A robust and tunable mitotic oscillator in artificial cells
Figures
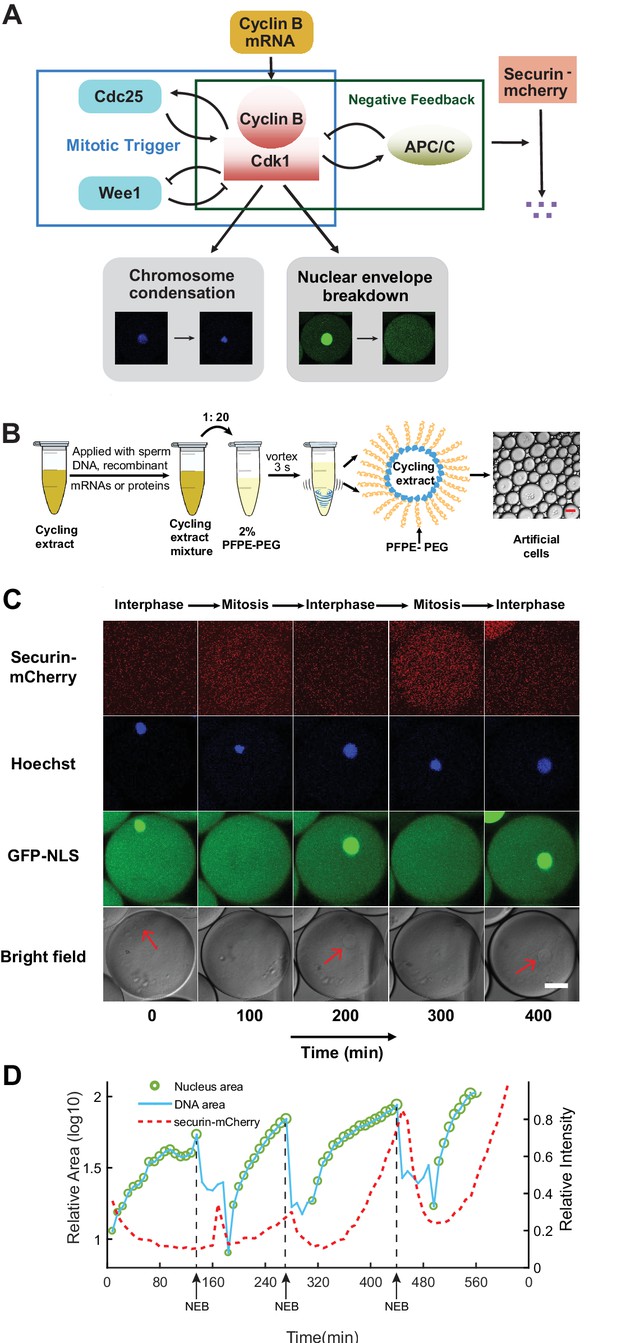
Reconstitution of an in vitro cell cycle clock and downstream mitotic events.
(A) Schematic view of a cell cycle oscillator that consists of coupled positive and negative feedback loops. The central regulator, cyclin B-Cdk1 complex activates its own activator, phosphatase Cdc25, forming a positive feedback loop, and inhibits its own inhibitor, kinase Wee1, forming a double negative feedback loop. Additionally, cyclinB-Cdk1 activates the E3 ubiquitin ligase APC/C, which targets cyclin B for degradation and completes a core negative feedback loop. Active APC/C also promotes the degradation of another substrate securin. Once cyclinB1-Cdk1 complex is activated, the circuit drives a set of mitotic events including chromosome condensation and nuclear envelope breakdown (NEB). (B) Experimental procedures. Cycling Xenopus extracts are supplemented with various combinations of recombinant proteins, mRNAs, and de-membraned sperm DNAs, which are encapsulated in 2% Perfluoropolyether-poly (ethylene glycol) (PFPE-PEG) oil microemulsions. Scale bar is 100 µm. (C) Snapshots of a droplet were taken periodically both in fluorescence channels (top three rows) and bright-field (the last row). The cyclic progression of the cell cycle clock and its downstream mitotic processes are simultaneously tracked by multiple fluorescence reporters. The clock regulator APC/C activity is reported by its substrate securin-mCherry, chromosomal morphology changes by the Hoechst stains, and NEB by GFP-NLS. Nuclear envelopes (red arrows) are also detectable on bright field images, matching the localization of GFP-NLS indicated nuclei. Scale bar is 30 µm. (D) Multi-channel measurements for the droplet in Figure 1C. The nucleus area (green circle) is calculated from the area of the nuclear envelope indicated by GFP-NLS, noting that the areas of the green circles are also scaled with the real areas calculated for the nuclei. DNA area curve (blue line) shows the chromosome area identified by Hoechst 33342 dye. Chromosome condensation happens almost at the same time as the nuclear envelope breaks down (black dashed line). The red dashed line represents the intensity of securin-mCherry over time, suggesting that degradation of the APC/C substrate lags behind NEB consistently at each cycle.
Reconstitution of cell cycle clock and mitotic events.
This movie corresponds to Figure 1A. Green fluorescence channel shows alternations of nuclear envelope breakdown and reformation indicated by GFP-NLS protein. Green circles disappear when nuclear envelopes break down and reappear when nuclei assemble again. Blue channel (Hoechst) shows chromosome morphology changes over time. Securin-mCherry protein oscillations are shown in the red channel. The last channel is bright field, from which we can see nuclear envelope. Scale bar is 50 µm. The time stamp gives the real time in hour:minute format. The movie is shown at a rate of 10 frames per sec.
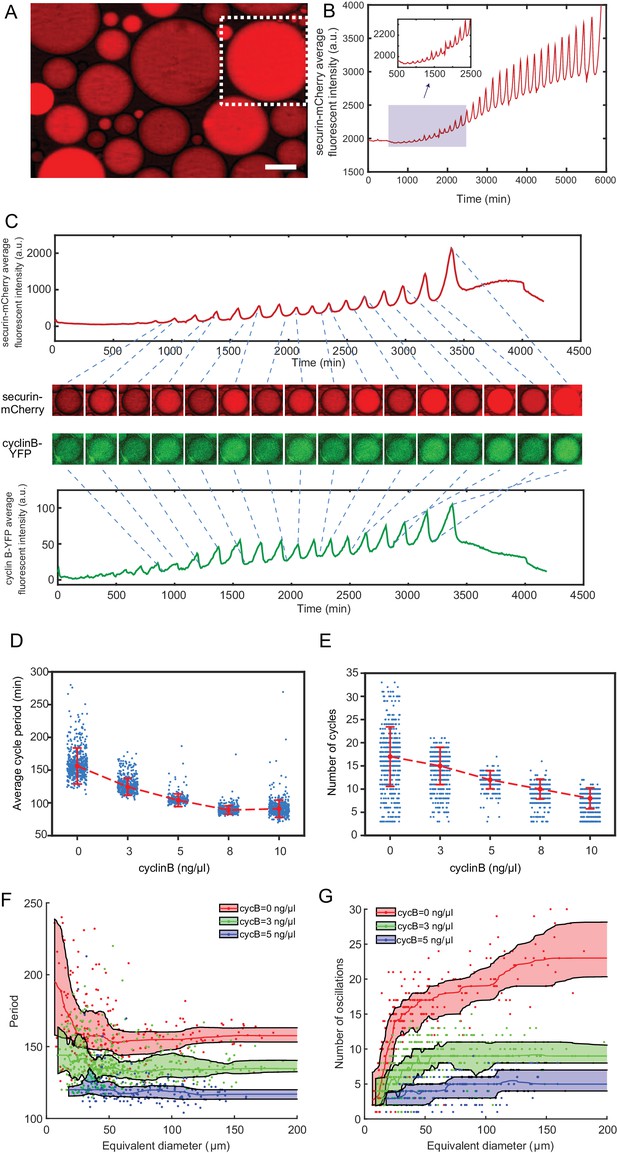
The minimal cell cycle oscillator is robust and tunable.
(A) Fluorescence image of securin-mCherry, a reporter for the cell cycle oscillator, in micro-emulsion droplets (scale bar, 100 µm). One example droplet (inside the white dotted framed square) is selected for time course analysis in Figure 2B. (B) The time course of securin-mCherry fluorescence intensity of the selected droplet from Figure 2A, indicating 32 undamped oscillations over a course of 100 hr. (C) Simultaneous measurements of fluorescence intensities of securin-mCherry (upper panel) and cyclin B-YFP (lower panel), showing sustained oscillations for about 58 hr. The mRNA concentrations of securin-mCherry and cyclin B-YFP are 10 ng/µL and 1 ng/µL. The series of mCherry and YFP images correspond to selected peaks and troughs in the time courses of fluorescence intensities. The two channels have coincident peaks and troughs for all cycles, suggesting that they both are reliable reporters for the cell cycle oscillator. (D, E) The oscillator is tunable in frequency (D) and number of cycles (E) as a function of the concentration of cyclin B mRNAs. Cyclin B not only functions as a substrate of APC/C but also binds to Cdk1 for its activation, functioning as an ‘input’ of the clock. In Figure 2D, the cell cycle periods are shortened by increasing the mRNA concentrations. In Figure 2E, the number of total cell cycles is reduced in response to increasing cyclin B mRNA concentrations. Each data point represents a single droplet that was collected from one of the loading replicates (see Materials and methods 7). Red dashed line connects medians at different conditions. Error bar indicates median absolute deviation (MAD). (F, G) Droplets with smaller diameters have larger periods on average and a wider distribution of periods (F), and exhibit smaller number of oscillations on average (G). Colored areas represent moving 25 percentiles to 75 percentiles and is smoothed using the LOWESS smoothing method (see Materials and methods 7). The equivalent diameter is defined as the diameter of a sphere that has an equal volume to that of a droplet, estimated by a volume formula in literature (Good et al., 2013). Note that these size effects are smaller for droplets with higher cyclin B mRNA concentrations.
-
Figure 2—source data 1
Source data for generating Figure 2D-E.
- https://doi.org/10.7554/eLife.33549.005
-
Figure 2—source data 2
Source data for generating Figure 2F-G.
- https://doi.org/10.7554/eLife.33549.006
Free-running in vitro cell cycles detected by securin-mCherry reporter.
This video is from the experiment with no demembranated sperm chromatin added. No nuclei are reconstituted in this experiment. An example time course is shown in Figure 2B. Securin-mCherry proteins undergo multiple oscillations of synthesis and degradation, and the extract activity lasts for days. The scale bar is 100 µm and the movie is shown at a rate of 25 frames per sec.
Tuning of the clock speed.
This video is related to Figure 2C. The clock period can be tuned by the level of its input signal, cyclin B mRNAs. The droplets shown in the movie are applied with 3 ng/µL of cyclin B1-YFP mRNAs. The YFP channel on the top shows oscillations from cyclin B1-YFP, the middle channel from securin-mCherry, and the bottom channel for the bright field. The scale bar is 100 µm and the movie is shown at a rate of 20 frames per sec.
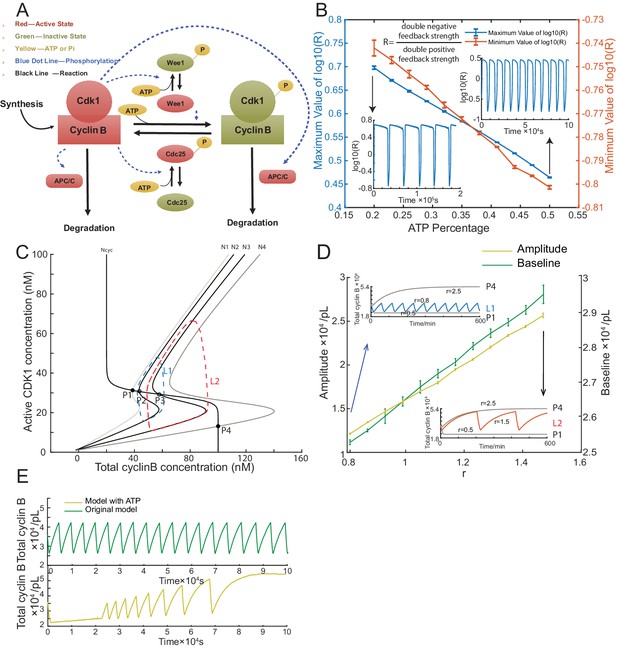
Simulation of cell cycle model with energy depletion.
(A) Schematic view of the cyclin B-Cdk1 oscillation system. Note that ATP is taken into consideration. Activated molecules are marked in red, inactivated molecules in green and ATP or Pi in yellow. Black line indicates a reaction and blue dotted line a phosphorylation. (B) Relationship between ATP percentage and R value, showing that decreasing the ATP concentration leads to a higher R value. Error bars represent ranges from three simulations. Two inserts represent the dynamics of R value over time when the ATP percentage [ATP]/([ATP]+[ADP]) is set as 0.2 (left) and 0.5 (right). The model is simulated using Gillespie algorithm. (C) Phase plots of the two-ODE model. Parameters for the cyclin B nullcline (Ncyc) (Yang and Ferrell, 2013) and the Cdk1 nullclines with a variety of values of r (N1, r = 0.5; N2, r = 0.8; N3, r = 1.5; N4, r = 2.5) were chosen based on previous experimental work (Pomerening et al., 2003; Sha et al., 2003). Note that the r here is a parameter and is different from R in Figure 3B. Two sample traces of limit cycle oscillations are plotted for r = 0.8 (L1) and r = 1.5 (L2), showing that a larger r value leads to a higher amplitude and baseline. In addition, r = 0.5 (N1) generates a low stable steady-state of cyclin B (P1), while r = 2.5 (N4) a high stable steady-state of cyclin B (P4). These stable steady-states are indicated by the intersections of the nullclines (filled circles). The unstable steady states are labeled with open circles (P2 and P3). (D) Relationship between the oscillation baseline and amplitude values and ATP concentration (positively correlated with r). Error bars indicate the ranges of 3 replicates. Inserts show two example time courses of total cyclin B with different r values (L1, r = 0.8; L2, r = 1.5), colors of which match the ones in Figure 3C. Simulation is done using Gillespie algorithm. (E) Time series of total cyclin B molecules from the model without ATP (top panel, green line) and with ATP (bottom panel, yellow line).
-
Figure 3—source data 1
Simulated data for generating Figure 3—figure supplement 1 using the energy depletion model.
- https://doi.org/10.7554/eLife.33549.011
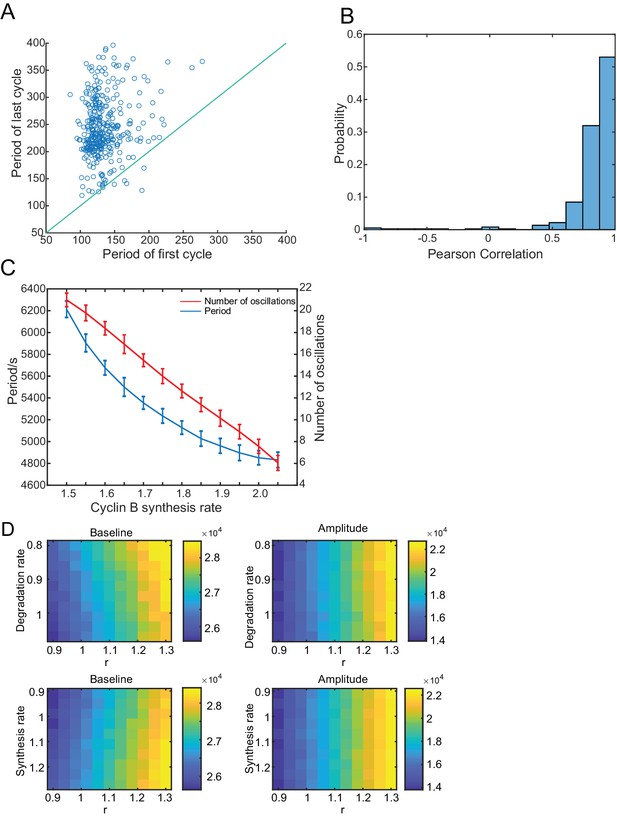
Analysis of the dependence of oscillations on the cyclin B synthesis and degradation rates.
(A) The period of the first cycle vs the last cycle in a droplet, showing that the period of the last cycle tends to be longer. Blue line indicates the same first and last periods. (B) Pearson correlation between the period of cycle and the index of cycle in a droplet. Positive correlation indicates that the cell cycle period tends to increase over time. (C) The period and number of oscillations decrease with an increasing cyclin B synthesis rate. Error bars represent standard deviations from 50 simulations. (D) Effects of synthesis and degradation rates of cyclin B as well as r on the baseline and amplitude of oscillations of total cyclin B. Color bar indicates baseline or amplitude of cyclin B (number of molecules per pL).
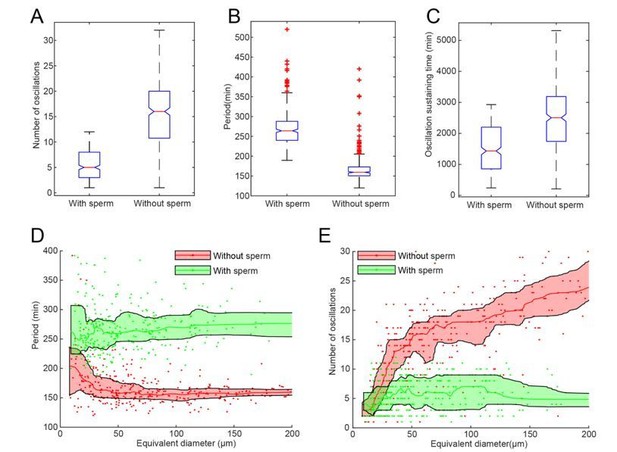
A-C Statistics of droplets with sperm DNA (N=320) and without sperm DNA (N=373).
All droplets were prepared in the same day from the same batch of frog eggs. The figures show that adding sperm DNA may cause significant difference in number of oscillations (A), period (B) and oscillation sustaining time (C). D-E. Droplet-size dependence of period (D) and number of oscillations (E) with nucleus effect, showing that adding sperm DNAs has a more significant impact on larger droplets.
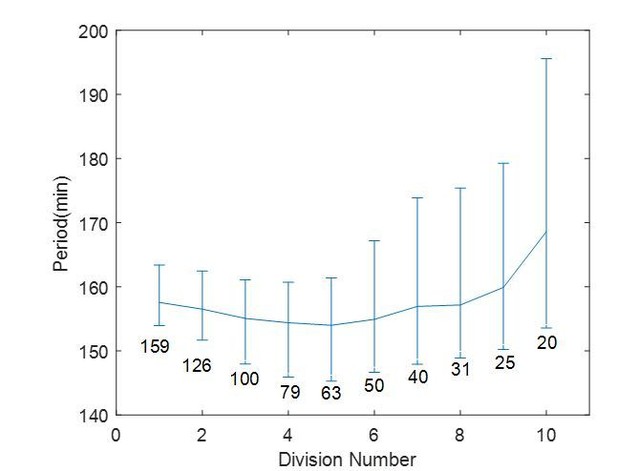
A thought experiment of a droplet of 200 µm in diameter undergoing ten divisions.
The cell cycle period is calculated for each division, with each corresponding diameter labeled on the figure. The error bar is showing 25th and 75th percentile as in Figure 2F in the main text.
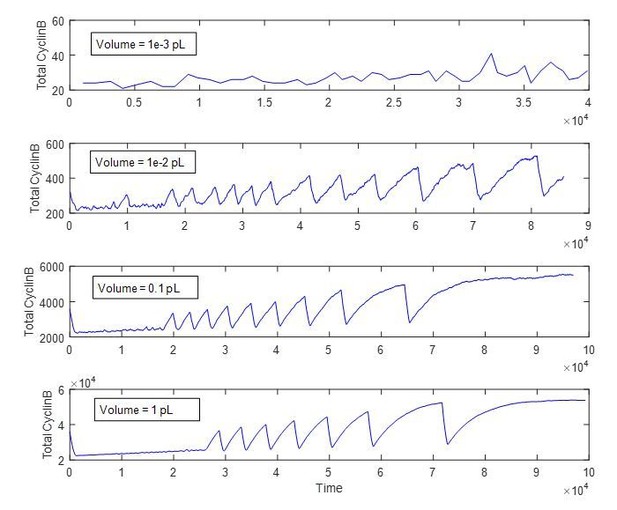
Reproduction of Figure 3E bottom panel from the main text with different reaction volumes.
https://doi.org/10.7554/eLife.33549.022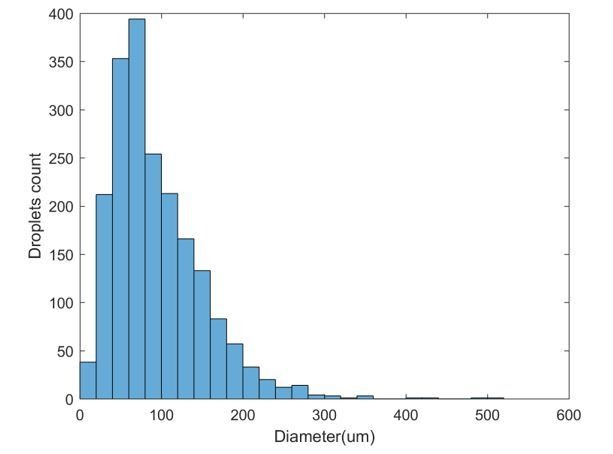
Size distribution for 1997 droplets obtained using a specific vortexing speed and duration.
https://doi.org/10.7554/eLife.33549.023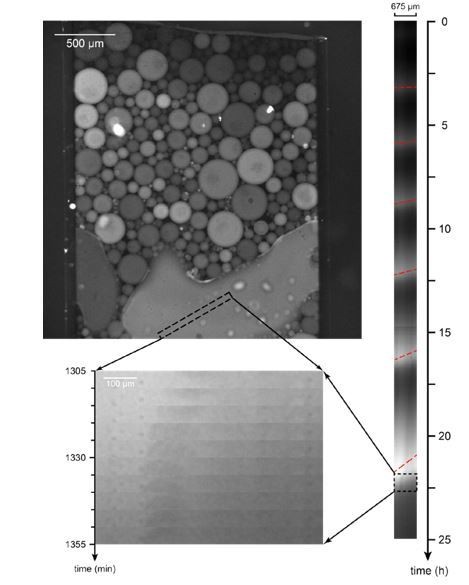
Mitotic trigger waves in a large droplet.
https://doi.org/10.7554/eLife.33549.024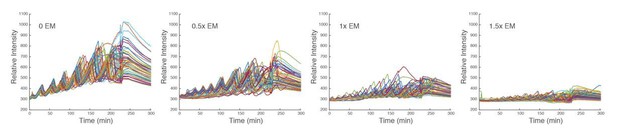
Time traces of individual droplets with different concentrations of energy mix.
Each condition has two duplicate experiments and only one is shown. It is evident that the amplitude, baseline, and period decrease as the level of energy mix increases.
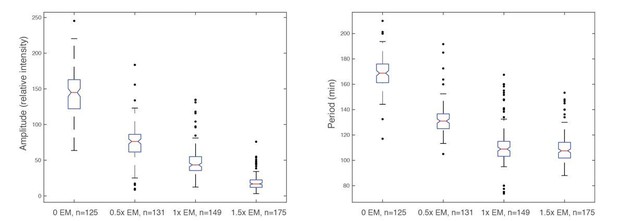
Amplitude (left panel) and period (right panel) decrease as energy mix concentration increases.
https://doi.org/10.7554/eLife.33549.026Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Biological sample (Xenopus laevis) | Xenopus eggs | Xenopus-I Inc. | RRID:NXR_0.0080 | |
| recombinant DNA reagent | securin-mCherry | this paper | Progentiors: PCR; Gateway vector pMTB2 | |
| recombinant DNA reagent | cyclin B1-YFP | this paper | ||
| recombinant DNA reagent | GST-GFP-NLS | PMID: 23863935 | Dr. James Ferrell's lab | |
| peptide, recombinant protein | GFP-NLS protein | this paper | BL21 (DE3)-T-1 competent cells | |
| commercial assay or kit | QIAprep spin miniprep kit | QIAGEN | Cat No.: 27104 | |
| commercial assay or kit | mMESSAGE mMACHINE SP6 Transcription Kit | Ambion | Cat No.: AM1340 | |
| Strain, strain background | BL21 (DE3)-T-1 competent celss | Sigma-Aldrich | Cat No.: B2935 | |
| chemical compound, drug | calcium ionophore | Sigma-Aldrich | Cat No.: A23187 | |
| chemical compound, drug | Hoechst 33342 | Sigma-Aldrich | Cat No.: B2261 | |
| chemical compound, drug | Trichloro (1H,1H,2H,2H- perfluorooctyl) silane | Sigma-Aldrich | Cat No.: 448931 | |
| chemical compound, drug | PFPE-PEG surfactant | Ran Biotechnologies | 008-FluoroSurfactant- 2wtH-50G | |
| other | GE Healthcare Glutathione Sepharose 4B beads | Sigma-Aldrich | Cat No.: GE17-0756-01 | |
| other | PD-10 column | Sigma-Aldrich | Cat No.: GE17-0851-01 | |
| other | VitroCom miniature hollow glass tubing | VitroCom | Cat No.: 5012 |
Reaction rates and stoichiometry of the two-ODE model.
https://doi.org/10.7554/eLife.33549.012| Reaction | Rate | Stoichiometry |
|---|---|---|
| Active Cdk1 Synthesis | ||
| Active Cdk1 to Inactive Cdk1 | ||
| Inactive Cdk1 to Active Cdk1 | ||
| Active Cdk1 Degradation | ||
| Inactive Cdk1 Degradation |
Reaction rates in the model considering ATP.
https://doi.org/10.7554/eLife.33549.013| Reaction | Rate |
|---|---|
| Active Cdk1 Synthesis | |
| Active Cdk1 to Inactive Cdk1 | |
| Inactive Cdk1 to Active Cdk1 | |
| Active Cdk1 Degradation | |
| Inactive Cdk1 Degradation |
Additional files
-
Source code 1
Source code to generate Figure 1D based on raw image file.
- https://doi.org/10.7554/eLife.33549.014
-
Source code 2
Source code to generate Figure 2D–E and Figure 2F–G using Figure 2—source data 1 and Figure 2—source data 2 respectively.
- https://doi.org/10.7554/eLife.33549.015
-
Source code 3
Source code to generate Figure 3B–E.
- https://doi.org/10.7554/eLife.33549.016
-
Source code 4
Source code to generate Figure 3—figure supplement 1A–B using Figure 3—source data 1.
- https://doi.org/10.7554/eLife.33549.017
-
Transparent reporting form
- https://doi.org/10.7554/eLife.33549.018





