Tandem riboswitches form a natural Boolean logic gate to control purine metabolism in bacteria
Figures
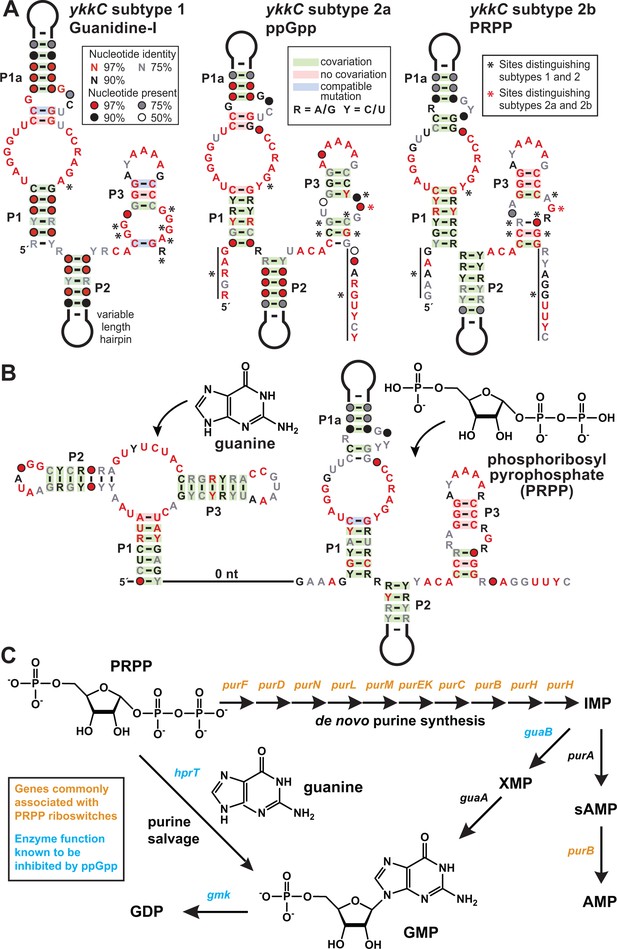
PRPP riboswitches and their regulatory network.
(A) Comparison of the consensus sequence and secondary structure models for the guanidine-I riboswitch aptamer (subtype 1, left) and the aptamers for ppGpp (subtype 2a, middle; ME Sherlock, N Sudarsan, RR Breaker, manuscript submitted), and PRPP (subtype 2b, right). For sequence alignments of the 257 unique PRPP riboswitch aptamers in Stockholm format, see Supplementary file 2. (B) Consensus sequence and secondary structure model for the 127 unique examples of adjacent guanidine-PRPP aptamers that form the corresponding tandem riboswitch architecture. For sequence alignments of the tandem guanine- PRPP riboswitch aptamers in Stockholm format, see Supplementary file 3. (C) Only genes in the purine biosynthetic pathway that are necessary for the production of adenosine nucleotides are commonly associated with PRPP riboswitches.
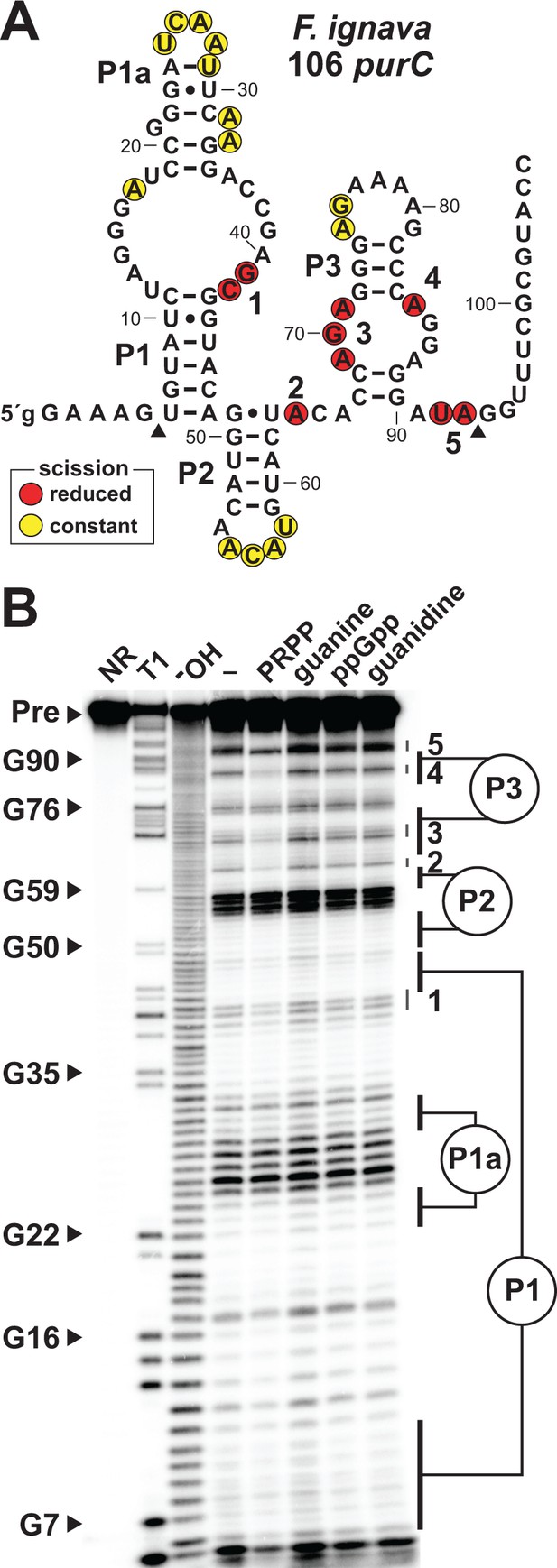
Ligand binding by a natural singlet PRPP riboswitch.
(A) Sequence and secondary structure of the 106 nucleotide RNA derived from the purC gene of F. ignava. Data collected in B were used to determine regions of constant or reduced scission upon the addition of PRPP to in-line probing assays. Lowercase g indicates a guanosine nucleotide added to enable efficient transcription by T7 RNA polymerase. Arrowheads demark the approximate range of nucleotides with in-line probing data available as depicted in B. (B) Polyacrylamide gel electrophoresis (PAGE) analysis of the spontaneous RNA cleavage products generated during in-line probing of 5′-32P-labeled 106 purC RNA. NR, T1 and –OH represent RNA undergoing no reaction, partial digest with RNase T1 (cleaves after guanosine nucleotides), or partial digest under alkaline conditions (cleaves after every nucleotide), respectively. Bands corresponding to selected G residues are annotated. In-line probing experiments contained either no ligand (–) or 1 mM of the compound indicated except guanine which was tested at 10 µM. Annotations 1 through 5identify prominent regions of the RNA that undergo structural stabilization in a PRPP-dependent manner. Data shown are representative of multiple experiments.
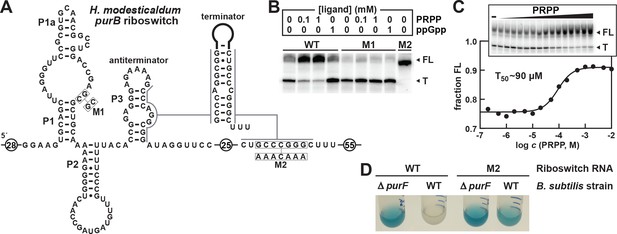
Transcription regulation and reporter gene expression by a natural singlet PRPP riboswitch.
(A) Sequence and secondary structure of the natural PRPP singlet riboswitch derived from the purB gene from H. modesticaldum. An alternative RNA structure is depicted in which the terminator stem forms followed by a U-rich tract, which only forms in the absence of PRPP. (B) PAGE analysis of a single-round transcription termination assay of WT and mutant H. modesticaldum purB riboswitches at the indicated ligand concentration. FL and T denote full length and terminated transcripts, respectively. Values for the fraction of full-length transcripts relative to the total transcription yield is listed for each reaction. (C) Plot of the fraction of full length WT H. modesticaldum purB RNA riboswitch transcripts contributing to the total number of transcripts (FL plus T) as a function of the logarithm (base 10) of the molar PRPP concentration. The concentration of PRPP required to cause half-maximal termination efficiency (T50) was determined by a sigmoidal curve fit (see METHODS). Inset: PAGE analysis of single round transcription termination assays of the WT H. modesticaldum purB RNA with either no ligand (–) or PRPP ranging from 200 nM to 10 mM. Data for replicates of this transcription termination assay are presented in Figure 3—figure supplement 1. (D) Reporter gene expression of WT and ΔpurF [BKE06490 (depicted) and BKK06490 (not shown)] B. subtilis cells containing wild-type (WT) or mutant (M2) H. modesticaldum purB riboswitch-lacZ reporter fusion constructs as described in A. Cells were grown in glucose minimal medium (GMM) containing 50 µg mL-1 X-gal.
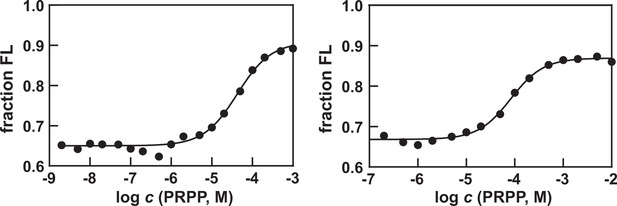
Replicate in vitro transcription termination assays with the H.
modesticaldum purB riboswitch construct. Assays are conducted as described for the data generated in Figure 3C. T50 values are estimated to be 40 μM (left) and 80 μM (right).
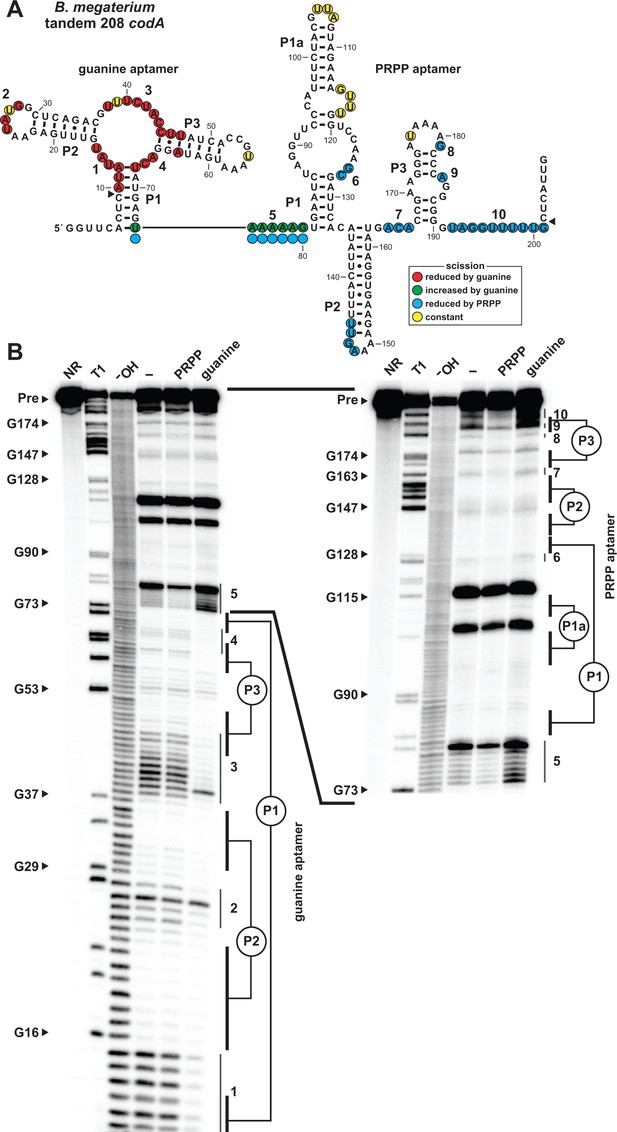
Ligand binding by tandem guanine and PRPP aptamers.
(A) Sequence and secondary structure of the 208 nucleotide RNA derived from the codA gene of B. megaterium. Data collected in B were used to determine regions of constant, increased, or reduced scission upon the addition of guanine or PRPP to in-line probing assays. Additional annotations are as described for Figure 4A. Note that the precise locations of regions experiencing strand scission are approximated after nucleotide 74, as guided by the related in-line probing data generated for the singlet PRPP aptamer depicted in Figure 2. (B) PAGE analysis of the spontaneous RNA cleavage products generated during in-line probing of 5′-32P-labeled 208 codA RNA. Annotations are described in the legend of Figure 2B. In-line probing experiments contained either no ligand (–), 1 mM PRPP or 10 µM guanine. Annotations 1 through 10 identify prominent regions of the RNA that undergo structural stabilization from either guanine or PRPP. Both gel images contain RNA from the same in-line probing reactions, but the gel in the image on the right was run for a longer time to increase resolution of the 3′ region of the RNA, which contains the regions of the PRPP aptamer involved in ligand recognition.
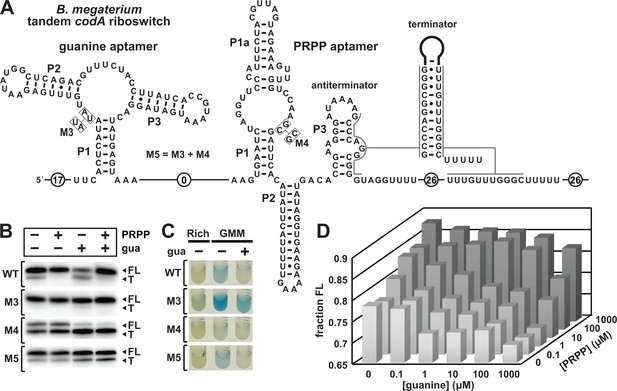
Guanine and PRPP competitively regulate transcript termination via a riboswitch integrating two aptamers.
(A) Sequence and secondary structure of the tandem guanine-PRPP riboswitch derived from the codA gene of B. megaterium. An alternative RNA structure is depicted to include the terminator stem followed by a U-rich tract, which forms based on the relative abundance of each ligand. (B) PAGE analyses of single-round transcription termination assays of WT and various mutant B. megaterium codA tandem riboswitches in the presence or absence of the two ligands. FL and T denote full length and terminated transcripts, respectively. (C) Reporter gene expression of B. subtilis containing wild-type (WT) and mutant (M3 through M5) B. megaterium tandem riboswitch-lacZ reporter fusion constructs as described in (A). Cells were grown in rich (lysogeny broth) medium or glucose minimal medium (GMM) containing either no (–) or 10 µM additional guanine (gua) and 50 µg mL-1 X-gal. (D) Plot of the fraction of full length B. megaterium codA tandem riboswitch transcripts contributing to the total number of transcripts as a function of both guanine and PRPP concentrations. Data shown are the average of three experiments (see Figure 5—figure supplement 1).
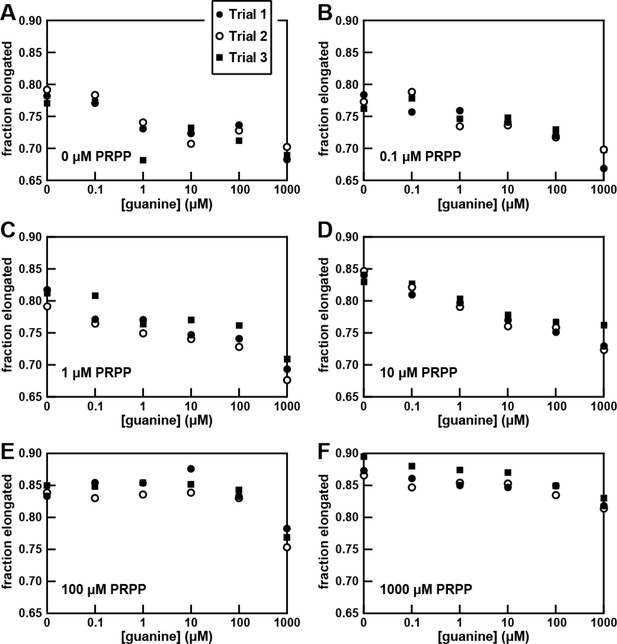
Comprehensive dataset for the in vitro transcription termination assays with the B. megaterium codA riboswitch construct.
Data for three separate transcription termination ‘trials’ are plotted for six conditions tested as noted in panel A. Panels (A) through (F) include the PRPP concentrations used in the assays in the lower left of each plot. Details are as described in the legend to Figure 5.
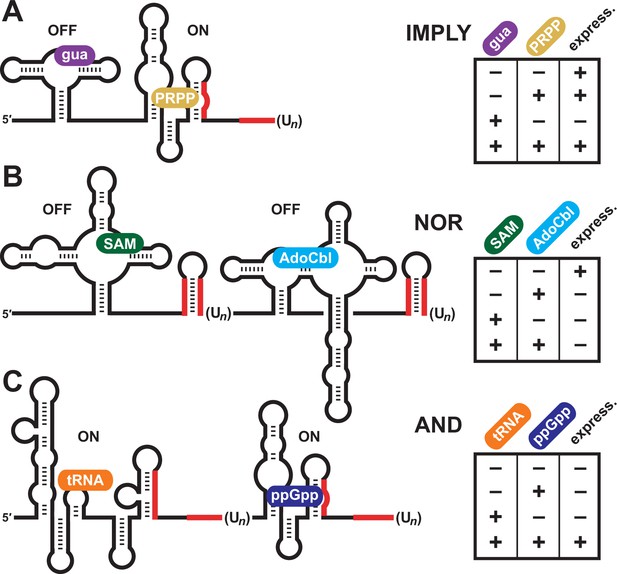
Natural examples of tandem riboswitches that approximate Boolean logic functions.
(A) Tandem guanine and PRPP aptamers form an IMPLY logic gate to determine the state of downstream gene expression (express.). (B) Tandem S-adenosylmethionine (SAM) and adenosylcobalamin (AdoCbl) riboswitches form a NOR logic gate. (C) Tandem T box RNA and ppGpp riboswitches form an AND logic gate. The T box and ppGpp riboswitch elements can occur in either order.
Additional files
-
Supplementary file 1
This file includes a table listing the DNA constructs used in the study.
- https://doi.org/10.7554/eLife.33908.011
-
Supplementary file 2
This file presents a sequence alignment in Stockholm format for individual PRPP riboswitch aptamers.
- https://doi.org/10.7554/eLife.33908.012
-
Supplementary file 3
This file presents a sequence alignment in Stockholm format for riboswitches that carry tandem guanine and PRPP aptamers.
- https://doi.org/10.7554/eLife.33908.013
-
Transparent reporting form
- https://doi.org/10.7554/eLife.33908.014





