Hyperactivation of ERK by multiple mechanisms is toxic to RTK-RAS mutation-driven lung adenocarcinoma cells
Figures
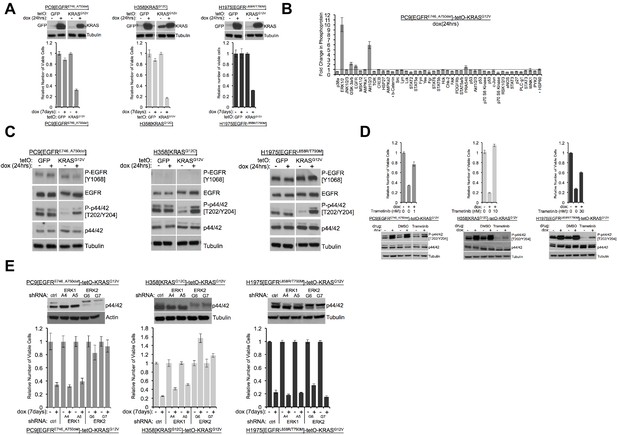
Induction of mutant KRAS reduces the numbers of viable lung cancer cells harboring KRAS or EGFR mutations, and the effects can be rescued by inhibiting ERK (A) Reduced numbers of viable LUAD cells after activation of KRAS.
Production of GFP or KRASG12V was induced by addition of 100 ng/mL dox in the indicated three cell lines as described in Methods. GFP and KRAS protein levels were measured by Western blotting 24 hr later. (top); tubulin served as a loading control. The numbers of viable cells, normalized to cells grown in the absence of dox (set to 1.0), were determined by measuring with Alamar blue six days later. Error bars represent standard deviations based on three replicates. (B) Induction of KRASG12V uniquely increases phosphorylation of ERK1/2 among several phosphoproteins. PC9-tetO-KRAS cells were treated with dox for 24 hr and cell lysates incubated on an array to detect phosphorylated proteins. Fold changes of phosphorylation compared with lysates from untreated cells (set to 1.0, dotted line) to treated cells is presented from a single antibody array. Error bars are derived from duplicate spots on antibody array. The detection of HSP60 and ß-catenin are of total protein, not phosphoprotein. (C) Phosphorylation of ERK occurs early after induction of mutant KRAS. Lysates prepared as described for panel (A) were probed for the indicated proteins by western blot. Loading control is the same as in A. (D) Drug-mediated inhibition of the MEK1/2 kinases ameliorates KRAS-induced loss of viable cells. Mutant KRAS was induced with dox in the three indicated cell lines in the absence and presence of trametinib at the indicated dose for 7 days. The relative number of viable cells was measured with Alamar blue. Error bars represent standard deviations determined from three samples grown under each set of conditions. Values are normalized to measurements of cells that received neither dox nor trametinib (bottom). Cells were treated with dox and with or without trametinib for 24 hr at the dose conferring rescue of numbers of viable cells. Lysates were probed for indicated proteins to confirm inhibition of MEK. (E) Reduction of ERK proteins with inhibitory small hairpin (sh) RNAs protects cells from loss of viability in response to induction of mutant KRAS. LUAD cell lines, transduced with the indicated shRNA targeted against ERK1 or ERK2, were assessed for levels of ERK proteins, p42 and p44, by Western blotting (top panels). The same lines were treated with dox for 7 days and the number of viable cells measured with Alamar blue. Values are normalized to numbers of viable cells of each type grown in the absence of dox (1.0), with error bars representing standard deviations among three replicates. Similar results were obtained from 2 or 3 independent experiments.
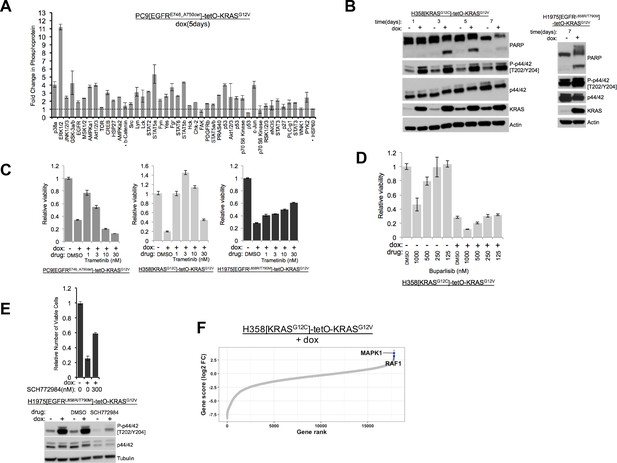
Letality induced by mutant KRAS induction is rescued by supression of ERK.
(A) Multiple proteins are phosphorylated after prolonged induction of mutant KRAS. Lysates from PC9-tetO-KRAS cells treated or not treated with dox for 5 days were incubated on an array to detect changes in phosphorylation of 43 proteins. HSP60 and β-catenin signals represent total protein content, not phosphoprotein. Fold-changes in dox-treated cells (compared with lysates from untreated cells [set to 1.0, dotted line] are shown from a single antibody array, with error bars derived from duplicate spots on the array. (B) Induction of KRASG12V increases phospho-ERK1/2 and cleaved PARP. Mutant KRAS was induced with doxycycline in cell lines and protein abundance measured as indicated over the course of 7 days (H358-based cell line) or at 7 days (H1975-based cell line). Results are representative of 2 independent experiments. (C) Trametinib-mediated rescue of mutant KRAS-induced toxicity. Extension of Figure 1D including a dose response of trametinib plus doxycycline. (D) PI3K inhibitor (buparlisib) fails to rescue mutant KRAS-induced toxicity in H358-tetO-KRAS cells. A dose response of buparlisib alone or in combination with doxycycline is shown for cells cultured for 7 days. The relative number of viable cells was measured with Alamar blue. Error bars represent standard deviations from three wells. Values were normalized to cells treated with only DMSO. (E) Drug-mediated inhibition of both ERK1 and ERK2 can rescue viability in H1975-tetO-KRAS cells. Cells were treated with SCH772984, an ERK inhibitor, and dox for 7 days. The relative number of viable cells was measured with Alamar blue. Error bars represent standard deviations determined from three samples grown under each set of conditions. Values were normalized to measurements of cells that did not receive either dox or SCH772984. H1975-tetO-KRAS cells were also treated with dox and SCH772984 for 24 hr at the dose (300 nM) conferring significant rescue of viable cells. Lysates were probed for indicated proteins to confirm inhibition of P-ERK1/2. (F) Genome wide CRISPR-Cas9 screen in H358-tetO-KRAS cells (grown in doxycycline) reveals a dependence on ERK2 (MAP3K1). The change in guide RNA abundance is shown. The positions of ERK2 and RAF1 sgRNA are highlighted, indicating that cells in which those genes are inactivated are enriched in the presence of doxycycline.
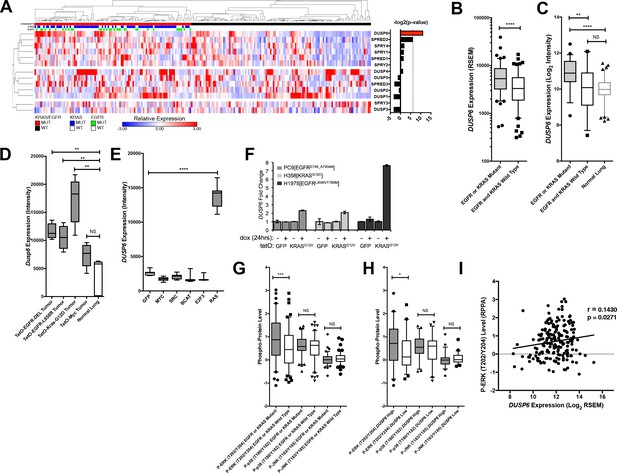
DUSP6 is the only negative feedback regulator significantly up-regulated in LUAD tumors with KRAS or EGFR mutations.
(A) Negative feedback regulators differentially expressed between clinical LUADs with or without EGFR or KRAS mutations (as indicated in green or blue, respectively, in the third and second horizontal bars). Expression levels for the indicated genes as determined by RNA-seq were compared between LUAD tumors with (n = 107, red) and without (n = 123, black) KRAS or EGFR mutations. In the heatmap, red indicates high relative expression and blue, low expression. Significance, as determined by two-tailed unpaired t-test with Bonferroni multiple testing correction, is indicated as the –log2(p-value). The significance threshold was set at a p-value < 0.01 and is indicated by the dotted line. Only DUSP6 surpassed this threshold. (B) DUSP6 is the main negative feedback regulator upregulated in LUADs with EGFR or KRAS mutations. Box plots show levels of DUSP6 RNA from samples in A. LUADs with EGFR or KRAS mutations (n = 107) express DUSP6 at higher levels than do LUADs with wildtype KRAS and EGFR (n = 123) in the TCGA dataset. (C) Validation of increased DUSP6 expression in LUADs with mutated KRAS or EGFR. In an independent internal dataset from the BCCA, LUADs with EGFR or KRAS mutations (n = 54) demonstrated higher expression of DUSP6 compared to LUADs in which both EGFR and KRAS were wild-type (n = 29) and to normal lung tissues (n = 83). (D) Dusp6 is upregulated in the lungs of mice with tumors induced by mutant EGFR or Kras transgenes. Tumor-bearing lung tissues from mice expressing EGFR or Kras oncogenes produce higher levels of Dusp6 RNA than do normal lung controls or tumor-bearing lungs from mice with a MYC transgene. (E) Increased DUSP6 RNA is specific to cells with oncogenic signaling through RAS. Human primary epithelial cells expressing a HRAS oncogene (n = 10 biological replicates) express DUSP6 at higher levels than control cells producing GFP (n = 10 biological replicates) whereas cells expressing known oncogenes other than RAS genes (MYC, SRC, B-Catenin, and E2F-3) do not. (F) DUSP6 RNA levels increase in PC9, H358 and H1975 cells expressing mutant KRAS. Dox was added to induce either GFP or the KRASG12V oncogene for 24 hr; DUSP6 RNA was measured by qPCR. (G–I) DUSP6 expression is associated with P-ERK levels. (G) LUADs with EGFR or KRAS mutations (n = 107) have higher P-ERK levels, but not P-p38 or P-JNK levels, than LUADs with wildtype KRAS and EGFR (n = 123) in the TCGA dataset. (H) LUADs with the highest DUSP6 RNA levels (n = 46) demonstrated higher P-ERK levels, but not P-p38 or P-JNK levels, than LUADs with the lowest DUSP6 RNA levels (n = 46). (I) DUSP6 RNA levels correlate with the levels of P-ERK in LUADs (n = 182). Pearson correlation coefficient (r) and p-value are indicated. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, NS = Not Significant.
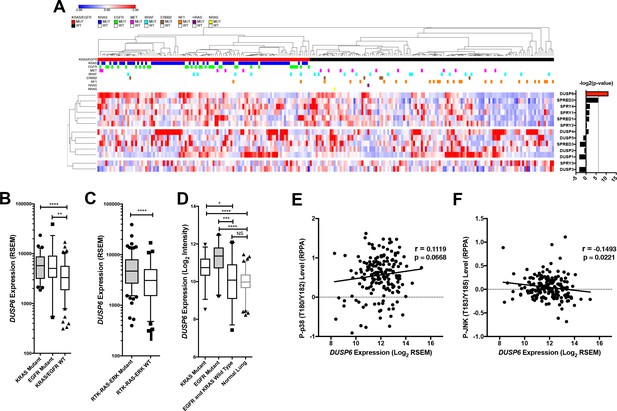
Negative feedback regulators are differentially expressed in clinical LUADs with or without EGFR or KRAS mutations.
(A) The heat map shown in Figure 2A is displayed with information about mutations affecting other genes encoding members of the RTK-RAS-ERK pathway; a color key is included at the top. (B) Box plots of levels of DUSP6 RNA from tumors represented in panel A. The data are from the same tumors analyzed in Figure 2B, but the LUADs with EGFR (n = 33) and KRAS (n = 75) mutations are plotted separately. Both groups have higher DUSP6 RNA levels than do LUADs with wildtype KRAS and EGFR (n = 123) in the TCGA dataset. (C) TCGA dataset with DUSP6 RNA levels in samples with RTK-RAS-ERK pathway mutations (EGFR, KRAS, BRAF, MET, ERBB2, NF1, NRAS, HRAS, n = 162) compared to those without (n = 68). (D) Box plots of DUSP6 RNA levels from British Columbia Cancer Agency (BCCA) LUADs as in Figure 2C, but with LUADs plotted separately as EGFR (n = 20) and KRAS (n = 34) mutants. Both groups demonstrated greater expression of DUSP6 than did LUADs in which both EGFR and KRAS were wild-type (n = 29) or normal lung tissues (n = 83). (E-F) Expression of DUSP6 is not positively correlated with levels of P-p38 or P-JNK in LUADs (n = 182). Pearson correlation coefficient (r) and p-value are indicated. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, NS = Not Significant.
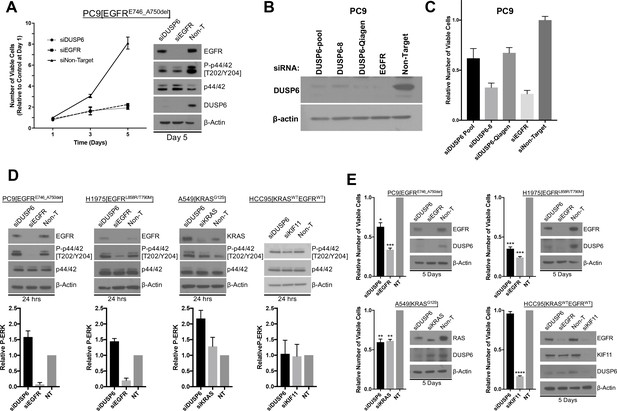
Knockdown of DUSP6 increases P-ERK and selectively inhibits LUAD cell lines with KRAS or EGFR mutations.
(A) Interference with DUSP6 RNA induces toxicity in PC9 cells. Pooled siRNAs for DUSP6, EGFR or a non-gene targeting control (Non-T) were transfected into PC9 cells (carrying an EGFR mutation) on day 0 and day 3, and the numbers of viable cells in each condition was measured with Alamar blue at the indicated time points and scaled to the Non-T condition at day 1 to measure the relative changes in numbers of viable cells. Experiments were done in biological triplicate with the average values presented ±SEM. Western blots were performed at the endpoint of the assay (day 5) to confirm reduced amounts of DUSP6 protein and measure levels of ERK and P-ERK (p42/44 and P-p42/44, respectively). (B–C) A siRNA that targeted the 5’ region of DUSP6 mRNA coding sequence (siDUSP6-Qiagen; different from siDUSP6-8 that targets the 3’ mRNA coding region), reduces levels of DUSP6 protein and decreases the numbers of viable cells. The indicated siRNAs (DUSP6-pool, DUSP6-8, DUSP6-Qiagen, EGFR and Non-Target) were delivered to PC9 cells, the levels of DUSP6 protein measured and the numbers of viable cells was determined as described for panel A. Experiments were done at least three times, and the average ±SEM is indicated for cell viability. (D) Interference with DUSP6 RNA acutely increases P-ERK levels. DUSP6 was knocked down in PC9 and H1975 cells (EGFR mutants), A549 cells (KRAS mutant), and HCC95 cells (KRAS and EGFR wild-type); levels of ERK and P-ERK were measured by Western blot 24 hr later. Relative P-ERK levels (ratio of phosphorylated to total levels normalized to actin) were determined by dosimetry and compared to the non-targeting control (NT) to quantify the relative increase after DUSP6 knockdown. Three independent western blots were performed and the average ±SEM is plotted. (E) Interference with DUSP6 RNA inhibits LUAD cell lines with activating mutations in genes encoding components of the EGFR/KRAS signaling pathway. Numbers of viable cells 5 days after knockdown of DUSP6 or knockdown of positive controls (EGFR, KRAS or KIF11) were assessed with Alamar blue and compared to the non-targeting controls to determine relative changes. Experiments were done in biological triplicate with the average values presented ±SEM. Western blots to monitor knockdown of target genes at Day 5 are also displayed. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, NS = Not Significant.
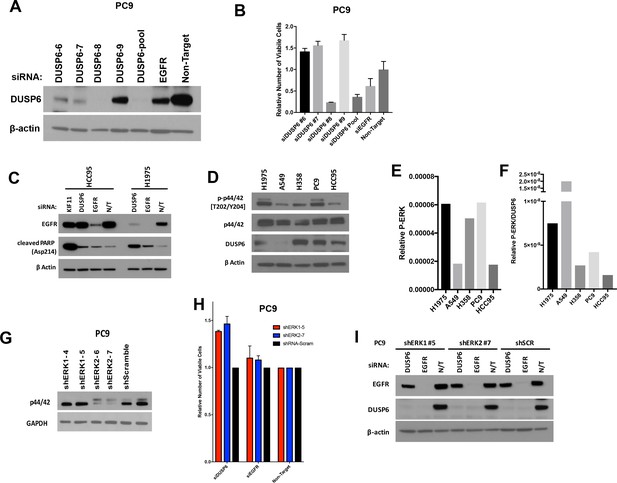
(A–B) Extensive knockdown of expression of DUSP6 with individual or pooled siRNAs is necessary to induce toxicity in PC9 cells.
Individual siRNAs that comprise the pool of siRNA from Dharmacon (DUSP6-6,−7,–8 and −9) were transfected on days 0 and 3 into PC9 cells (carrying an EGFR mutation) at the same final concentrations as in Figure 3. Levels of DUSP6 protein were compared on day 5 in cells that received the individual DUSP6 siRNAs, DUSP6 siRNA pool, EGFR siRNA pool or a non-targeting control (Non-Target). (A) The number of viable cells was also measured with Alamar blue at day 5 and scaled relative to the sample that received the non-targeting siRNA to measure the relative change in viability. Experiments were done with at least three biological replicates; the average ±SEM indicated. (B) Only the DUSP6 siRNA pool and the siRNA DUSP6-8 reduced DUSP6 protein to nearly undetectable levels, with a concurrent decrease in viable cells. Conversely, less extensive knockdown of DUSP6, as seen after introduction of the other individual siRNAs (DUSP6-6,−7 and −9), was associated with an increase in viable cells. Experiments were done with biological triplicates; the average values presented ±SEM. (C) Interference with DUSP6 RNA specifically induces cleaved PARP in cells with RTK-RAS-ERK pathway mutations. Decreased numbers of viable cells were observed after knockdown of DUSP6 in cells with EGFR or KRAS mutations - but not those with wild-type versions of these genes - as in Figure 3C. As evidence that these effects are mediated in part by apoptosis, cleaved PARP is induced in EGFR mutant H1975 but not KRAS/EGFR wild-type HCC95 cells when DUSP6 is inhibited. KIF11 and EGFR siRNAs serve as positive controls for induction of apoptosis in HCC95 and H1975 cell lines, respectively. Western blots were performed at day 5 after transfections at day 0 and 3 as described above. (D–F) Comparison of basal levels of P-ERK and DUSP6. Western blots were performed with extracts of cell lines with (H1975, A549, H358 and PC9) and without (HCC95) EGFR or KRAS mutations. (D) Relative P-ERK levels (E) and P-ERK/DUSP6 levels (F) were determined and plotted for each line. Cell lines which display decreased numbers of viable cells after knockdown of DUSP6 have greater relative P-ERK and/or P-ERK/DUSP6 levels than those that do not show decreased numbers of viable cells. (G–I) ERK inhibition partially rescues PC9 cells from the toxic effects of DUSP6 knockdown. Lentiviral vectors containing shRNAs for either ERK1 or ERK2 were transduced into PC9 cells and puromycin treatment was used to establish stable cell lines as described in Figure 1. Resulting knockdown of ERK1 or ERK2 compared to scramble shRNA containing control cells was confirmed by western blot (G) PC9 cells with decreased ERK demonstrated increased relative numbers of viable cells after DUSP6 siRNA-mediated knockdown compared to control cells receiving scrambled siRNA, whereas no difference was observed after knockdown of EGFR. Experiments were done in at least biological triplicate with the average ±SEM indicated. (H) Knockdown of the intended target was confirmed by western blot (I) in stable cell lines. All experiments were performed as in Figure 3; measured with Alamar blue and western blots were performed at the end of the experiments (day 5).
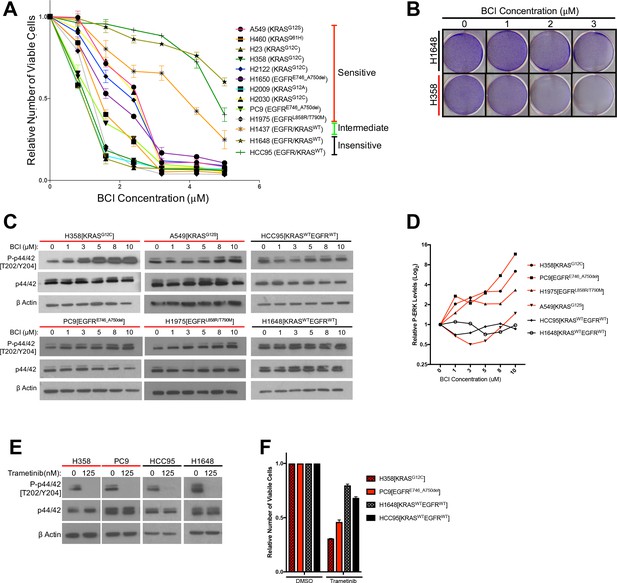
Treatment with the DUSP6 inhibitor BCI selectively kills LUAD cell lines with KRAS or EGFR mutation, implying a dependence on ERK-mediated signaling.
(A–B) BCI induces toxicity specifically in lung cancer cell lines with mutations in genes encoding components in the EGFR-KRAS-ERK pathway. (A) Eleven lung cancer cell lines were treated with increasing doses of BCI for 72 hr based on the reported effective activity of the drug (Shojaee et al., 2015). Cell lines could be assigned to three distinct groups: sensitive (red), intermediate (green) and insensitive (black). All sensitive cell lines contained either EGFR or KRAS mutations; the intermediate and insensitive cell lines were wild-type for genes encoding components of the EGFR-KRAS-ERK signaling pathway (as determined by the Sanger Cell Line Project and the Cancer Cell Line Encyclopedia [Barretina et al., 2012]). Experiments were done in biological duplicate with the average values presented ±SEM. (B) Crystal Violet stain of cells plated in the indicated doses of BCI or control (0 = 0.1% DMSO) for 72 hr. Sensitive cells with a KRAS mutation (H358 cells; denoted with red underlining) show a more pronounced decrease in cell number than do cells without oncogenic mutations in genes encoding components of the EGFR-KRAS-ERK pathway (H1648 cells; black underlining). Experiments were done in biological duplicate with a representative image shown. (C) BCI increases P-ERK levels specifically in BCI-sensitive cell lines. Sensitive lines (H358, PC9, H1975 and A549; red underlining) and insensitive lines (HCC95 and H1648; black underlining) were treated with the indicated doses of BCI or vehicle control (0.1% DMSO) for 30 min, and the levels of ERK (p44/p42) and P-ERK (P-p44/42 T202/Y204) assessed by Western blot. P-ERK appeared in the sensitive cells at low doses of BCI, but P-ERK levels did not increase in the insensitive cells at the tested doses of BCI. (D) Dosimetry plots from the experiment shown in panel. (C) (E–F) Cell lines sensitive to BCI are also dependent on P-ERK for survival. BCI-sensitive cells with oncogenic mutations in EGFR or KRAS (PC9 and H358, respectively; red underlining) and BCI-insensitive cells (H1648 and HCC95; black underlining) were treated with the indicated doses of the MEK inhibitor trametinib for 72 hr; viable cells were measured with Alamar blue and compared to cells receiving the vehicle control (0 = 0.1% DMSO). (E) Treatment with trametinib decreased P-ERK levels as determined by western blot. (F) The reduction in P-ERK corresponded to a greater decrease in viable cells in BCI-sensitive lines (red coloring), compared to BCI-insensitive cell lines (black coloring).
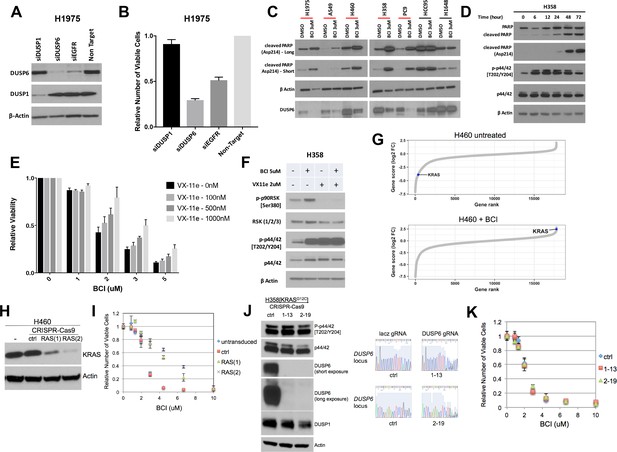
(A–B) Knockdown of DUSP6, but not DUSP1, decreases viability of LUAD cells.
DUSP1 and DUSP6 siRNAs were introduced into H1975 cells as described in Figure 1C. On day 5, western blots were performed to confirm knockdown of appropriate proteins (A) and measured with Alamar blue was used to count viable cells, (B) relative to cells receiving non-targeting control siRNAs. Reduction of DUSP6, but not of DUSP1, decreases viable cells, suggesting DUSP6 is the primary mediator of BCI-induced toxicity. Experiments were done in at least biological triplicate, with the average ±SEM indicated. (C) BCI induces cleaved PARP specifically in lung cancer cell lines with mutations in genes encoding components in the EGFR-KRAS-ERK pathway. A subset of cell lines from Figures 4A–5 categorized as sensitive (red line) and two insensitive (black line) - were treated with 3 uM BCI for 72 hr; induction of cleaved PARP was assessed by Western blot. Cleaved PARP is increased only in sensitive cell lines containing mutations in EGFR or KRAS. (D) Time course of cleaved PARP after BCI treatment in H358 cells in relation to pERK induction. 3 uM BCI increased P-ERK followed by cleavage of PARP, as determined by western blot at the indicated time points. (E–F) Decreased ERK activity partially rescues LUAD cells from BCI-induced toxicity. H358 cells received the indicated doses of BCI and the ERK inhibitor VX-11e for 72 hr; numbers of viable cells were determined by Alamar blue as in Figure 4A. Values for each line exposed to the VX-11e/BCI combination were normalized to results obtained only with VX-11e. Experiments were done in at least biological triplicate, with the average ±SEM indicated. Treatment of H358 cells with VX-11e decreased toxicity induced by BCI in a dose dependent manner, (E) corresponding to a decrease in downstream ERK activity as indicated by western blot for the ERK target RSK. (F) (G) Genome wide CRISPR-Cas9 screen in H460 cells reveals a dependence on KRAS for BCI sensitivity. The changes in abundance of guide RNAs are shown, revealing that a guide RNA targeting KRAS is depleted in control cells and enriched in the presence of BCI. (H) Validation of CRISPR-Cas9 screen. Two separate guide RNAs targeting KRAS (labeled 1 and 2) and a control gRNA targeting lacz (ctrl) were independently introduced into H460 cells, along with Cas9 in a lentiCRISPR v2 vector. Cell lines carrying these modifications and control cells were evaluated for KRAS depletion by western blot. The same cell lines were evaluated for their sensitivity to BCI in a dose response curve (I). Viable cell numbers are plotted relative to each line in the absence of BCI (set to 1.0) (J) H358 cells deficient in DUSP6 are responsive to BCI. Clones derived from H358 cells carrying a control (ctrl) guide RNA or two independent DUSP6 guides (1-13, 2-19) were evaluated by western blot for abundance of the indicated protein (left) and for successful targeting of the DUSP6 locus by DNA sequencing (right). DUSP6 protein is absent in the two clones. (J) H358 cells deficient for DUSP6 and cells targeted with a control gRNA were evaluated for sensitivity to BCI in a dose response curve. (K) Viable cell numbers are plotted relative to each independent line in the absence of BCI (set to 1.0). Results are representative of 3 independent experiments.
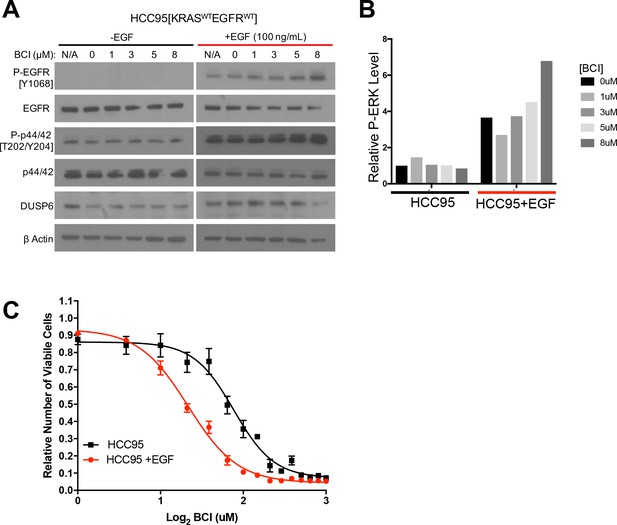
EGF-mediated activation of ERK signaling leads to dependence on DUSP6.
(A) EGF increases P-ERK in HCC95 cells. BCI- insensitive HCC95 cells were grown in the presence and absence of EGF (100 ng/mL) and increasing doses of BCI; levels of the indicated proteins were assessed in cell lysates by Western blotting. EGF increased the levels of P-EGFR and P-ERK, and levels of P-ERK were further increased by BCI. (B) Relative P-ERK levels (ratio of phosphorylated to total levels normalized to actin) were determined by dosimetry and compared to the vehicle controls (0 BCI = 0.1% DMSO) to quantify the relative increase after BCI treatment from the gels in A. (C) Increase of P-ERK promotes sensitivity of lung cancer cell lines without KRAS or EGFR mutations to BCI. BCI- insensitive HCC95 cells were treated with 100 ng/mL of EGF for 10 days and then grown in medium containing escalating doses on BCI with continued EGF. Viable cells were measured 72 hr later with Alamar blue and compared to the vehicle controls (in 0.1% DMSO) to assess the relative change in numbers of viable cells. Experiments were done in biological triplicate with the average values presented ±SEM. The EGF-treated cells (red line) showed increased sensitivity (decreased viable cells at lower BCI conditions) than those without EGF treatment (black line). (B–C).
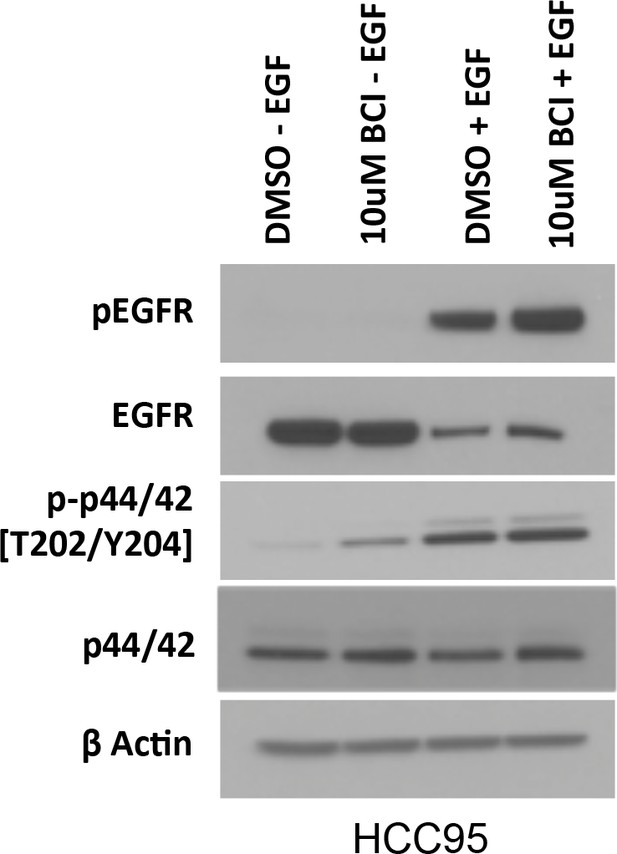
Protein lysates from conditions indicated in Figure 5A were subjected to electrophoresis on the same gel to directly compare p-EGFR and P-ERK levels in EGF-treated and untreated HCC95 cells.
https://doi.org/10.7554/eLife.33718.011Additional files
-
Supplementary file 1
Table containing the log2 fold change values for all sgRNAs from CRISPR-Cas9 screens.
- https://doi.org/10.7554/eLife.33718.012
-
Transparent reporting form
- https://doi.org/10.7554/eLife.33718.013





