Early structural and functional plasticity alterations in a susceptibility period of DYT1 dystonia mouse striatum
Figures
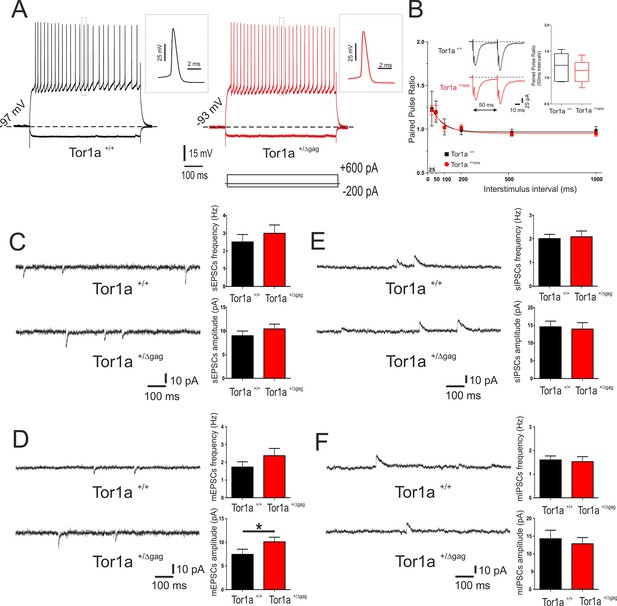
Electrophysiological and synaptic properties of striatal SPNs.
(A) Superimposed traces showing voltage responses to both depolarizing (+600 pA) and hyperpolarizing (−200 pA) current steps in SPNs recorded from P26 Tor1a+/+ (black) and Tor1a+/Δgag (red) mice. The insets display single action potentials (amplitude: Tor1a+/+69.62 ± 1.14 mV, N = 11, n = 11; Tor1a+/Δgag66.65 ± 1.68 mV, N = 8, n = 11; Student’s t test p>0.05). (B) Summary plot of paired-pulse ratio values showing similar facilitation in both genotypes. Each data point represents mean ± SEM. P26 Tor1a+/+ mice N = 3, 25 ms: 1.24 ± 0.20, n = 5; 50 ms: 1.20 ± 0.12, n = 5, Student’s t test p<0.05; P26 Tor1a+/Δgag mice N = 3, 25 ms: 1.22 ± 0.05, n = 5; 50 ms: 1.19 ± 0.08, n = 5; Student’s t test p<0.05. Insets represent sample traces showing facilitation at ISI = 50 ms in both genotypes. (C) Representative sEPSCs recordings in PTX from SPNs of P26 Tor1a+/+ and Tor1a+/Δgag mice. HP: −70 mV. The summary plots show no significant difference between genotypes in sEPSCs frequency and amplitude (Student’s t test p>0.05). (D) Representative whole-cell recordings in PTX plus TTX of mEPSC from P26 Tor1a+/+ and Tor1a+/Δgag SPNs. HP: −70 mV. Plots show a significant difference in the amplitude of mEPSCs recorded from Tor1a+/Δgag mice compared to wild-types (Tor1a+/+, 7.45 ± 1.09, N = 9, n = 9; Tor1a+/Δgag, 10.11 ± 0.97, N = 8, n = 9; Student’s t test *p<0.05). (E) Representative recordings in MK-801 and CNQX of sIPSCs from P26 Tor1a+/+ and Tor1a+/Δgag SPNs. HP:+10 mV. The summary plots show no significant difference in sIPSC frequency and amplitude (Student’s t test p>0.05). (F) Representative traces of mIPSCs recorded in MK-801, CNQX and TTX. HP:+10 mV. The summary plots show no difference in frequency and amplitude between genotypes (Student’s t test p>0.05). Data are presented as mean ± SEM.
-
Figure 1—source data 1
Electrophysiological and synaptic properties of striatal SPNs.
- https://doi.org/10.7554/eLife.33331.003
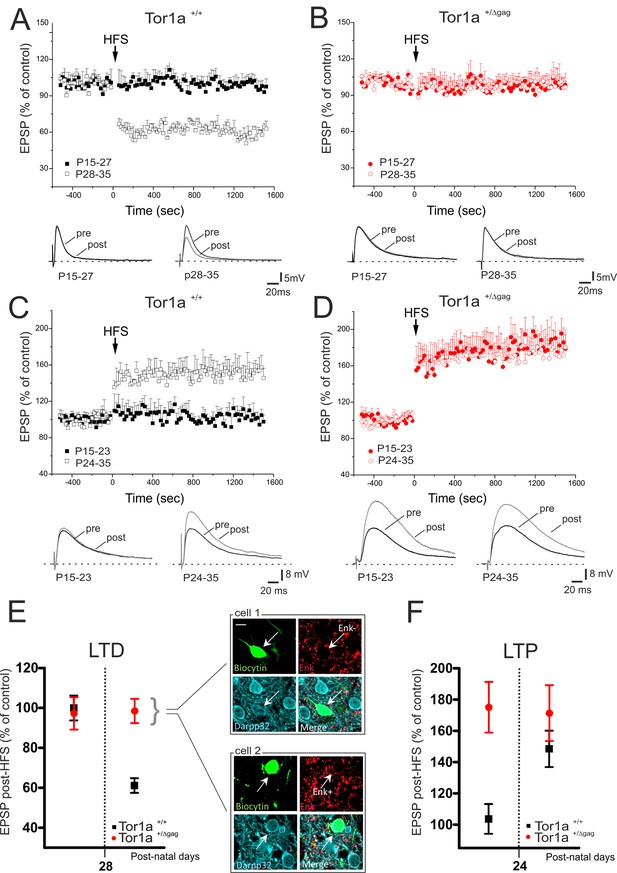
Altered developmental profile of corticostriatal long-term synaptic plasticity expression in Tor1a+/Δgag mice.
(A) (Top) Developmental time-course of LTD expression in Tor1a+/+ mice. HFS protocol (arrow) induces LTD in SPNs recorded from Tor1a+/+ mice after P28 (59.63 ± 2.63% of control; N = 8, n = 8; paired Student’s t test p<0.05), but not from P15 to P27 (99.46 ± 4.65, N = 9, n = 10; paired Student’s t test p>0.05). (Bottom) Representative EPSP traces recorded before (pre) and 20 min after (post) HFS protocol delivery. (B) (Top) In Tor1a+/Δgag mice, HFS protocol fails to induce any LTD, irrespective of the postnatal age (P15-27, 96.85 ± 11.35% of control; N = 8, n = 12; P28-35, 100.29 ± 4.16% of control, N = 8, n = 12; paired Student’s t test p>0.05). (Bottom) Representative traces of EPSPs recorded pre- and post-HFS. (C) (Top) Time-course of corticostriatal LTP expression during postnatal development in Tor1a+/+ mice. HFS of corticostriatal afferents (arrow) induces LTP expression in Tor1a+/+ mice after P24 (148.80 ± 15.39% of control; N = 6, n = 10; paired Student’s t test p<0.05), but not at P15-23 (104.68 ± 8.99% of control; N = 6, n = 10; paired Student’s t test p>0.05). (Bottom) Sample EPSPs recorded pre- and post-HFS protocol in Tor1a+/+ mice. (D) (Top) SPNs recorded from Tor1a+/Δgag mice exhibit a premature LTP (P15-23, 174.68 ± 22.59% of control; N = 6, n = 10; P24-35, 172.35 ± 11.06% of control; N = 9, n = 10; paired Student’s t test p<0.05). (Bottom) EPSP traces recorded pre- and post-LTP induction. (E) Mean plot comparing LTD expression at different postnatal days in Tor1a+/+ and Tor1a+/Δgag SPNs. (Inset) Confocal imaging of two SPNs recorded from Tor1a+/Δgag slices filled with biocytin (green) and immunolabelled for ENK (red) and DARPP-32 (cyano), marker of SPNs. Both ENK-positive and ENK-negative biocytin-labeled SPNs showed lack of LTD (scale bar: 10 µm). (F) Mean plot comparing LTP expression at different postnatal days in Tor1a+/+ and Tor1a+/Δgag SPNs. Values are presented as mean ± SEM.
-
Figure 2—source data 1
Altered developmental profile of corticostriatal long-term synaptic plasticity expression in Tor1a+/Δgag mice.
- https://doi.org/10.7554/eLife.33331.005
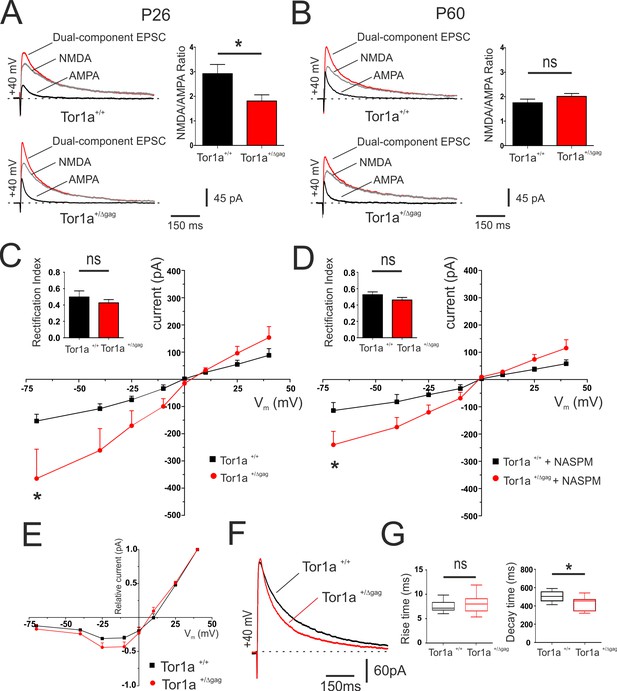
Electrophysiological characterization of AMPAR and NMDAR currents at corticostriatal synapses of SPNs in both Tor1a+/+ and Tor1a+/Δgag mice.
(A) (Left) Representative EPSCs traces recorded at HP=+40 mV from SPNs of juvenile Tor1a+/+ and Tor1a+/Δgag mice. The NMDAR antagonist MK-801 isolates the AMPAR-mediated EPSC component (black trace), while the NMDAR-EPSC (grey trace) is obtained by digital subtraction of the AMPAR EPSC from the dual-component EPSC (red). (Right) Summary plot of NMDA/AMPA current ratio calculated in SPNs from P26 Tor1a+/+ and Tor1a+/Δgag mice. A significant decrease of NMDA/AMPA ratio was detected in P26 Tor1a+/Δgag mice, compared to Tor1a+/+ (Tor1a+/+, 2.92 ± 0.38, N = 3, n = 8; Tor1a+/Δgag, 1.81 ± 0.25, N = 3, n = 6; Student’s t test, p<0.05). (B) (Left) Representative EPSCs traces recorded at HP =+40 mV from SPNs of adult Tor1a+/+ and Tor1a+/Δgag mice. (Right) Summary plot of NMDA/AMPA current ratio showing no significant difference between genotypes (Tor1a+/+, 1.75 ± 0.15, N = 3, n = 7; Tor1a+/Δgag, 2.01 ± 0.12, N = 3, n = 7; Student’s t test, p>0.05). (C) AMPAR-mediated currents recorded at different HP in P26 Tor1a+/+ and Tor1a+/Δgag SPNs. The IV relationship shows a significant increase in the current recorded at more hyperpolarized range from P26 Tor1a+/Δgag SPNs (HP=−70 mV: two-way ANOVA, *p<0.01). (Left) Summary plot of rectification index values of P26 Tor1a+/+ and Tor1a+/Δgag SPNs (Tor1a+/+, 0.50 ± 0.07, n = 7; Tor1a+/Δgag, 0.43 ± 0.04, n = 8; Student’s t test p>0.05). (D) AMPAR-mediated currents recorded in the presence of the GluA2-lacking AMPAR antagonist NASPM at P26. HP =−70 mV; to-way ANOVA, *p<0.01). (Left) Summary plots of the rectification index measured at P26 (Tor1a+/+, 0.53 ± 0.04, n = 5, N = 6; Tor1a+/Δgag, 0.46 ± 0.03, n = 7; Student’s t test, p>0.05). (E) Normalized IV relationships of NMDAR-mediated currents show no difference between genotypes at P26 (two-way ANOVA, p>0.05). (F) Representative NMDA-mediated EPSCs recorded at HP =+40 mV from P26 SPNs. (G) Summary plots display rise and decay time of NMDA-EPSCs recorded at HP =+40 mV in SPNs from P26 Tor1a+/+ and Tor1a+/Δgag mice (rise time: Tor1a+/+, 7.78 ± 0.42, n = 9; Tor1a+/Δgag, 9.23 ± 1.37, n = 7; Student’s t test p>0.05; decay time: Tor1a+/+, 502.50 ± 20.06, n = 9; Tor1a+/Δgag, 422.10 ± 30.15, n = 7, Student’s t test, *p<0.05). Values are presented as mean ± SEM.
-
Figure 3—source data 1
Electrophysiological characterization of AMPAR and NMDAR currents at corticostriatal synapses of SPNs in both Tor1a+/+ and Tor1a+/Δgag mice.
- https://doi.org/10.7554/eLife.33331.007

Molecular analysis of the SPNs postsynaptic compartment in P26 and P60 Tor1a+/Δgag compared to age-matched wild-type mice.
WB analyses were performed on the post-synaptic TIF fraction in a minimum of three different animals per genotype. (A) WB analysis for GluN2A, GluN2B, PSD-95 and tubulin in P26 (left panel) and P60 (right panel) Tor1a+/Δgag and age-matched Tor1a+/+ mice. (C) WB analysis for GluA1, GluA1p845, GluA2 and tubulin in P26 (left panel) and P60 (right panel) Tor1a+/Δgag and age-matched Tor1a+/+ mice. (B,D) The histogram shows the quantification of protein levels following normalization on tubulin (P26 Tor1a+/Δgag compared to Tor1a+/+, GluA1: 142.8 ± 9.8%, n = 5, p<0.05; GluA1-p845: 200.9 ± 36.6%, n = 5, p<0.05; GluA2: 175.1 ± 16.6%, n = 5, p<0.05; GluN2A: 197.3 ± 34.0%, n = 5, p<0.05; P60 Tor1a+/Δgag GluA1: 90.0 ± 23.4%, n = 5, p>0.05; GluA1-p845: 77.7 ± 14.2%, n = 5, p>0.05; GluA2: 103.2 ± 16.2%, n = 5, p>0.05; GluN2A: 88.8 ± 18.0%, n = 5,p>0.05). All values are mean ± SEM expressed as % of Tor1a+/+ mice.
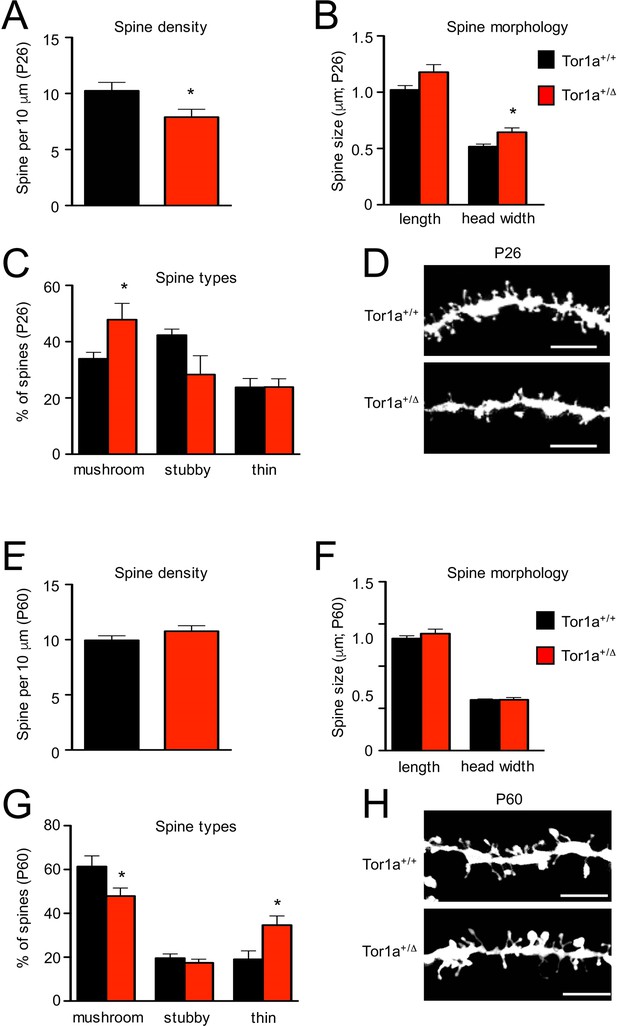
Analysis of dendritic spines morphology in P26 and P60 Tor1a+/Δgag compared to age-matched Tor1a+/+mice.
(A) Histogram representing dendritic spine density in P26 Tor1a+/Δgag and Tor1a+/+ mice (Tor1a+/+, 10.25 ± 0.75 spines/10 μm, n = 10; Tor1a+/Δgag, 7.89 ± 0.70 spines/10 μm, n = 10; unpaired Student’s t test *p<0.05). (B,C) Histograms showing the quantification of dendritic spine size (B, spine length and head width) and dendritic spine type (C, mushroom, stubby, thin) in P26 Tor1a+/Δgag compared to Tor1a+/+ mice (dendritic spine width Tor1a+/+, 0.51 ± 0.02 μm, n = 10; Tor1a+/ Δgag, 0.64 ± 0.04 μm, n = 10, unpaired Student’s t-test *p<0.05; mushroom-type spines Tor1a+/+, 33.92 ± 2.32%, n = 10; Tor1a+/Δgag, 47.81 ± 5.79%, n = 10, unpaired Student’s t-test *p<0.05). (D) Representative images show dendrites of P26 Tor1a+/Δgag and Tor1a+/+ mice. (E) Histogram representing dendritic spine density in P60 Tor1a+/Δgag and Tor1a+/+ mice (Tor1a+/+, 9.94 ± 0.41 spines/10 μm, n = 10; Tor1a+/ Δgag, 10.76 ± 0.50 spines/10 μm, n = 10; unpaired Student’s t-test p>0.05). (F,G) Histograms showing the quantification of dendritic spine size (F, spine length and head width) and dendritic spine type (G, mushroom, stubby, thin) in P60 Tor1a+/Δgag, compared to Tor1a+/+ mice (spine width Tor1a+/+, 0.600 ± 0.012 μm, n = 10; Tor1a+/Δgag, 0.602 ± 0.027 μm, n = 10; p>0.05; mushroom-type spines Tor1a+/+, 61.40 ± 4.81%, n = 10; Tor1a+/Δgag, 47.92 ± 3.67%, n = 10; *p<0.05; thin spines Tor1a+/+, 19.04 ± 3.85%, n = 10; Tor1a+/ Δgag, 34.64 ± 4.16%, n = 10; *p<0.05; unpaired Student’s t-test). (H) Representative images show dendrites of P60 Tor1a+/Δgag and Tor1a+/+ mice. Data were collected in a minimum of three different animals per genotype.
-
Figure 5—source data 1
Analysis of dendritic spines morphology in P26 Tor1a+/Δgag compared to age-matched Tor1a+/+ mice.
- https://doi.org/10.7554/eLife.33331.010
-
Figure 5—source data 2
Analysis of dendritic spines morphology in P60 Tor1a+/Δgag compared to age-matched Tor1a+/+ mice.
- https://doi.org/10.7554/eLife.33331.011
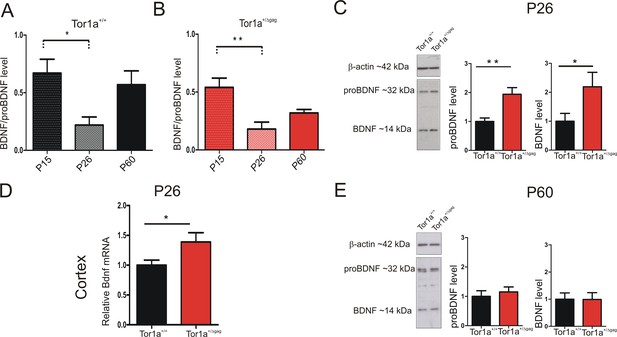
BDNF protein expression in the striatum of Tor1a+/+and Tor1a+/Δgag mice.
(A, B) Striatal BDNF protein expression in Tor1a+/+ and Tor1a+/Δgag mice at postnatal stages (P15, P26, P60). The graphs show the quantification of BDNF/proBDNF ratio at the various ages. Data are represented as mean ± SEM (Tor1a+/+ P15: 0.67 ± 0.12, N = 4; P26: 0.22 ± 0.08, N = 4; P60: 0.57 ± 0.14, N = 3; Tor1a+/Δgag P15: 0.54 ± 0.08, N = 4; P26: 0.18 ± 0.06, N = 4; P60: 0.32 ± 0.03, N = 4; one-way ANOVA, *p<0.05; **p<0.01). (C) (Left) Representative WB of proBDNF and BDNF protein levels relative to β-actin in striatal extracts (30 μg) derived from P26 Tor1a+/+ and Tor1a+/Δgag mice. (Right) The graphs show the quantitative analysis. The amount of proBDNF and BDNF was quantified relative to β-actin and normalized to wild-type mice. Data are represented as mean ± SEM (proBDNF Tor1a+/+ 1.00 ± 0.12, n = 10; Tor1a+/Δgag1.95 ± 0.29, n = 8; BDNF Tor1a+/+: 1.00 ± 0.28, n = 8, Tor1a+/Δgag2.19 ± 0.50, n = 8, Student’s t test: *p<0.05; **p<0.01). (D) Bdnf mRNA is upregulated in the cortex of Tor1a+/Δgag determined by qRT-PCR. The 2-ΔΔCt method was used to determine the relative expression, and all of the values are expressed relative to the levels of the wild-type mice as mean ± SEM (Tor1a+/+ 1.000 ± 0.084, n = 10; Tor1a+/Δgag1.399 ± 0.163, n = 8; Student’s t test: *p<0.05). (E) (Left) Representative Western blots of proBDNF and BDNF proteins relative to β-actin in striatal extracts (15 μg) derived from Tor1a+/+ and Tor1a+/Δgag adult mice. (Right) The graphs show the quantitative analysis. The amount of proBDNF and BDNF was quantified relative to β-actin and normalized to wild-type mice. Data are represented as mean ± SEM (proBDNF Tor1a+/+ 1.00 ± 0.19, n = 7, Tor1a+/Δgag1.15 ± 0.17, n = 7, p>0.05; BDNF Tor1a+/+: 1.00 ± 0.23 n = 7, Tor1a+/Δgag0.99 ± 0.25, n = 7, Student’s t test: p>0.05).
-
Figure 6—source data 1
BDNF protein expression in the striatum of Tor1a+/+ and Tor1a+/Δgag mice.
- https://doi.org/10.7554/eLife.33331.013
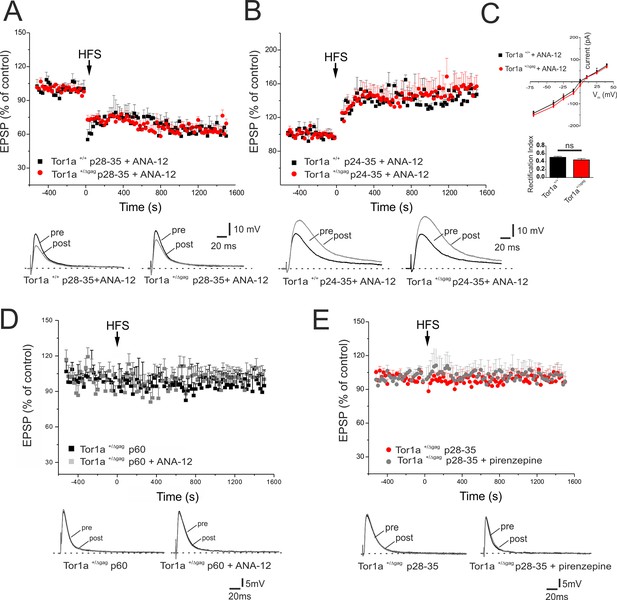
In vivo ANA-12 treatment rescues synaptic plasticity deficits in juvenile Tor1a+/Δgag mice.
(A) Time-course of corticostriatal LTD in juvenile Tor1a+/+ and Tor1a+/Δgag mice (P28-35): after in vivo treatment with the TrkB antagonist ANA-12, the HFS protocol (arrow) induces corticostriatal LTD expression in juvenile Tor1a+/Δgag mice (Tor1a+/+ P28-35, 65.31 ± 1.44% of control; N = 3, n = 12, p<0.05; Tor1a+/Δgag P28-35, 63.41 ± 4.39% of control; N = 3, n = 10; paired Student’s t test p<0.05). (Bottom) Representative EPSPs recorded before (pre) and 20 min after (post) HFS protocol. (B) Time-course of corticostriatal LTP after in vivo ANA-12 treatment: LTP displays a physiological amplitude in SPNs from in P24-35 Tor1a+/Δgag compared to wild-type littermates (Tor1a+/+ P24-35, 144.55 ± 2.67% of control; N = 3, n = 8; Tor1a+/Δgag P24-35, 148.11 ± 10.55% of control; N = 3, n = 9; Tor1a+/Δgag vs. Tor1a+/+ Student’s t test p>0.05). (Bottom) Sample traces recorded pre and post LTP induction. (C) AMPAR-mediated currents recorded from P26 SPNs at HP from −70 mV to + 40 mV after in vivo treatment of Tor1a+/+ and Tor1a+/Δgag mice with ANA-12. The treatment normalizes the current-voltage relationship in Tor1a+/Δgag neurons (HP=−70 mV: 2-way ANOVA p>0.05) and the rectification index (Tor1a+/+, 0.51 ± 0.03, N = 3, n = 3; Tor1a+/Δgag, 0.45 ± 0.04, N = 3, n = 5; Student’s t test p>0.05) (D) In vivo treatment with ANA-12 does not restore corticostriatal LTD in adult (P60) SPNs recorded from Tor1a+/Δgag mice (vehicle: 95.66 ± 9.09% of control, N = 3, n = 8; ANA-12: 98.75 ± 11% of control, N = 3, n = 4; paired Student’s t test p>0.05). (E) Slice pre-treatment with pirenzepine (100 nM) does not rescue LTD expression in P28-35 Tor1a+/Δgag SPNs (vehicle: 101.54 ± 1.07% of control, N = 3, n = 3; pirenzepine: 100.34 ± 8.96% of control; N = 3, n = 3; paired Student’s t test p>0.05). (Bottom) Superimposed traces of EPSPs recorded pre and 20 min post HFS delivery.
-
Figure 7—source data 1
In vivo ANA-12 treatment rescues synaptic plasticity deficits in juvenile Tor1a+/Δgag mice.
- https://doi.org/10.7554/eLife.33331.015
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Mus musculus) | Tor1a | MGI:1353568 | Gene ID: 30931 | official full name: torsin family 1, member A (torsin A) |
| Strain, strain background (M. musculus) | C57BL/6J mice | Charles River | catalog number B6JSIFE10SZ - C57BL/6J SPF/VAF; RRID:IMSR_JAX:000664 | |
| Genetic reagent (M. musculus) | heterozygous knock-in Tor1a+/Δgag | Goodchild et al. (2005) | - | maintained on the C57BL/6J background |
| Antibody | monoclonal anti-PSD-95 | Neuromab | clone (k28/43) - catalog number 75–028; RRID:AB_2292909 | dilution 1:2000 in I-Block |
| Antibody | monoclonal anti-GluN2B | Neuromab | clone 59/20 - catalog number 75–097; RRID:AB_10673405 | dilution 1:1000 in I-Block |
| Antibody | polyclonal anti-GluA1 | Merck Millipore | catalog number AB1504; RRID:AB_2113602 | dilution 1:1000 in I-Block |
| Antibody | polyclonal anti-phospho-GluA1 (Ser845) | Merck Millipore | catalog number 04–1073; RRID:AB_1977219 | dilution 1:1000 in I-Block |
| Antibody | polyclonal anti-GluN2A | Sigma-Aldrich | catalog number M264 RRID:AB_260485 | dilution 1:1000 in I-Block |
| Antibody | monoclonal anti-GluA2 | Neuromab | clone L21/32 - catalog number 75–002; RRID:AB_2232661 | dilution 1:1000 in I-Block |
| Antibody | monoclonal anti-α-tubulin | Sigma-Aldrich | clone DM1A - catalog number T9026; RRID:AB_477593 | dilution 1:5000 in I-Block |
| Antibody | goat anti-DARPP-32 | R and D system | catalog number AF6259; RRID:AB_10641854 | dilution 1:500 in I-Block |
| Antibody | mouse anti-Enkephalin | Millipore | catalog number MAB350; RRID:AB_2268028 | dilution 1:1000 in I-Block |
| Antibody | mouse anti-β-actin | Sigma Aldrich | catalog number A5441; RRID:AB_476744 | dilution 1:20000 in I-Block |
| Commercial assay or kit | Clarity Western ECL Substrate | BioRad | - | reagent used to visualize protein bands with Chemidoc Imaging System |
| Commercial assay or kit | ECL reagent | GEHealthcare | catalog number GERPN2232 | reagent used to visualize protein bands with membranes were exposed to film |
| Commercial assay or kit | TRI-reagent | Sigma Aldrich | catalog number T9424 | reagent used to RNA extraction |
| Commercial assay or kit | DNAase I | Invitrogen | catalog number AMPD1-1KT | reagent used for elimination of DNA from RNA |
| Commercial assay or kit | Transcriptor First Strand cDNA Synthesis Kit | Roche | catalog number04379012001 | reagent used to reverse transcribe RNA |
| Commercial assay or kit | Extract-N-Amp™ Tissue PCR Kit | SIGMA | catalog number XNAT2 | genotyping primers UP- AGT CTG TGG CTG GCT CTC C; Low- CCT CAG GCTGCT CAC AAC C |
| Chemical compound, drug | ANA-12 | Sigma-Aldrich | catalog number SML0209 | in vivo administration |
| Chemical compound, drug | CNQX disodium salt | Tocris | catalog number 0190/10 | application in bath during electrophysiology analysis |
| Chemical compound, drug | (+)-MK 801 maleate | Tocris | catalog number 0924/10 | application in bath during electrophysiology analysis |
| Chemical compound, drug | Tetrodotoxin citrate (TTX) | Tocris | catalog number 1069/1 | application in bath during electrophysiology analysis |
| Chemical compound, drug | Picrotoxin | Tocris | catalog number 1128/1 | application in bath during electrophysiology analysis |
| Chemical compound, drug | Biocytin | Tocris | catalog number 3349/10 | electrodes filled with biocytin, versatile marker used for neuroanatomical investigations of neuron IHC |
| Chemical compound, drug | Naspm trihydrochloride | Tocris | catalog number 2766/10 | application in bath during electrophysiology analysis |
| Software, algorithm | ImageLab | BioRad | - | software used for quantification of protein bands in western blotting experiments |
| Software, algorithm | ImageJ software | NIH;Schneider et al. (2012) | RRID:SCR_003070 | software used for the quantification of protein bands in western blotting and confocal laser scanning microscope |
| Software, algorithm | ClampFit 9 | pClamp | Molecular Devices; RRID:SCR_011323 | data analysis |
| Software, algorithm | Origin 8.0 | Microcal | RRID:SCR_002815 | data analysis |
| Software, algorithm | Prism 5.3 | GraphPad | RRID:SCR_002798 | data analysis |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.33331.016





