sample (Mm) | parkin KO brain cytosol | this paper | see In organello ubiquitination assays | |
| antibody | anti-actin | Millipore | MAB1501 | |
| antibody | anti-B-III-tubulin | Sigma | T8660 | |
| antibody | anti-MAVS | Enzo | ALX-210–929 C100 | |
| antibody | anti-cytochrome c | BD | 556432 | |
| antibody | anti-GFP | Abcam | ab6673 | IP |
| antibody | anti-GFP | Invitrogen | A6455 | WB |
| antibody | anti-Grp78 | Santa Cruz | sc-376768 | |
| antibody | anti-HA | Abcam | ab9134 | |
| antibody | anti-HK1 | Cell Signaling | 2024S | |
| antibody | anti-Mfn1 | Santa Cruz | sc-50330 | |
| antibody | anti-Mfn2 | Sigma | M6319 | WB in Figure 3—figure supplement 2D |
| antibody | anti-Mfn2 | Cell Signaling | 9482 | all other assays (IF, WB, IP) |
| antibody | anti-CIV-COXI | Abcam | ab14705 | |
| antibody | anti-p62 | Progen | GP62-C | |
| antibody | anti-PDH E1a | Abcam | ab110330 | |
| antibody | anti-PDH E2/E3bp | Abcam | ab110333 | |
| antibody | anti-PDI | Abcam | ab2792 | |
| antibody | anti-PINK1 | Cell Signaling | 6946 | |
| antibody | anti-pS65 Ub | Millipore | ABS1513-I | |
| antibody | anti-Rab11A | Cell Signaling | 2413 | |
| antibody | anti-Miro1 | Sigma | HPA010687 | |
| antibody | anti-CII-SDHA | Abcam | ab14715 | |
| antibody | anti-Stx17 | ProteinTech | 17815–1-AP | |
| antibody | anti-TH | Pel-Freez | P40101-150 | |
| antibody | anti-TIM23 | BD | 611222 | |
| antibody | anti-TOM20 | Santa Cruz | sc-11414 | |
| antibody | anti-TOM70 | Santa Cruz | sc-390545 | |
| antibody | anti-Ub [FK2] | Enzo | BML-PW8810 | IF |
| antibody | anti-Ub [P4D1] | Santa Cruz | sc-8017 | WB |
| antibody | anti-CIII-core2 | Abcam | ab14745 | |
| antibody | anti-CIII-Rieske | Abcam | ab14746 | |
| antibody | anti-p97 | Abcam | ab11433 | |
| antibody | anti-VDAC1 | Abcam | ab14734 | |
| recombinant protein (Rattus norvegicus) | GST-R0RBR WT | PMID 23661642 | ||
| recombinant protein (Rn) | GST-R0RBR A320R | this paper | see Plasmids and transfection | |
| recombinant protein (Rn) | GST-parkin WT | PMID 28276439 | ||
| recombinant protein (Rn) | GST-parkin C431A | PMID 28276439 | ||
| recombinant protein (Hs) | UbcH7 | PMID 28276439 | ||
| recombinant protein (Hs) | UBE1 | BostonBiochem | E-305 | |
| recombinant protein (Hs) | Ubiquitin | BostonBiochem | U-100H | |
| recombinant protein (Hs) | Usp2 catalytic domain | BostonBiochem | E-504 | |
| recombinant protein (Tribolium castaneum) | TcPINK1 | PMID 24784582 | ||
| recombinant protein (Hs) | GST-4xUb G76V | PMID 23670163 | ||
| recombinant protein (Mm) | His-p97 | PMID 19506019 | ||
| commercial assay or kit | QuikChange II site-directed mutagenesis kit | Agilent | 200523 | |
| commercial assay or kit | BCA protein assay | ThermoFisher | 23227 | |
| chemical compound, drug | CCCP | Sigma | C2759 | |
| chemical compound, drug | MG132 | Sigma | M8699 | |
| chemical compound, drug | Hoechst 33342 | ThermoFisher | H3570 | |
| chemical compound, drug | NMS-873 | ApexBio | B2168 | |
| software, algorithm | BioTools | Bruker | ||
| software, algorithm | MASCOT | Matrix Science | ||
| software, algorithm | Data Analysis | Bruker | ||
| software, algorithm | ImagJ | NIH | ||
| software, algorithm | PyMOL | Schrodinger | ||
| software, algorithm | Excel | Microsoft | ||
| software, algorithm | Prism | GraphPad |
Additional files
-
Supplementary file 1
MS identification of selective Ub and pUb interactors.
Table depicting GST-4xUb interactors that are selective for S65-phosphorylated (top) or unphosphorylated (bottom) Ub. p97-related data (shaded in yellow) are also depicted in Figure 6—figure supplement 1C.
- https://doi.org/10.7554/eLife.32866.028
-
Transparent reporting form
- https://doi.org/10.7554/eLife.32866.029





