Nicotinamide adenine dinucleotide is transported into mammalian mitochondria
Figures
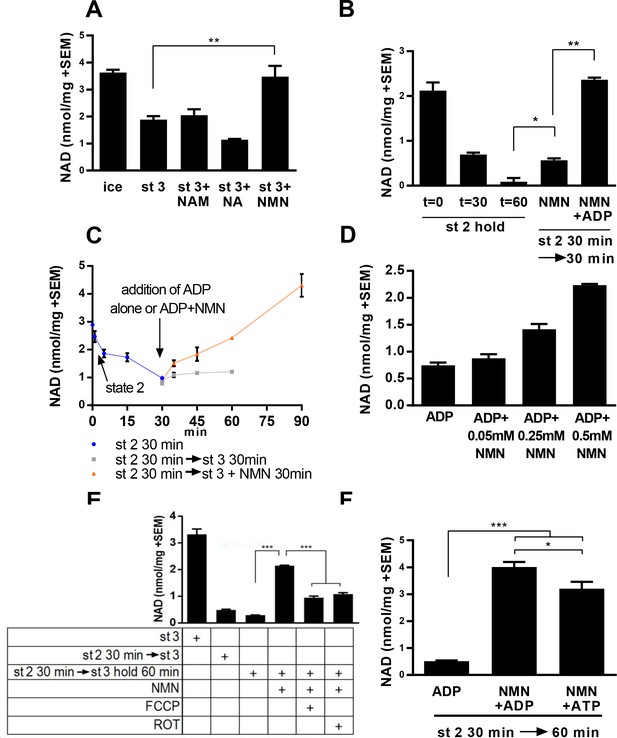
Mitochondria synthesize NAD from nicotinamide mononucleotide.
(A) Mitochondria isolated from murine skeletal muscle were maintained for 30 min at 37°C with shaking in respiratory state 3 (MirO5 respiration buffer containing 10 mM Pyruvate, 5 mM Malate, 12.5 mM ADP) supplemented with 0.5 mM NAM, NA, or NMN. (N = 2–4). (B) Mitochondria initially held for 30 min in state 2 (MirO5 respiration buffer containing 10 mM Pyruvate, 5 mM Malate; 37°C with shaking) were then supplemented with NMN alone or NMN + ADP and incubated for an additional 30 min at 37°C. (N = 2). (C) Time course of mitochondrial NAD levels before and after addition of NMN or NMN + ADP. (N = 2). (D) Isolated mitochondria were held in state 2 for 30 min before adding ADP to stimulate state 3 respiration for 60 min in the presence of increasing amounts of NMN added concomitantly with ADP. (N = 2). (E) Isolated mitochondria were maintained in state 2 at 37°C with shaking for 30 min and then transitioned to state three in the absence or presence of NMN (0.5 mM), FCCP (4 µM), or rotenone (ROT; 0.5 µM) and incubating for an additional 60 min at 37°C with shaking. (N = 4). (F) Isolated mitochondria were held in state 2 for 30 min before being supplemented with NMN alone, NMN + ADP or NMN + ATP and incubated for an additional 30 min at 37°C (N = 2–4). The data shown are means ± SEM from two or more biological replicates, each measured in technical duplicate and are representative of three independent experiments. (*, p<0.05; **, p<0.001; ***, p<0.0001; 2-tailed, unpaired Student’s t-test).
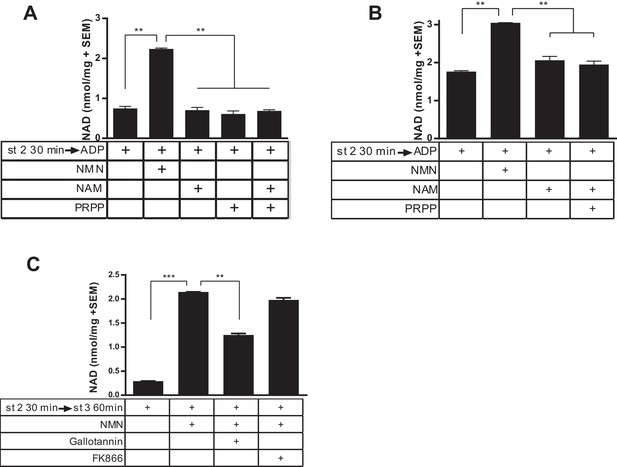
Isolated mitochondria do not produce NAD from nicotinamide.
(A) Mitochondria isolated from murine skeletal muscle were held in respiratory state 2 for 30 min at 37°C with shaking before addition of ADP (state 3) and incubation for 60 min in the absence or presence of the precursors NMN, NAM, PRPP, or NAM and PRPP (0.5 mM). (N = 2). Results are representative of two independent experiments. (B) Mitochondria isolated from murine liver were held in state 2 at 37°C for 30 min with shaking before the addition of ADP (state 3) in the presence or absence of 0.5 mM NMN, NAM or NAM and PRPP. (N = 2). (C) Muscle mitochondria were maintained in state 2 for 30 min before the addition of ADP (state 3) and further incubated at 37°C for 60 min in the presence or absence of NMN and inhibitors Gallotannin (100 µM) or FK866 (10 nM). (N = 4). (*, p<0.05; **, p<0.005; ***, p<0.0001; 2-tailed, unpaired Student’s t-test).
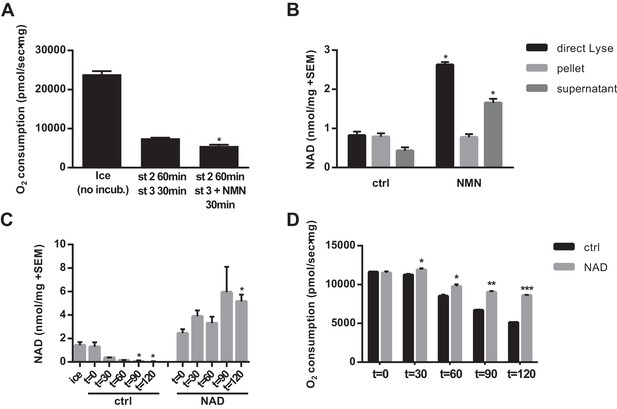
NAD synthesized from NMN by isolated mitochondria remains outside of the organelles.
(A) State three coupled mitochondrial oxygen consumption. Isolated skeletal muscle mitochondria were extracted directly (ice) or maintained in respiratory state 2 (MirO5 respiration buffer containing 10 mM Pyruvate, 5 mM Malate) at 37°C with shaking for 60 min before addition of 12.5 mM ADP (state 3) with or without 0.5 mM NMN and incubated an additional 30 min at 37°C before being measured. (N = 2–3). (B) Isolated skeletal muscle mitochondria were maintained in state 2 at 37°C with shaking for 30 min before addition of ADP (state 3) with or without NMN and incubated an additional 60 min. The mitochondrial suspension was then either lysed directly in 0.6M perchloric acid (final concentration) or centrifuged at 10,000 x g for 2 min at 4°C to collect the supernatant and (subsequently washed) pellet which were then extracted with perchloric acid. (N = 2). (C and D) Isolated skeletal muscle mitochondria were maintained in state 2 at 37°C with shaking with or without 10 mM NAD. At the indicated time points, aliquots were removed from the pooled mitochondrial suspensions and centrifuged to separate the pellet and supernatant (C), or analyzed for state three respiratory capacity using high-resolution respirometry (D). (N = 2). Results are expressed as mean ±SEM and are representative of three independent experiments. (*, p<0.05; **, p<0.005; ***, p<0.0001; unpaired Student’s t-test).
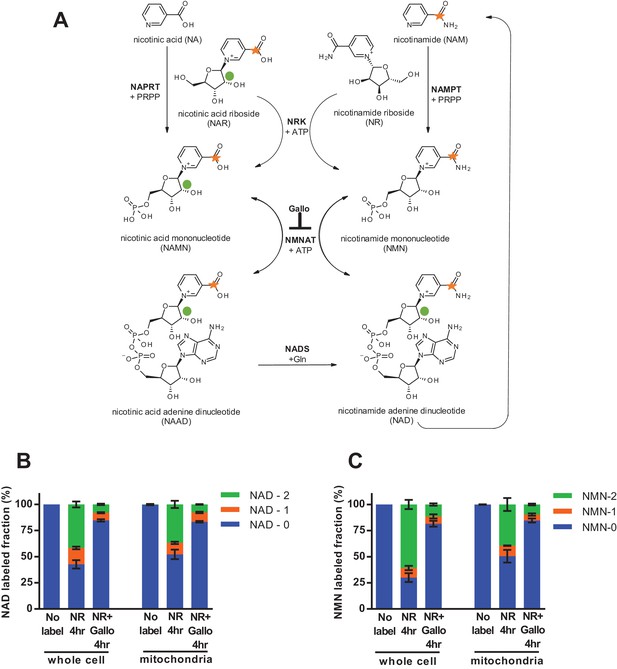
Nicotinamide riboside is incorporated intact into mitochondrial NAD.
(A) Isotopically-labeled nicotinamide riboside (NR) or nicotinic acid (NAR) was synthesized to contain a C-13 on the pyridine carboxyl group and a deuterium on the ribose moiety (NAR labeling shown). (B) Fractional labeling of NAD in C2C12 whole cell lysate and isolated mitochondria following 4 hr of incubation with double-labeled NR with or without the NMNAT inhibitor, gallotannin (Gallo; 100 µM). (No label group, N = 1; NR 4 hr group, N = 4; NR + Gallo 4 hr group, N = 3). (C) Fractional labeling of NMN found in C2C12 whole cell lysate and isolated mitochondria following 4 hr of incubation with double-labeled NR with or without gallotannin (Gallo; 100 µM). (No label group, N = 1; NR 4 hr group, N = 4; NR + Gallo 4 hr group, N = 3). Data shown are means ± SEM and are representative of 2 independent experiments.
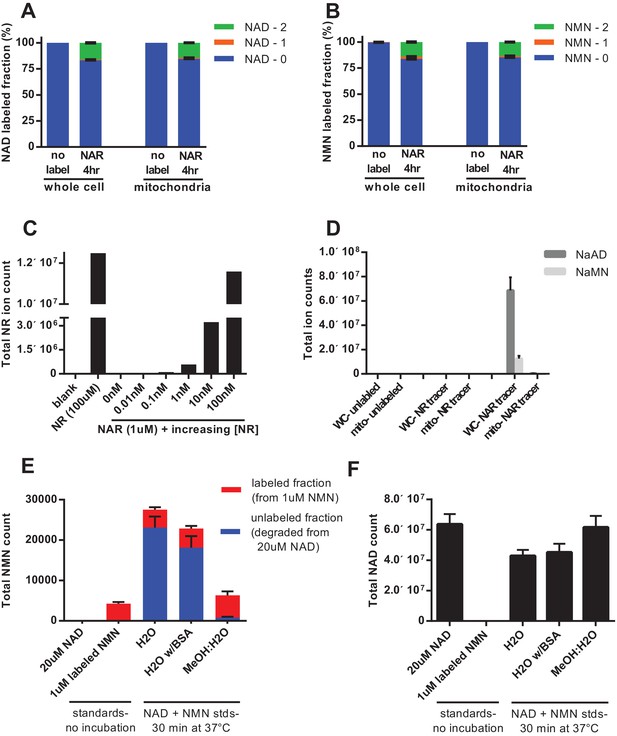
Nicotinic acid riboside is incorporated intact into mitochondrial NAD.
(A) Fractional labeling of NAD in C2C12 whole cell lysates and isolated mitochondria following 4 hr of incubation with doubly-labeled NAR. (N = 5). (B) Fractional labeling of NMN in C2C12 whole cell lysates and isolated mitochondria following 4 hr of incubation with doubly-labeled NAR. (N = 5). (C) Confirmation of the lack of NR contamination in NAR. 1 µM NAR was combined with increasing concentrations of NR (0–100 nM) to demonstrate that NR is absent in the NAR and readily detected by this methodology. (Single measurements). (D) Total ion counts for NAAD and NAMN in whole cell lysates and mitochondrial isolates from differentiated C2C12 cells treated with isotopically-labeled NR or NAR tracers for 4 hr. Results expressed as means ± SEM. (N = 3). (E) Incubation of NAD at 37°C in water, but not 80% methanol results in substantial degradation to NMN. Blue bars show unlabeled NMN resulting from degradation from a 20 µM NAD standard spike; Red bars indicate labeled NMN from spiked-in standard (1 µM, dual labeled). (N = 3). (F) NAD total ion count measured in parallel from same samples in (E). (N = 3).
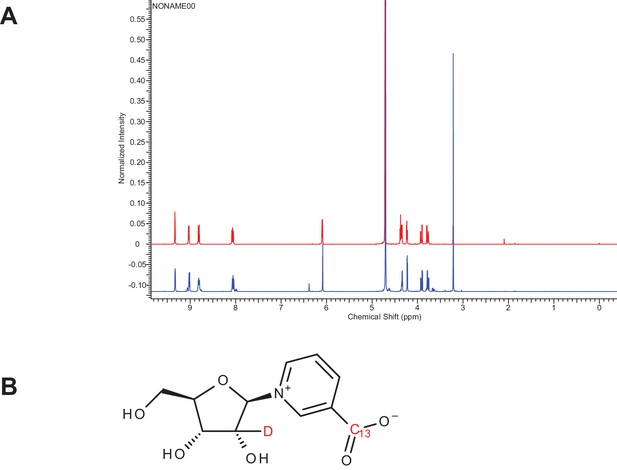
Characterization of doubly-labeled nicotinic acid riboside.
(A) NMR spectra confirming the NAR synthesis product. Red NMR trace represents unlabeled β–NAR. Blue NMR trace represents double-labeled NAR (~15% ɑ-NAR based on integration). (B) β–NAR-deuterium labeled on ribose and C-13 labeled on the nicotinic acid moiety.
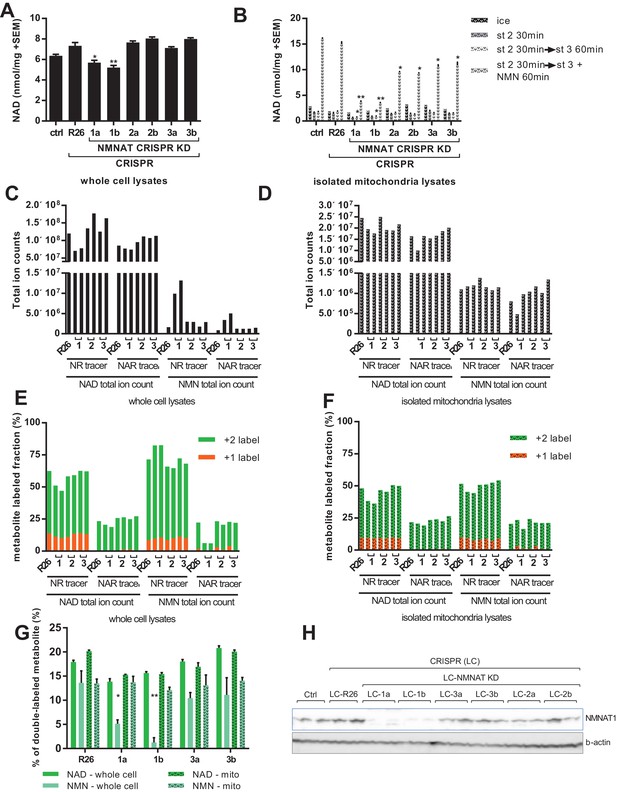
Labeling of mitochondrial NAD tracks that of total NAD, but not of total NMN.
For all panels, data representing whole cells are depicted as solid bars, whereas data from isolated mitochondria are shown with a stippled pattern. (A) Differentiated C2C12 parental and LentiCRISPR transgenic myotubes were analyzed for NAD content. The cells are as follows: ctrl- parental line with no vector; R26- vector control; 1a and 1b- two separate guide RNAs targeting NMNAT1; 2a and 2b- two separate guide RNAs targeting NMNAT2; 3a and 3b- two separate guide RNAs targeting NMNAT3. (N = 3). (B) Mitochondria isolated from differentiated C2C12 cells were held in state 2 (MirO5 respiration buffer containing 10 mM Pyruvate, 5 mM Malate) at 37°C with shaking for 30 min. They were then collected and lysed in perchloric acid immediately, or transitioned into state three by adding ADP (12.5 mM, final concentration) with or without supplementation with NMN (0.5 mM, final concentration) and maintained for 60 min at 37°C with shaking before collection. (N = 2–4). (C–D) Total ion counts for NAD and NMN in extracts from C2C12 LentiCRISPR whole cells (C) and isolated mitochondria (D) following a 4 hr incubation with isotopically-labeled NR or NAR tracer. (Single measuements). (E–F) Fractional labeling of metabolites (NAD and NMN) measured in C2C12 LentiCRISPR whole cells (E) and isolated mitochondria (F) after a 4 hr incubation with isotopically-labeled NR or NAR tracer. (Single measurements). (G) Fractions of double-labeled NAD and NMN measured in C2C12 LentiCRISPR whole cell and mitochondrial lysates following 4 hr incubation with isotope-labeled NAR (means ± SEM). (N = 3). (H) Immunoblot confirming NMNAT1 knockout in CRISPR C2C12 cell line. (*, p<0.05; **, p<0.001; 2-tailed, unpaired Student’s t-test versus R26).
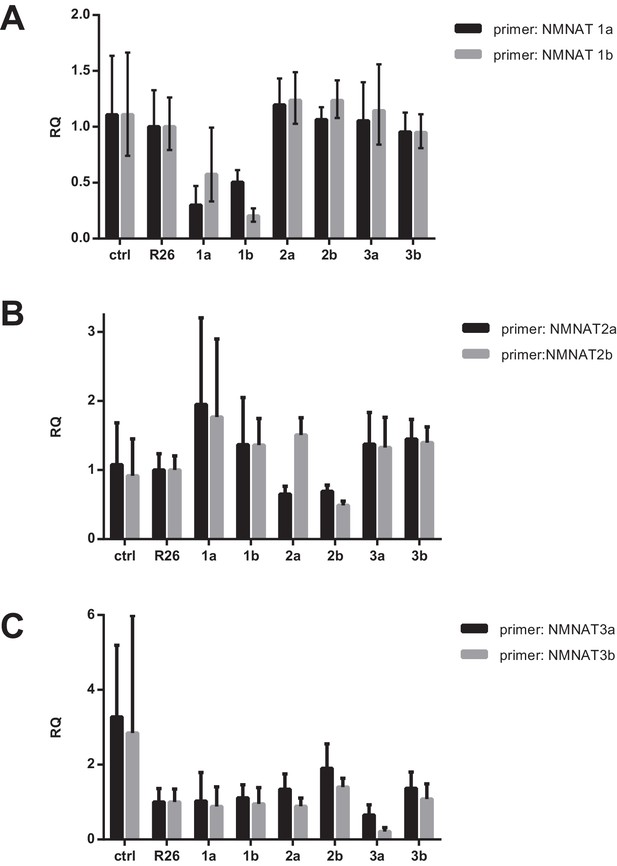
mRNA expression of NMNAT isoforms after CRISPR targeting.
Total RNA was extracted from differentiated cells in duplicate using Trizol (Invitrogen) according to the manufacturer’s instructions. Subsequently, cDNA was synthesized using a high capacity cDNA reverse transcription kit (ABI). RT-PCR was performed using Power SYBR Green PCR master mix (ABI) on the Quantstudio 7 Flex RT-PCR system (ABI). (A–C) For all assays, the plots show gene expression values relative to the reference sample, R26 (control), and are normalized to the control gene 36B4 (Rplp0). For each NMNAT isoform, two distinct gRNAs were generated (a and b) near the 5’ end, which are separated by a short sequence (~40 bp) and the correspondingly named primers amplify a region that is just downstream of the target site. The cells are named as follows: ctrl - parental line with no vector; R26 - control; 1a and 1b - two separate guide RNAs targeting NMNAT1; 2a and 2b - two separate guide RNAs targeting NMNAT2; 3a and 3b - two separate guide RNAs targeting NMNAT3.
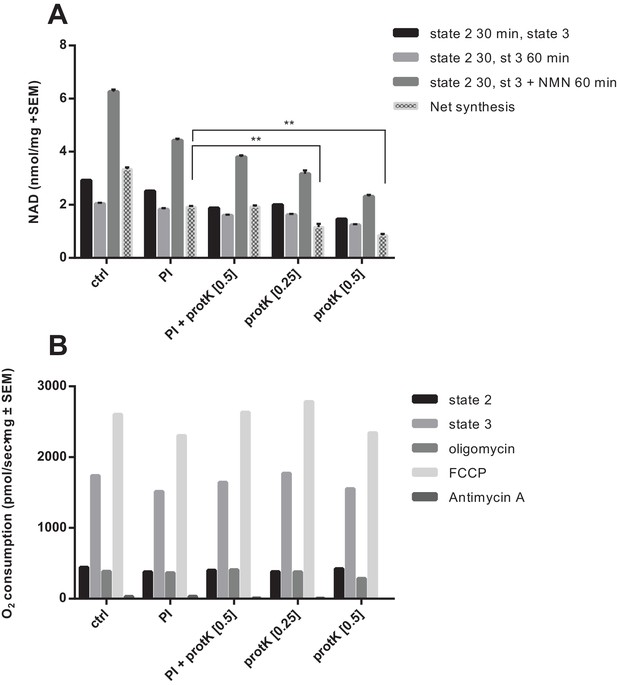
Partial digestion of mitochondrial preps with proteinase K impairs NAD synthesis, but not respiration.
Isolated murine skeletal muscle mitochondria were diluted in MiRO5 to equal concentrations (1.1 mg/mL) and were incubated on ice for 30 min with the following treatments: ctrl - diluted in MiRO5 only; PI - incubated in MiRO5 only for 30 min followed by addition of protease inhibitor (1:100 dilution; Sigma); PI + protK [0.5] - incubated with both protease inhibitor (1:100) and proteinase K (0.5 mg/mL; Roche); protK [0.25] - incubated with proteinase K (0.25 mg/mL) followed by the addition of protease inhibitor; protK [0.5] - incubated with proteinase K (0.5 mg/mL) followed by the addition of protease inhibitor. (A) Treatment with proteinase K significantly impaired the ability of the mitochondria to generate NAD from NMN. (N = 2). (B) Proteinase K-treated does not impair mitochondrial OXPHOS activity. (N = 1).
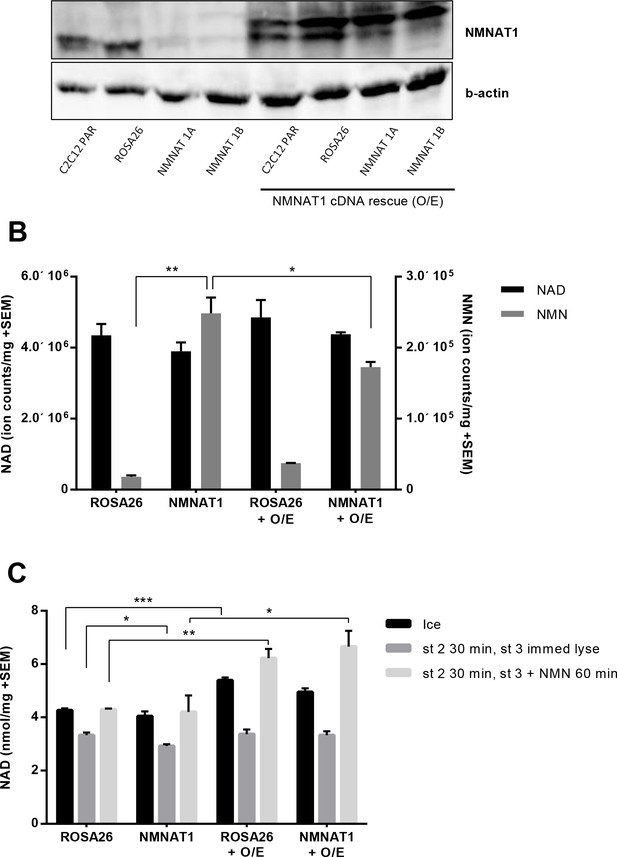
Overexpressing human NMNAT1 decreases NMN accumulation in NMNAT1 targeted cells and enhances the ability of mitochondrial preps to synthesize NAD from NMN.
NMNAT1 protein was restored by introduction of a viral vector containing the human form of the NMNAT1 gene in order to prevent targeting by the mouse-specific NMNAT1 gRNA. The gateway compatible lentiviral vector, pLX304 (clone ID:HsCD00434593, DNASU, Arizona), was co-transfected with the pMD2-G envelope and psPAX2 packaging vectors into 293 cells using Fugene six transfection reagent (Promega). The consequent lentivirus was collected as described and was used to infect the C2C12 CRISPR cells. (A) Expression of human NMNAT1 in C2C12 cells lacking the murine isoform and the Rosa26 control line. (B) Human NMNAT1 reduced NMN accumulation in CRISPR-targeted cells. (C) Human NMNAT1 cDNA enhanced the ability of mitochondrial preps to synthesize NAD from NMN. Three biological replicates were used for the experiments reported in panels B and C.
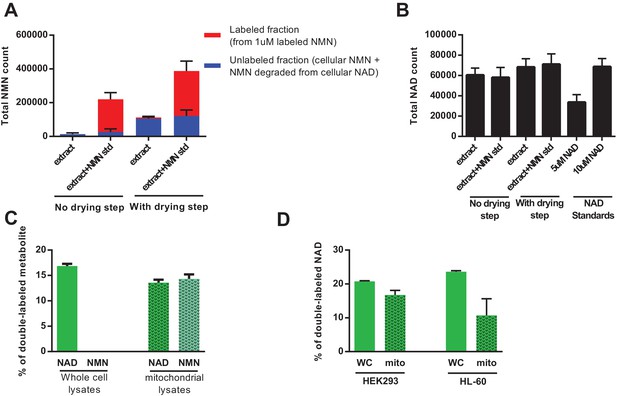
Direct injection of methanolic extracts reveals preferential labeling and mitochondrial uptake of NAD over NMN.
(A) NMN concentration and labeling in differentiated C2C12 cells extracted with −80°C 80:20 methanol:water, analyzed either by hydrophilic interaction chromatography (no drying step), or dried under N2, re-suspended in water and analyzed by reversed-phase ion-pairing chromatography (with drying step). Blue bars show unlabeled NMN resulting from intracellular NMN + NAD degradation after the drying/resuspension step; Red bars indicate labeled NMN from spiked-in standard (1 µM, dual labeled). (N = 2). (B) NAD total ion count measured in parallel from same samples in (A). (N = 2). (C) NAD and NMN labeling (mass + 2 fraction) in differentiated C2C12 cells treated with dual labeled NAR for 4 hr (whole cell vs. isolated mitochondria, N = 3). Data are compiled from three biological replicates and are displayed as means ± SEM. (D) NAD labeling (mass +2 fraction) in the human cell lines HEK293 and HL-60 following a 4 hr incubation with dual labeled NAR (whole cell vs. isolated mitochondria, N = 3). Data are compiled from three biological replicates and are displayed as means ± SEM.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Mus musculus) | C2C12; myotubes; myoblasts; | ATCC | ATCC CRL-1772 | Mouse myoblast, mycoplasma negative |
| Cell line (Homo sapiens) | HL-60 | ATCC | ATCC CCL-240 | Human leukemia, authenticated by STR profiling, mycoplasma negative |
| Cell line (Homo sapiens) | 293; HEK293 | Gift from Morris Birnbaum’s lab | Human embryonic kidney, authenticated by STR profiling, mycoplasma negative | |
| Strain, strain background (Mus musculus) | C57BL/6 mice | The Jackson Laboratory | 000664 | C57BL/6J |
| Recombinant DNA reagent | LentiCRISPR v2 (Lentiviral vector) | Addgene | 52961 | |
| Recombinant DNA reagent | psPAX2 (Lentiviral packaging plasmid) | Addgene | 12260 | |
| Recombinant DNA reagent | pMD2.G (Lentiviral envelope expressing plasmid) | Addgene | 12259 | |
| Recombinant DNA reagent | pLX304 (Gateway Lentiviral vector) | DNASU plasmid repository | NMNAT1 | Clone ID: HsCD00434593 |
| Chemical compound, drug | ATP | Sigma-Aldrich | A2383 | |
| Chemical compound, drug | Protease | Sigma-Aldrich | P5380 | Protease from Bacillus lichenformis |
| Chemical compound, drug | ADP | Sigma-Aldrich | A2754 | |
| Chemical compound, drug | Pyruvate | Sigma-Aldrich | P2255 | |
| Chemical compound, drug | Malate | Sigma-Aldrich | M1000 | |
| Chemical compound, drug | B-NMN | Sigma-Aldrich | N3501 100 MG | |
| Chemical compound, drug | PRPP | Sigma-Aldrich | P8296 | |
| Chemical compound, drug | FCCP | Sigma-Aldrich | C2920-10MG | |
| Chemical compound, drug | Oligomycin | Sigma-Aldrich | O4876-5MG | |
| Chemical compound, drug | Protease inhibitor cocktail (Sigma); PI | Sigma-Aldrich | P8340 | Protease inhibitor cocktail solution |
| Chemical compound, drug | Alcohol dehydrogenase | Sigma-Aldrich | A3263-150KU | |
| Chemical compound, drug | Diaphorase | Sigma-Aldrich | D5540-500UN | |
| Chemical compound, drug | Resazurin | Sigma-Aldrich | R7017 | |
| Chemical compound, drug | Flavin mononucleotide | Sigma-Aldrich | F6750 | |
| Chemical compound, drug | Nicotinamide; NAM | Sigma-Aldrich | 72345 | |
| Chemical compound, drug | Hexadinitrine | Sigma-Aldrich | 107689 | |
| Chemical compound, drug | Nicotinic acid; NA | Sigma-Aldrich | N4126 | |
| Chemical compound, drug | Perchloric Acid | Sigma-Aldrich | 244252 | |
| Chemical compound, drug | NAD; Nicotinamide adenine dinucleotide | Roche | 101127965001 | |
| Chemical compound, drug | Proteinase K; ProtK | Roche | 03115887001 | |
| Chemical compound, drug | Protease inhibitor cocktail (Roche) | Roche | 11697498001 | Complete protease inhibitor cocktail tablets |
| Chemical compound, drug | BSA | Roche | 03117057001 | Bovine serum albumin Fraction V, heat shock, fatty acid free |
| Chemical compound, drug | Puromycin | ThermoFisher Scientific | A111380-03 | 10 mg/mL stock |
| Chemical compound, drug | Blasticidin | ThermoFisher Scientific | R21001 | |
| Chemical compound, drug | Gallotannin | Enzo Life Sciences | ALX-270–418 G001 | |
| Chemical compound, drug | Insulin | Novo-Nordisk Novolin N | U-100 | 100 units/mL; recombinant DNA origin |
| Chemical compound, drug | Fugene 6 | Promega | E2691 | |
| Chemical compound, drug | NR; isotope-labeled nicotinamide riboside | PMID: 27508874 | ||
| Chemical compound, drug | NAR; isotope-labeled nicotinic acid riboside | this paper | ||
| Commercial assay or kit | Micro BCA Protein Assay Kit | Thermo Fisher Scientific | 23235 | |
| Commercial assay or kit | SuperSignal West femto kit | Thermo Fisher Scientific | 34095 | |
| Antibody | anti-NMNAT1 (rabbit polyclonal) | Gift from Lee Kraus, Zhang et al. (2009) PMID: 19478080 | ||
| Antibody | anti-VDAC (rabbit monoclonal) | Abcam | ab154856 | [EPR10852(B)] |
| Antibody | anti-B-actin HRP (mouse monoclonal) | Abcam | ab20272 | [mAbcam 8226] |
| Antibody | Secondary antibody | GE Healthcare Life Sciences | NA934 | Amersham ECL anti-rabbit IgG, HRP-linked whole Ab (from donkey) |
| Antibody | Secondary antibody | GE Healthcare Life Sciences | NA931 | Amersham ECL anti-mouse IgG, HRP-linked whole Ab (from sheep) |
Additional files
-
Supplementary file 1
Table of gRNA sequences cloned into LentiCRISPR v2 vector backbone.
Two independent guides were used to target each NMNAT isoform, and were compared to a control targeting the ROSA26 locus.
- https://doi.org/10.7554/eLife.33246.013
-
Supplementary file 2
Primer sequences used to detect NMNAT transcripts.
Each primer pair amplifies a region just downstream of the guide RNA that bears the corresponding name (see Supplementary file 1).
- https://doi.org/10.7554/eLife.33246.014
-
Transparent reporting form
- https://doi.org/10.7554/eLife.33246.015





