Environmental stimuli shape microglial plasticity in glioma
Figures
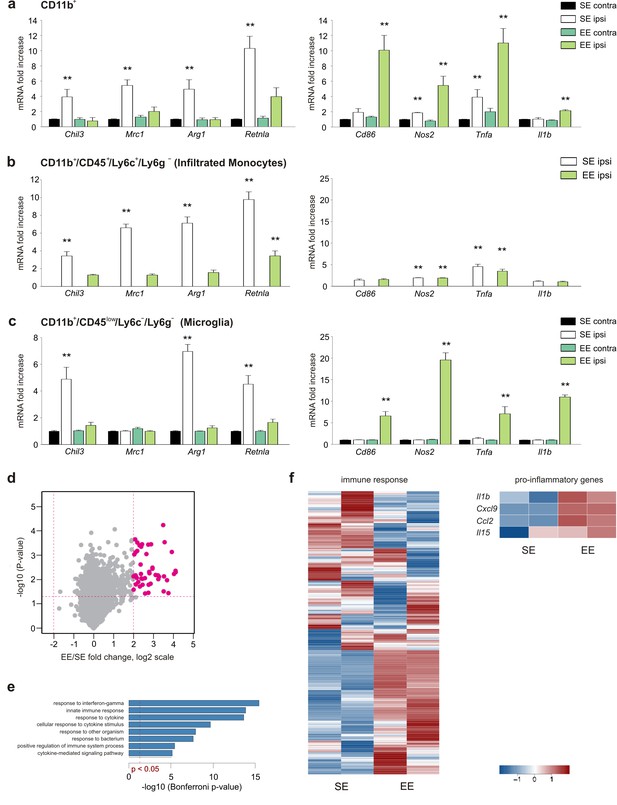
EE modulates myeloid cell phenotype.
(a) RT-PCR of anti- (Chil3, Mrc1, Arg1, Retnla) and pro-inflammatory (Cd86, Nos2, Tnfa, Il1b) genes in CD11b + cells sorted from ILH and CLH of GL261-bearing mice housed in EE or SE. Data are the mean ± S.E.M., **p<0.01 versus CLH by one-way ANOVA; n = 6. (b–c) RT-PCR of anti- (Chil3, Mrc1, Arg1, Retnla) and pro-inflammatory (Cd86, Nos2, Tnfa, Il1b) genes in CD45low/CD11b+/Ly6c–/Ly6g– and CD45+/CD11b+/Ly6c+/Ly6g–cells sorted from ILH and CLH of GL261-bearing mice housed in EE or SE. Data are the mean ± S.E.M., **p<0.01 versus CLH (taken from CD11b + cells in [a]), by one-way ANOVA; n = 5. (d) Comparison of gene expression profiles resulting from EE (n = 2) compared to the SE exposure (n = 2) in microglia from ILH. Each point on the scatterplot represents a gene. The X-axis shows expression ratio (EE/SE) on the log2 scale. Y-axis presents corresponding p-values (–log10[p-value]). Genes that are significantly upregulated after EE exposure (t-test p-value<0.05, fold-change >4) are marked by large purple dots. Global gene expression was determined using Affymetrix microarrays. (e) Results of Gene Ontology over-representation analysis (Bonferroni corrected p-values <0.05) of the genes that are upregulated in microglia after EE exposure (d). X -axis corresponds to –log10-transformed p-values. (f) Expression profile of genes associated with immune response according to the Gene Ontology database and manually selected pro-inflammatory genes.
-
Figure 1—source data 1
Raw data and detailed statistical analysis report.
Raw data of Gene Ontology over-representation analysis (Bonferroni corrected p-values <0.05) of the genes that are upregulated in microglia after EE exposure.Image: raw data refer to Figure 1 data.
- https://doi.org/10.7554/eLife.33415.004
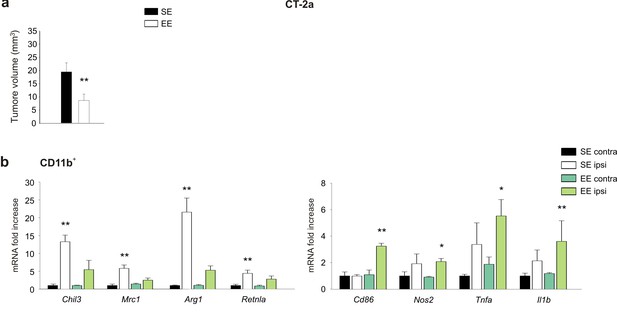
EE modulates tumor size and myeloid cell phenotype in mice injected with CT-2A cells.
(a) The mean tumor volumes at 17 days after implantation of CT-2a cells in mice housed in SE or EE, as indicated. n = 5 **p=0.007, Student’s t-test. (b) RT-PCR analysis of anti- (Chil3, Mrc1, Arg1, Retnla) and pro-inflammatory (Cd86, Nos, Tnfa, Il1bβ) genes in CD11b+ cells sorted from ILH and CLH of CT-2a-bearing mice housed in EE or SE. Data are the mean ± S.E.M., *p<0.05 **p<0.01 versus CLH by one-way ANOVA; n = 5.
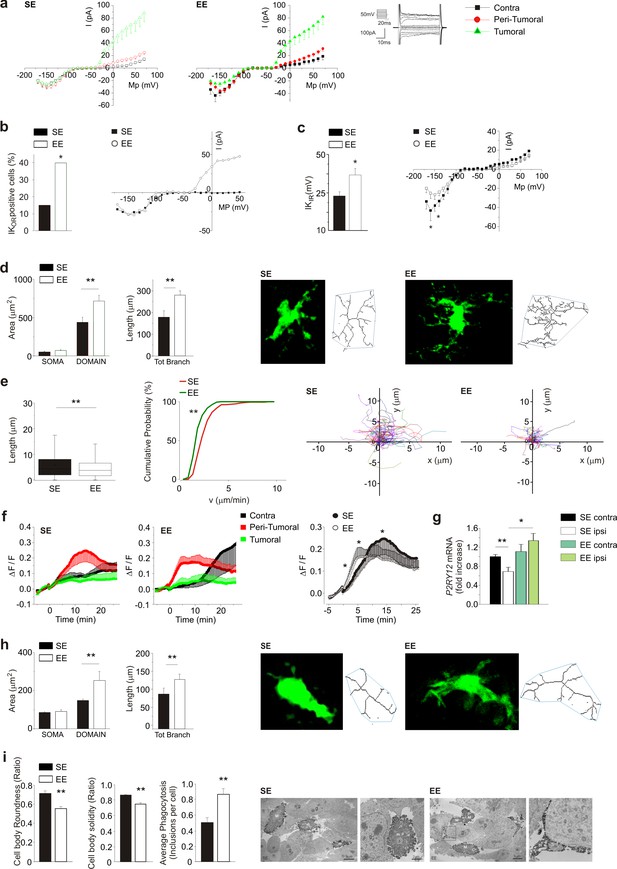
Effect of EE on myeloid cell morphology.
(a) Left: current/voltage relationship of microglia cells in response to voltage steps stimulation (steps from −170 to +70 mV, only one out of two steps are shown; holding potential −70 mV) in CLH (n = 38/9 mice), peritumoral area (n = 60/9 mice) and inside the tumor (n = 57/9 mice) of SE housed, GL261-bearing mice. Right: Current/voltage relationship of microglia cells in CLH (n = 27/9 mice), peritumoral area (n = 57/9 mice) and inside the tumor (n = 64/9 mice) of EE mice. (b) Percentage of GFP+-cells expressing Kor currents in the peritumoral area in SE and EE mice (∗p<0.05, z-test). Representative current/voltage relationships are shown on the right. (c) Amplitude of Kir current expressed by GFP+ cells in the peritumoral area in SE and EE mice (∗p<0.05, z-test). Representative current/voltage relationships are shown on the right. (d) Left: quantification of area of the soma and scanning domain of GFP+ cells measured by ImageJ in slices from GL261-bearing mice housed in SE or EE, as indicated (15 cells, 6 slices, 4 mice per condition, **p=0.0034, t-test). Center: total branch length of GFP+ cells in SE and EE (**p=0.0046, t-test). Right: representative images of maximum intensity projections of a confocal z-stack imaging on peritumoral area of GFP+ cells in SE and EE mice, converted to binary images and then skeletonized by the Analyze Skeleton plugin in Image J (green lines). (e) Length (left, **p=1.35E-24) and cumulative probability histogram of mean velocity (right, p=0.0053) of spontaneous (basal) movement of all single processes of the peritumoral area measured in SE (red, n = 177 tracks, 6 mice) and in EE (green, n = 210 tracks, 6 mice). Right images show reconstruction of basal process migration tracks in SE and EE. Individual tracks were aligned to the origin. (f) Time course of fluorescent ratio (ΔF/F) measured in a circle (10 μm radius) centered on the tip of the ATP puff pipette, in CLH (black, n = 9/9, SE; n = 7/9, EE), peritumoral (red, n = 11/9, SE; n = 12/9, EE) and intra-tumoral (green, n = 9/9, SE; n = 11/9, EE) areas of slices from Cx3cr1+/GFP mice housed in SE or EE. Note that the fluorescence increases around the pipette tip only in the peritumoral area (p<0.05; one-way ANOVA). Right: time course of fluorescence ratio evaluated in the peritumoral area of Cx3cr1+/GFP mice housed in SE (red, n = 11) and EE (black, n = 12) (p=0.041 at 3 min; p=0.035 at 8 min; p=0.018 at 15 min; t-test). (g) RT-PCR of P2RY12 gene in CD11b+ cells sorted from ILH and CLH of GL261-bearing mice, housed in SE or EE. Data are the mean ± S.E.M., *p<0.05 **p<0.01 versus CLH by one-way ANOVA, n = 4. (h) Representative SE and EE z-projections of GFP+ cells (skeletonized as above) into the tumoral area of Cx3cr1+/GFP mice. Left: 13 cells, 6 slices, 4 mice per condition, **p<0.01, Student’s t-test. Note that the scanning domain in the tumoral area was significantly smaller than that in the peritumoral area (p=0.021; t-test). Right: bar chart reporting the morphometric analysis of microglia branches (total branch length) of SE and EE microglia p=0.0031, t-test). (i) Ultrastructural analysis of cell body circularity and solidity, as well as average number of phagocytic inclusions in SE versus EE myeloid cells. Representative pictures showing IBA1-stained myeloid cells from both conditions are also provided. M = microglial cell body; m = microglial process; s = secretion granule; in = phagocytic inclusion; g = glioma cell. Data are the mean ± S.E.M., **p<0.01 by unpaired t-test; n = 25 cells from two animals in SE and n = 45 cells from three animals in EE.
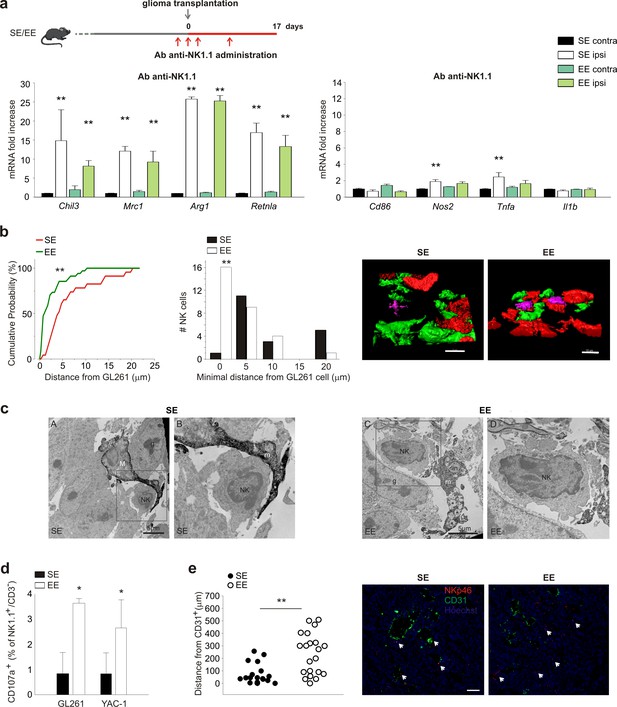
NK cells are modulated by EE.
(a) RT-PCR of anti- (Chil3, Mrc1, Arg1, Retnla) and pro-inflammatory (Cd86, Nos2, Tnfa, Il1b) genes in CD11b+ cells sorted from ILH and CLH in vehicle and NK1.1 Ab-treated GL261-bearing mice, housed in SE or EE. Scheme of Ab-NK1.1 administration above. Data are the mean ± S.E.M., **p<0.01 versus CLH by one-way ANOVA; n = 6. (b) Cumulative distributions of distances of GL261 3D iso-surface from NK cells iso-surface, in brain slices from SE- and EE-housed mice. Note that in SE-housed mice, NK cells are significantly more distant from glioma cells than they are in EE (SE n = 14, 6 slices, 4 mice; EE n = 16, 6 slices/4 mice); **p<0.01, Kolmogorov-Smirnov test). Top: representative 3D reconstruction, by Imaris software, of GFP+ cells, Tag-RFP GL261 cells, and NK1.1-positive cells (Alexa Fluor 633 conjugated secondary Ab) in the tumoral area of brain from SE- (left) and EE- (right) housed mice. (Scale bar 10 µm.) (c) Ultrastructural evidence of direct contacts between microglia and NK cells inside the tumor. Examples of microglial processes wrapping around NK cells are provided for both SE and EE. In EE, the increased prevalence of phagocytic inclusions containing intact elements (in; myelinated axon being internalized) or other types of debris that are being digested (*) is shown. Filopodia protruding from the NK cell body where it touches a microglial process are observed in the images. M = microglial cell body; m = microglial process; NK = NK cell; in = phagocytic inclusion; g = glioma cell. (d) NK cells, isolated from the brains of EE and SE GL261-bearing mice, were incubated with GL261 or YAC-1 cells, and degranulation was assessed by FACS analysis of CD107a+ cells. Average values ± SD of CD107a+ cell frequency upon GL261 or YAC-1 cell co-incubations, minus blanks (degranulation in the absence of targets, in three independent experiments). Student’s t-test, *p=0.011. (e) Mean distances of NK cells from endothelial cells in vessels (CD31+ cells) 17 days after GL261 cell transplantation in SE or EE conditions (n = 4 mice per condition; **p=0.001 Student’s t-test, scale bar 0.1 mm). Representative immunofluorescence is shown on the right.
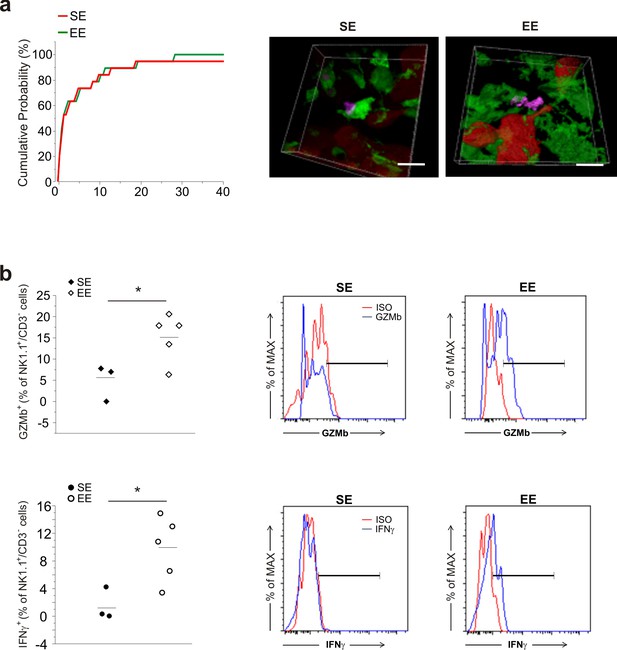
NK cells distance from GFP+ cells and NK cell activation in EE.
(a) Cumulative distributions of distances of GFP-positive cells 3D isosurface from NK cells iso-surface, in brain slices from SE and EE GL261-bearing mice. Note similar distances in both conditions (SE: n = 14, 6 slices, 4 mice; EE: n = 16, 6 slices, 4 mice). (b) Frequency of GZMb+ and IFN-γ+ cells in the CD3–/NK1.1+ cell populations isolated from the brains of SE or EE mice (n = 3–5; for GZMb *p=0.028 Student’s t-test, for IFN-γ *p=0.029 Student’s t-test.) Representative FACS analyses are shown on the right.
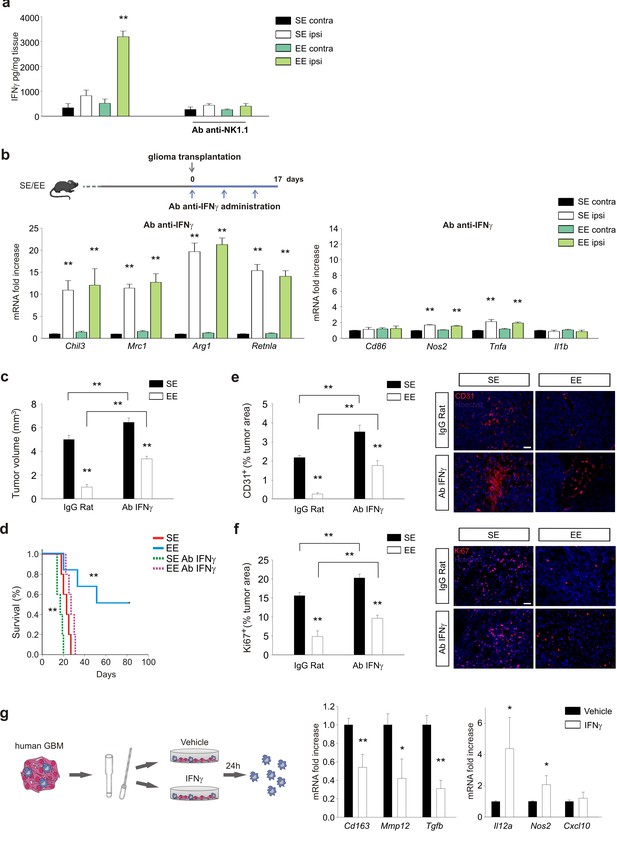
IFN-γ mediates the effects of EE on myeloid cells.
(a) Expression of IFN-γ in CLH and ILH of GL261-bearing mice treated with vehicle or Ab anti-NK1.1 as indicated, and housed in SE or EE. (n = 5–4, **p<0.01, one-way ANOVA.) (b) RT-PCR of anti- and pro-inflammatory genes in CD11b+ cells sorted from ILH and CLH in vehicle and Ab-anti-IFN-γ treated GL261-bearing mice, housed in SE or EE. Above: scheme of Ab-anti-IFN-γ administration. Data are the mean ± S.E.M., **p<0.01 versus CLH, one-way ANOVA; n = 4. (c) Analysis of GL261 tumor volume (expressed in mm3 ± s.e.m.) in IgG Rat or Ab-anti-IFN-γ-treated mice, housed in SE or EE; n = 5; **p<0.01, two-way ANOVA). (d) Kaplan–Meier analyses of GL261-transplanted mice exposed to SE or EE upon vehicle or Ab-anti-IFN-γ treatment; n = 5; log-rank test **p<0.01. (e) Quantification of CD31+ cells (mean ± S.E.M. of CD31+area as % of the tumor area, **p<0.05, one-way ANOVA, n = 4 mice per condition) 17 days after GL261 transplantation in mice treated with IgG-Rat or Ab-anti-IFN-γ, as indicated. Representative immunofluorescences are shown on the right. (f) Quantification of Ki67+ cells (mean ± s.e.m. of Ki67+ area as % of the tumor area, **p<0.05, one-way ANOVA, n = 4 mice per condition) 17 days after GL261 transplantation in mice treated with IgG-Rat or Ab-anti-IFN-γ, as indicated. Representative immunofluorescences are shown on the right. (g) RT-PCR of human pro- (Cxcl10, Nos2 and Il12a) and anti-inflammatory (Cd163, Mmp12 and Tgfb) genes in CD11b+ cells sorted from patient-derived GBM tissue, after tissue treatment with IFN-γ (20 ng/ml, 24 hr) or vehicle. Above: scheme of human GBM treatment. Data are the mean ± S.E.M., for Nos2 and Il12a *p=0.029, for Tgfb and mmp12**p=0.002, for cd163 p=<0.001 versus vehicle, Student’s t-test, n = 3–6.
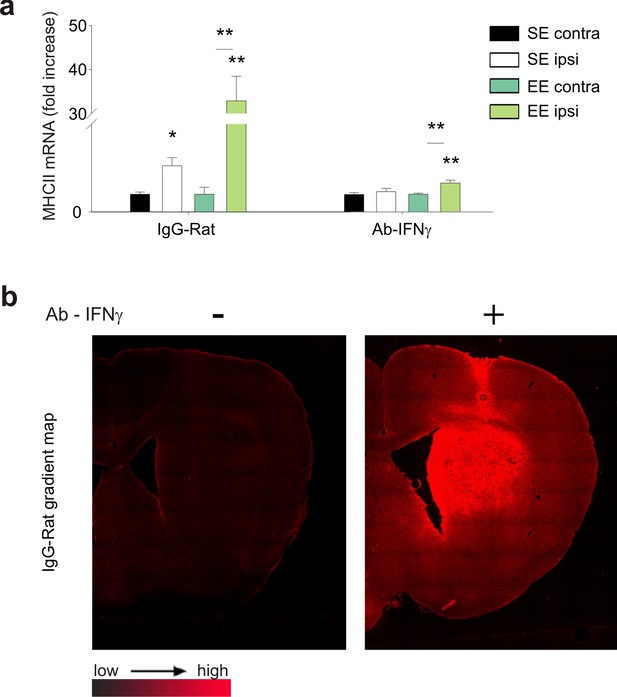
Control experiments to verify the efficacy of Ab-IFN-γ treatment.
(a) RT-PCR of mhcII mRNA expression in CD11b+ cells isolated from ILH and CLH cerebral hemispheres of Ab anti-IFN-γ-treated GL261-bearing mice, housed in SE or EE. Data are expressed as the mean ± S.E.M., *p<0.05 **p<0.01 versus CLH, one-way ANOVA, n = 4. (b) Representative immunofluorescence images of rat IgG in the ILH of vehicle- (left) or Ab-anti-IFN-γ-treated (right) GL261-bearing mice.
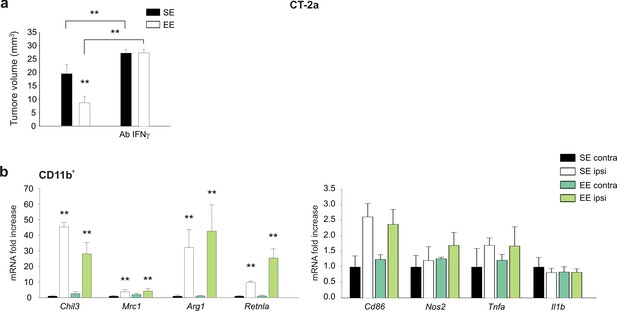
Effect of Ab-IFN-γ on tumor size and gene expression of myeloid cells in EE mice injected with CT-2A cells.
(a) Analysis of tumor volume (expressed as mm3 ± s.e.m.) in CT-2a-bearing mice treated with IgG (rat) or Ab-anti-IFN-γ, housed in SE or EE; n = 5; **p<0.01, two-way ANOVA). (b) RT-PCR of anti- and pro-inflammatory genes in CD11b+ cells sorted from ILH and CLH in vehicle- and Ab-anti-IFN-γ-treated CT-2a-bearing mice, housed in SE or EE. Above: scheme of Ab-anti-IFN-γ administration. Data are the mean ± S.E.M., **p<0.01 versus CLH, one-way ANOVA; n = 3.
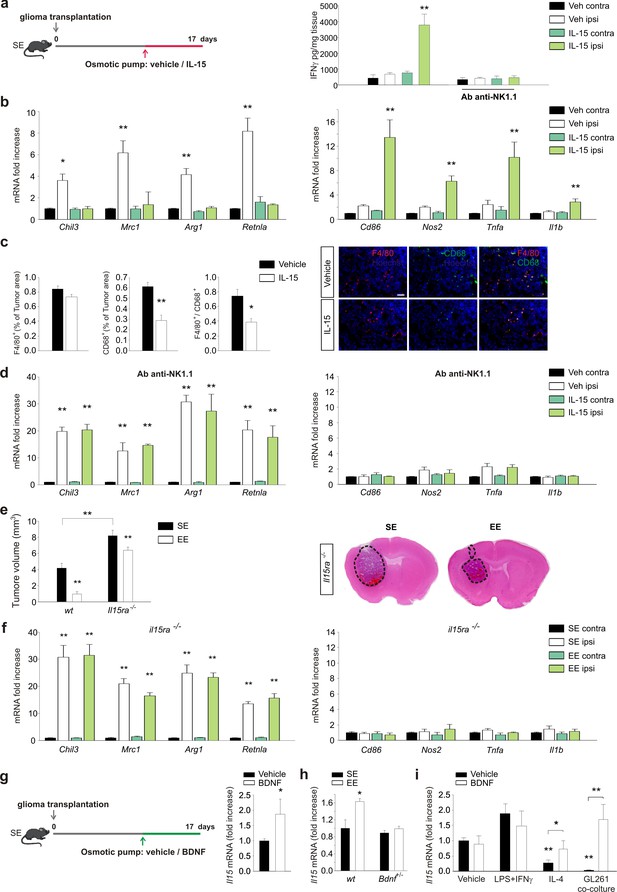
IL-15 is involved in the effects of EE on brain tumor.
(a) Left: mice housed in SE were infused for 7 days in the striatum, with vehicle or IL-15 through micro-osmotic pumps, starting 10 days after GL261 cell transplantation, as described in the scheme. Right: expression of IFN-γ in CLH and ILH of GL261-bearing mice treated with vehicle or Ab anti-NK1.1 as indicated, implanted with osmotic pumps releasing IL-15 or vehicle (n = 5, **p<0.01, one-way ANOVA). (b) RT-PCR of anti- (Chil3, Mrc1, Arg1, Retnla) and pro-inflammatory (Cd86, Nos2, Tnfa, Il1b) genes in CD11b+ cells sorted from ILH and CLH of GL261-bearing mice treated with IL-15 or vehicle. Data are expressed as mean ± S.E.M., *p<0.05 **p<0.01 versus CLH, one-way ANOVA; n = 4. (c) Myeloid cell activation (CD68) and infiltration (F4/80) in glioma mass in mice treated with IL-15 or vehicle, as shown in (a), analyzed at the end of treatment (17 days after glioma transplantation). Graph bars represent the mean (± S.E.M.) area expressed as percentage of total tumor area. Representative immunofluorescences are shown on the right (scale bar, 100 μm) (**p=0.002 *p=0.011 Student’s t-test; n = 4 mice per conditions). (d) RT-PCR of anti- and pro-inflammatory genes in CD11b+ cells isolated from ILH and CLH of GL261-bearing mice in vehicle and Ab-anti-NK1.1-treated mice, upon IL-15 or vehicle infusion, as shown in (a). Data are expressed as the mean ± S.E.M., **p<0.01 versus CLH, one-way ANOVA; n = 4. (e) Analysis of tumor volumes (expressed as mm3 ± S.E.M.) in wt or Il15ra–/– mice, housed in SE or EE; n = 5; **p<0.01, two-way ANOVA). Representative coronal sections are shown on the right. (f) RT-PCR of anti- and pro-inflammatory genes in CD11b+ cells sorted from ILH and CLH in Il15ra–/– mice, housed in SE or EE. Data are the means ± S.E.M., **p<0.01 versus CLH, one-way ANOVA; n = 4–5. (g) RT-PCR of Il15 mRNA in CD11b+ cells sorted from ILH and CLH of GL261-bearing mice treated with BDNF or vehicle. Data are the mean ± S.E.M., *p<0.05 versus CLH, one-way ANOVA; n = 5. Above: scheme of striatal infusion of vehicle or BDNF with micro-osmotic pumps starting 10 days after glioma cell transplantation and lasting 7 days, in SE mice. (h) RT-PCR of Il15 mRNA in CD11b+ cells isolated from ILH and CLH in wt or Bdnf+/– GL261 bearing mice, housed in SE or EE. Data are the mean ± S.E.M., *p<0.05 versus CLH, one-way ANOVA, n = 4. (i) RT-PCR of Il15 mRNA in primary mouse microglia stimulated with vehicle, LPS + IFNγ or IL-4 or in co-culture with GL261 for 24 h, in the presence or absence of BDNF. Data are expressed as the mean ± S.E.M., *p<0.05 **p<0.01, one-way ANOVA, n = 10.
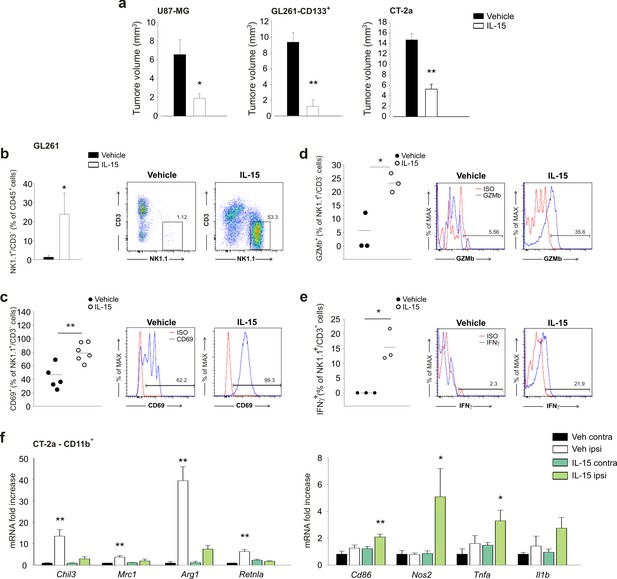
Effect of IL-15 treatment on tumor size, gene expression of myeloid cells and NK cell activation in EE mice injected with different glioma cells.
(a) Mean tumor volumes (expressed as mm3 ± s.e.m.), 17 days after implantation of human U87MG, purified CD133 +GL261, or murine CT-2a glioma cells into the striatum of SCID or C57BL/6 mice, upon IL-15 or vehicle infusion, as indicated; *p=0.026, **p<0.001, **p=0.003. Student’s t-test. n = 4–5. (b) CD3–/NK1.1+ cell frequency in the ILH upon IL-15 infusion (n = 6; *p=0.04 Student’s t-test). Representative FACS analysis is shown on the right. (c–e) Percentage of CD69+, GZMb+ and IFN-γ+ cells in the CD3–/NK1.1+ cell populations isolated from the brain of vehicle- or IL-15-treated GL261-bearing mice (n = 3–6; **p=0.003, *p=0.015, *p=0.013 Student’s t-test). Representative FACS analyses are shown on the right. (f) RT-PCR analysis of anti- (Chil3, Mrc1, Arg1, Retnla) and pro-inflammatory (Cd86, Nos2, Tnfa, Il1b) genes in CD11b+ cells sorted from ILH and CLH of CT-2a-bearing mice treated with IL-15 or vehicle. Data are expressed as mean ± S.E.M., *p<0.05 **p<0.01 versus CLH, one-way ANOVA; n = 4.
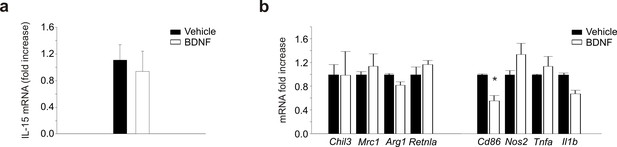
Gene expression in glioma cells and myeloid cells treated with BDNF.
(a) RT-PCR of il-15 mRNA in GL261 cells treated with BDNF or vehicle for 24 hr. (b) RT-PCR of anti- (Chil3, Mrc1, Arg1, Retnla) and pro-inflammatory (Cd86, Nos2, Tnfa, Il1b) genes in primary mouse microglia treated with BDNF (50 ng/ml, 24 hr) or vehicle. Data are the mean ± S.E.M., *p<0.05 versus vehicle, one-way ANOVA; n = 4.
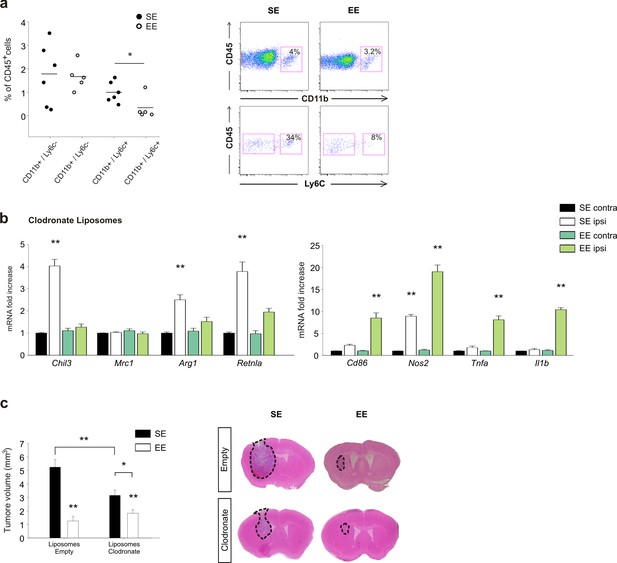
Effects of EE on mice treated with clodronate liposomes.
(a) Percentage of CD11b+ and Ly6c+ cells in the total CD45+ cell population obtained from the brain of EE or SE GL261-bearing mice (n = 5–6; *p=0.012, Student’s t-test). Representative FACS analyses are shown below. (b) RT-PCR of anti- and pro-inflammatory genes in CD11b+ cells sorted from ILH and CLH in empty or clodronate-filled liposome-treated mice, housed in SE or EE. Data are the mean ± S.E.M., **p<0.01 versus CLH by one-way ANOVA; n = 4. (c) Analysis of tumor volume (expressed as mm3 ± S.E.M.) in liposome filled with clodronate or empty liposomes-treated GL261-bearing mice, housed in SE or EE; n = 5; *p<0.05 **p<0.01, two-way ANOVA). Representative coronal section are shown on the right.
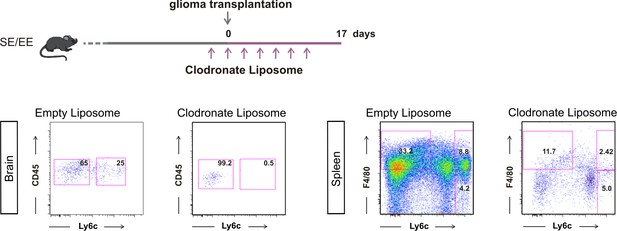
Representative FACS analysis to verify CD45+/Ly6c + cell depletion in the brains of GL261-bearing mice housed in EE or SE, upon clodronate liposome treatment; and F4/80+/Ly6c + cells in the spleen.
On top, scheme of liposome administration.
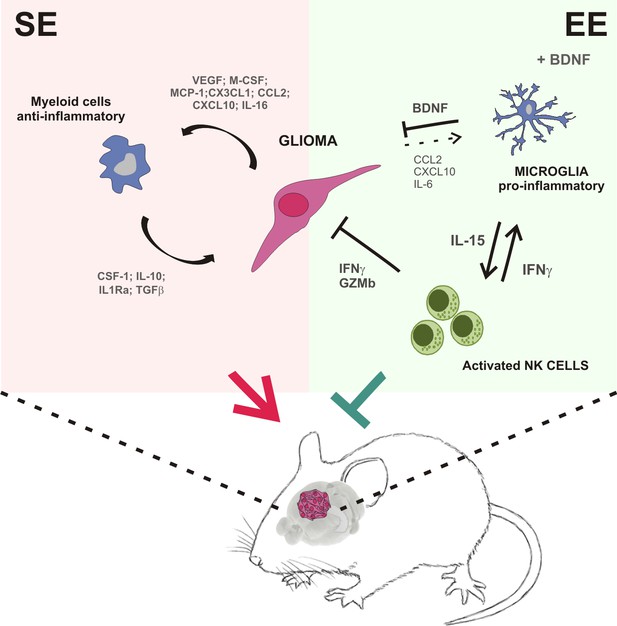
Summary of the events activated in EE-housed mice with glioma.
https://doi.org/10.7554/eLife.33415.016Tables
Housing conditions in enriched environment (EE).
https://doi.org/10.7554/eLife.33415.017| Variables | Value |
|---|---|
| EE cage size (cm) | 36 x54 x19 |
| EE cage composition | polycarbonate |
| Control cage | polycarbonate |
| EE floor space/mouse (cm 2) | 195 |
| # Mice/EE cage | 10 |
| Stimulating toys/objects in EE cage | 2 running wheels, tunnels, 2 refuges 1 swing, with nesting material |
| Objects varied regularly? | Yes |
| Strain of mice | C57BL/6 |
| Sex of mice | Male |
| EE, control cages in same room? | Yes |
| Lighting | 12 hours on/off |
| Temp (degrees C) | 22 ± 1 |
| Bedding | Sawdust |
| Humidity control? | No |
| Cleaning schedule | Once a week |
| Food based on wheat, oats, meat, soy and milk? | Yes (14% protein, 5% fat, 3041 kcal ME/kg) |
| Microbiota endemic in animal facility | Norovirus, Helicobacter |
| Age of mice put in cage initially | 3 weeks |
| # weeks habituation | 5 weeks |
| Tumor injected | GL261 |
| Route injected | intrastriatal |
| # Cells injected | 7.5×104 |
| Mouse handling frequency | Every 3 days |
| Statistical significance in tumor size? | Yes |
Passive properties of patched microglia: membrane capacitance (Cm), resting membrane potential (RP) and membrane resistance (Rm) were measured as described in the methods.
https://doi.org/10.7554/eLife.33415.018| Cm (pF) | Rm (MΩ) | RP (mV) | N | |
|---|---|---|---|---|
| Tum SE | 26 ± 2 | 1.5 ± 0.1 | –45 ± 3 | 57 |
| Tum EE | 26 ± 1 | 2.1 ± 0.2 | –46 ± 2 | 64 |
| Peri SE | 27 ± 1 | 1.9 ± 0.2 | –43 ± 4 | 60 |
| Peri EE | 25 ± 1 | 2.2 ± 0.2 | –49 ± 3 | 57 |
| CLH SE | 18 ± 3 | 2.7 ± 0.4 | –53 ± 3 | 38 |
| CLH EE | 20 ± 1 | 2.4 ± 0.3 | –47 ± 4 | 27 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.33415.019





