Dual tRNA mimicry in the Cricket Paralysis Virus IRES uncovers an unexpected similarity with the Hepatitis C Virus IRES
Figures
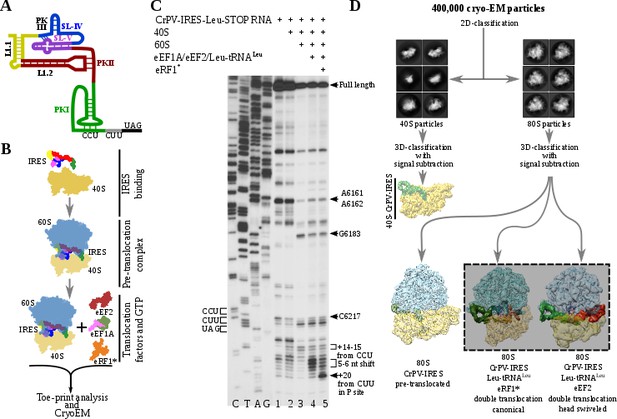
Experimental set up and cryoEM image processing workflow.
(A) Secondary structure scheme of CPV-IRES highlighting the modular architecture consisting of three pseudoknots (PKI, PKII and PKIII). (B) Diagram showing the in vitro reaction set up with purified components used for the toe-printing assays as well as for cryoEM. (C) Toe-print analysis of ribosomal complexes assembled with components indicated on top. Toe-prints corresponding to the pre-translocated complex are labeled as +14–15 nt from CCU. Toe-prints corresponding to the double translocated complex without and with eRF1* are marked as 5-6nt shift and +20 nt from CUU, respectively. Additional toe-prints previously attributed to different CrPV-IRES/ribosome contacts are seen, in agreement with previous reports (Muhs et al., 2015; Pestova et al., 2004). (D) CryoEM data processing workflow employed to resolve the high compositional and conformational heterogeneity of the in vitro reconstituted complexes described in B. Four populations were resolved and refined to high resolution, including two exhibiting clear density for a double translocated CrPV-IRES (squared).
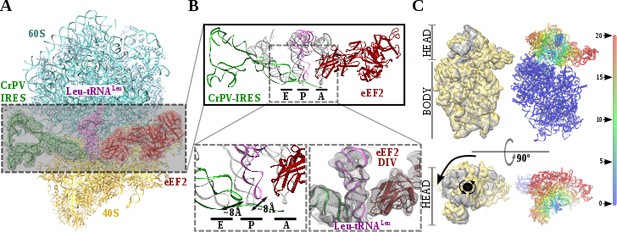
Structure of a double translocated CrPV-IRES intermediate with eEF2.
(A) Overview of a mammalian ribosome with double translocated CrPV-IRES (green), aminoacyl-tRNA (purple) and eEF2 (red). (B) Top, detailed view of the ribosomal sites E, P and A in the structure with a double translocated CrPV-IRES with eEF2. Canonical tRNAs (from PDB ID 4V5C) are depicted as semi-transparent grey cartoons. Bottom left, zoomed view highlighting the displacement of both aminoacyl-tRNA and CrPV-IRES PKI from canonical positions. Domain IV of eEF2 occupies the A site. Bottom right, final experimental densities for the ribosome ligands described at 7 Å. (C) On the left, two views of the experimental density for the 40S of the two double translocated reconstructions where it can be appreciated the swiveled configuration of the 40S head in the complex with eEF2 (grey). For comparative purposes, the map for the eRF1* containing complex (yellow) has been low pass filtered to a similar resolution as the eEF2 containing complex (grey). Right, atomic refined model colored according to the root-mean squared displacement (RMSD) with red and blue indicating the highest and lowest values, respectively (in Ångstroms).
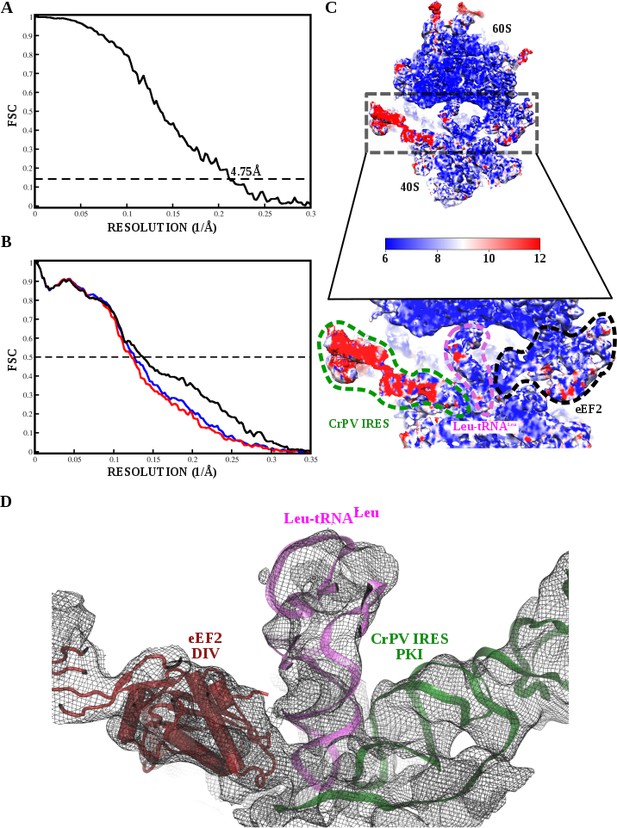
Fourier Shell Correlation curves and local resolution estimation for the 80S/CrPV-IRES/Leu-tRNALeu/eEF2 complex.
(A) Fourier Shell Correlation (FSC) computed for the two half maps of the final subset of particles after classification. The resolution is estimated to be 4.75 Å using the 0.143 criterion. (B) Map-versus-model cross validation FSC. The final model was validated using standard procedures. FSC of the refined model against half map 1 (blue) overlaps with the FSC against half map 2 (red, not included in the refinement). The black curve corresponds to the FSC of the final model against the final map. (C) Slice through the final, unsharpened map colored according to the local resolution as reported by RESMAP. (D) Due to the flexibility of the CrPV-IRES a Gaussian filter was applied to the map limiting the resolution to 7 Å. The final density is shown in D where it can be appreciated features corresponding to that resolution.
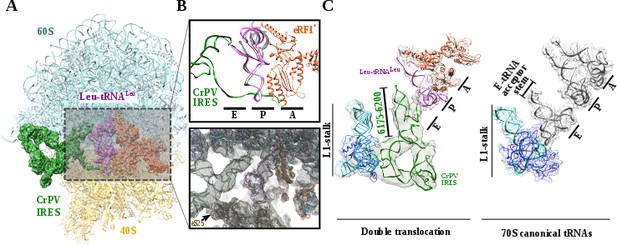
Structure of a double translocated CrPV-IRES.
(A) Overview of a mammalian ribosome with double translocated CrPV-IRES (green), aminoacyl-tRNA (purple) and eRF1* (orange). (B) Top, detailed view of the ribosomal E, P and A sites with a canonical configuration for the aminoacyl-tRNA in the P site, eRF1* in the A site and a disassembled CrPV-IRES PK I in the E site (green). Canonical P site tRNA (from PDB ID 4V5C) is depicted as semi-transparent grey cartoon. Bottom, large field of view of the final unsharpened map obtained for this reconstruction focused on the area described. (C) The position of the three ligands in the double translocated complex with eRF1* in comparison with canonical tRNAs. The L1 stalk is depicted in cyan.
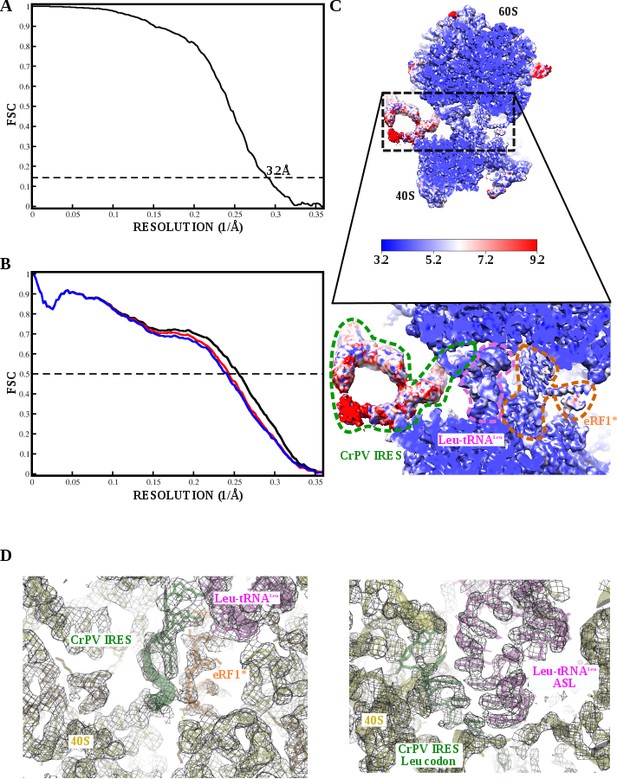
Fourier Shell Correlation curves and local resolution estimation for the 80S/CrPV-IRES/Leu-tRNALeu/eRF1* complex.
(A) Fourier Shell Correlation (FSC) computed for the two half maps of the final subset of particles after classification. The resolution is estimated to be 3.2 Å using the 0.143 criterion. (B) Map-versus-model cross validation FSC. Thefinal model was validated using standard procedures. FSC of the refined model against half map 1 (blue) overlaps with the FSC against half map 2 (red, not included in the refinement). The black curve corresponds to the FSC of the final model against the final map. (C) Slice through the final, unsharpened map colored according to the local resolution as reported by RESMAP.(D) Close-up views of the final density for the A site (left) and P site (right). Clear densities for individual bases could be observed.
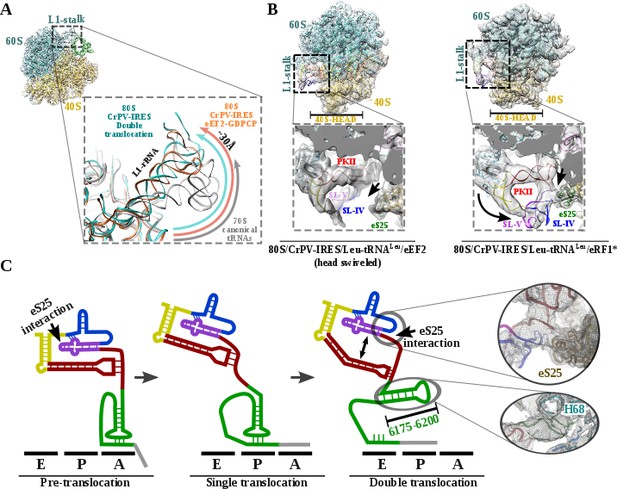
L1 stalk position and conformational change on double translocated CrPV-IRES.
(A) Top left, overview of the double translocated ribosome complex with eRF1* with the L1 stalk region highlighted. Main view, L1 stalk in the double translocated complex (cyan) is displaced from the position acquired in a complex with canonical tRNAs (grey) with a magnitude of approximately 30 Å. This displacement is similar to the one reported for the pre-translocated complex with eEF2 and a non-hydrolyzable GTP analog (orange)(Murray et al., 2016). (B) Conformational transition observed in CrPV-IRES upon back-swiveling of the 40S head. Left, due to a swiveled 40S head configuration in the double translocated complex with eEF2, SL-IV and SL-V remain solvent exposed, as in the single translocated complex (Muhs et al., 2015), and detached from the ribosomal protein eS25 (green). Right, once the head of the 40S relocates to its non-swiveled position, CrPV-IRES acquires a new conformation involving a new interaction with eS25 (green). (C) Scheme showing the secondary structure of CrPV-IRES in the pre-translocated state (left), after a single translocation (center) and after the double translocation (right). Arrows indicate the repositioning of PKII and PKIII as well as the new interaction established with ribosomal protein eS25. On the right, close up views of the final unsharped map obtained for this reconstruction for the regions indicated by circles.
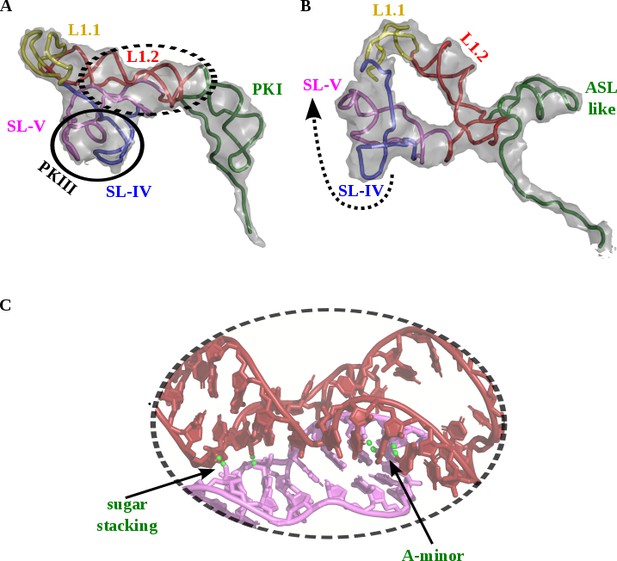
Structural transition observed in CrPV-IRESupon double translocation.
(A) Pre-translocation conformation of the CrPV-IRES (PDB ID 5IT9). PKI (green) is mimicking a cognate tRNA/mRNA pair and the PKIII (circled) and PKII (red) are in interacting distance as shown in C. (B) In the double translocated conformation, the PKI is disassembled and PKII and PKIII are no longer in interacting distance. PKIII (arrow) suffers a pronounce displacement. (C) In the pre-translocation state, a network of non-covalent interactions involving sugar-sugar stacking interactions as well as A-minor interactions.
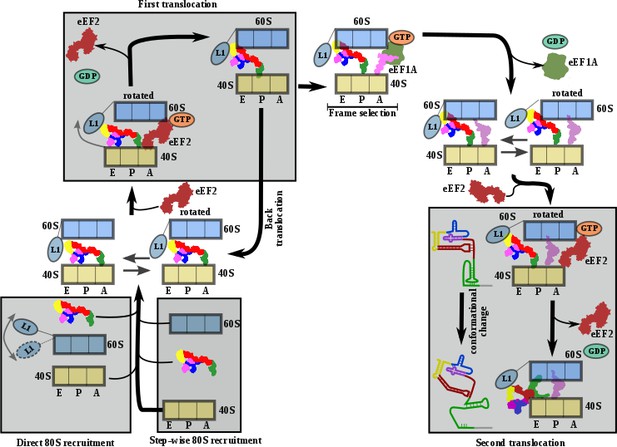
Comprehensive model describing CrPV-IRES strategy to hijack host ribosomes.
Bottom left, CrPV-IRES can directly recruit 80S to assemble a binary 80S/CrPV-IRES complex which, in its pre-translocation state, oscillates between rotated and non-rotated configurations of the 40S. However, a step-wise pre-translocation complex formation involving an initial interaction with the 40S, followed by recruitment of 60S, is more efficient and is favored. Top left, first translocation event involving the displacement of CrPV-IRES PKI from the A site in order for the first aminoacyl-tRNA to be delivered as a ternary complex with eEF1A and GTP. In the absence of an A site ligand this state is unstable and prone to back translocation (Muhs et al., 2015). According to smFRET studies, the frame is not defined until the first condon/anticodon interaction is established (Petrov et al., 2016). Top right, presumably a single translocated complex with A site aminoacyl-tRNA alternates between rotated and non-rotated configurations of the 40S as a bonafide pre-translocation complex with two tRNAs. Bottom right, binding of eEF2 in its GTP form assists in the translocation of CrPV-IRES and the first aminoacyl-tRNA which is achieved through a conformational change in the CrPV-IRES involving the disassembling of the PKI and reorientation of PKII and PKIII.
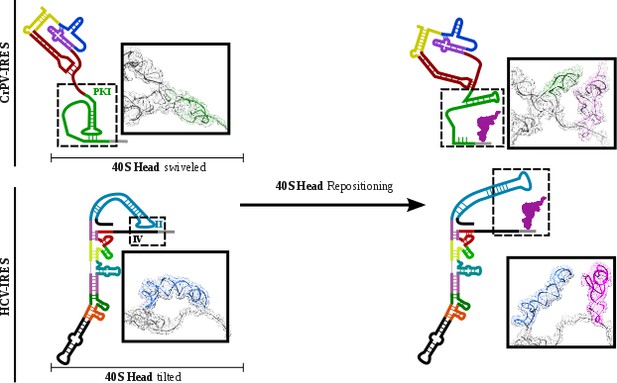
CrPV-IRES and HCV-IRES experiments a similar structural transition upon first aminoacyl-tRNA delivery to the ribosomal P site.
The structural transition experienced by the CrPV-IRES upon delivery of the first aminoacyl-tRNA to the P site (top) is similar to the one described for the HCV IRES (bottom). CrPV-IRES PKI remains assembled and in the vicinity of the E site of the 40S due to a swiveled configuration of the 40S head. The HCV-IRES maintains a similar internal interaction of domain II by a tilted configuration of the 40S head. In case of both IRESs, a reconfiguration involving back-positioning of the 40S head plus the placement of a structural element of the IRES in the vicinity of the 60S E site, facilitates the delivery of the first aminoacyl-tRNA to the P site of the ribosome, finalizing the initiation stage of translation.
Videos
Conformational changes experienced by the CrPV-IRES along its movement through the ribosome.
CrPV-IRES binds initially to the ribosome inserting the PKI (green) in the A site (PDBID 5IT9, [Murray et al., 2016]). After a first translocation, PKI is placed in the P site (PDBID 4D61, Muhs et al., 2015) and a second translocation event induces its disassembly (this work). Molecular transitions have been approximated by a linear morph using Chimera (Pettersen et al., 2004).
Tables
Data collection, model refinement and validation statistics.
https://doi.org/10.7554/eLife.34062.004| Data collection | ||
|---|---|---|
| Voltage (KV) | 300 | |
| Defocus range (μm) | −0.5/–3 | |
| Pixel size (Å/pixel) | 1.08 | |
| Electron dose (e-/Å2) | 64 | |
| Images collected | 16,303 | |
| Model Refinement | ||
| CrPV-IRES/eRF1* | CrPV-IRES/eEF2 | |
| Program/Protocol | Refmac5/Reciprocal space | Phenix/Real space |
| Resolution: | ||
| FSC 0.143 (Å) | 3.2 | 4.75 |
| Used in refinement (Å) | 3.8 | 7 |
| Map sharpening (Å) | −91.86 | −113.6 |
| Average B-factors (Å) | 169.58 | 394.12 |
| R.m.s deviations: | ||
| Bonds (Å) | 0.011 | 0.0033 |
| Angles (o) | 1.58 | 0.83 |
| Validation | ||
| Molprobity score | 2.81 | 1.50 |
| Clashcore, all atoms | 5.74 | 3.86 |
| Favored rotamers (%) | 88.51 | 99.7 |
| Ramachandran plot: | ||
| Outliers (%) | 3 | 0.02 |
| Favored (%) | 83.23 | 95.3 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.34062.014





