The Peptidisc, a simple method for stabilizing membrane proteins in detergent-free solution
Figures

Solubility test of NSP and NSPr.
(A) Peptide models computed by the 3D-hydrophobic moment peptide calculator. The direction of hydrophobic moment is indicated by a red line. Peptides are oriented with their N to C-terminus from bottom (red) to top (blue). (B) Turbidity measurement of peptide suspension. The absorbance of light at 550 nm for NSP (blue squares, 15 mg/mL) and NSPr (red circles, 25 mg/mL), re-suspended in distilled water (dH2O) were compared to a dH2O control (green triangles). (C) Calculated electropotential and hydrophobic moment of peptide variants. Calculations were performed using the 3D-hydrophobic moment peptide calculator as described in Materials and methods.
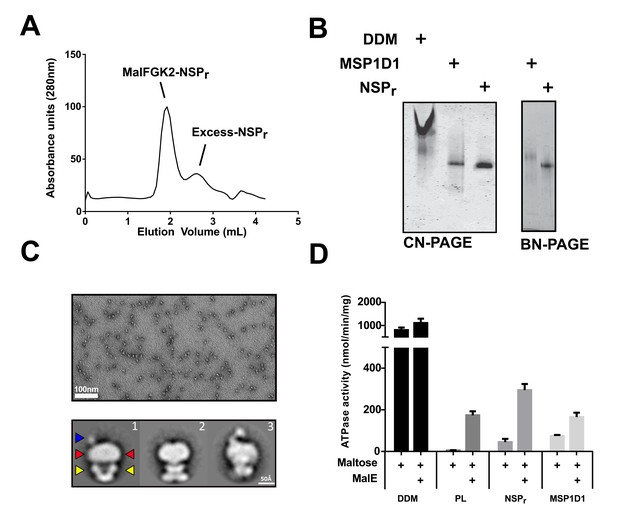
The ‘on-column’ reconstitution of MalFGK2.
(A) Typical size-exclusion chromatography of MalFGK2 in peptidisc (MalFGK2-NSPr) using the ‘on-column’ method. (B) CN-PAGE and BN-PAGE analysis of MalFGK2 in detergent micelle (DDM), nanodisc (MSP1D1), and peptidisc (NSPr). (C) Top panel: Field of view of particles stained with uranyl formate. Bottom panel: Selected class averages representing three characteristic views of MalFGK2 in peptidisc. The nucleotide-binding domains (MalK2), the transmembrane domain (MalFG), and periplasmic P2-loop are indicated with yellow, red and blue arrows, respectively. (D) Maltose-dependent ATPase activity of MalFGK2 (0.5 µM) reconstituted in detergent (DDM), proteoliposomes (PL), peptidiscs (NSPr), and nanodiscs (MSP1D1) obtained at 30°C in the presence or absence of MalE (2.5 µM). Error bars represent standard deviations from three separate experiments.
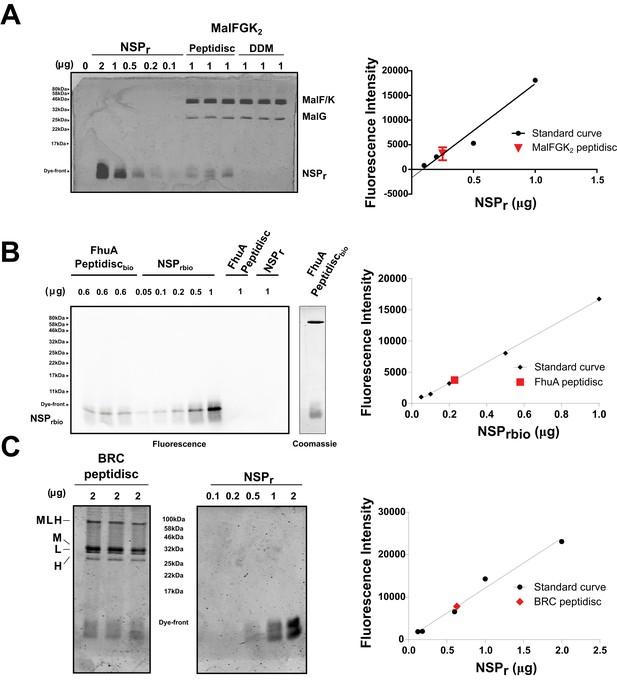
Quantification of NSPr in peptidiscs.
(A) Left panel; 15% SDS-PAGE analysis of MalFGK2 in peptidisc or DDM. NSPr runs at the bottom of the gel and can be visualized with Coomassie blue staining. Dye fluorescence was measured on a LICOR Odyssey scanner and quantified by Image J. Right panel; Standard curve derived from NSPr titration measurement (black dots), and average intensity of NSPr fluorescence from MalFGK2 peptidisc (red dot). (B) Left Panel: Western Blot of FhuA-peptidisc reconstituted into NSPrbio, and visualized by incubation with Streptavidin-Alexa 680. Fluorescence of the Alexa 680 dye was measured on a LICOR Odyssey scanner (700 nm, excitation 680 nm) and quantified in Image J. Right Panel: Standard curve as in A. (C) 15% SDS-PAGE analysis of BRC in peptidisc. The MLH subunits of BRC partially resist denaturation by SDS, resulting in a higher molecular weight band located above the single subunits. Each gel was repeated in triplicate with independent standard curves to calculate the values reported in Table 1.
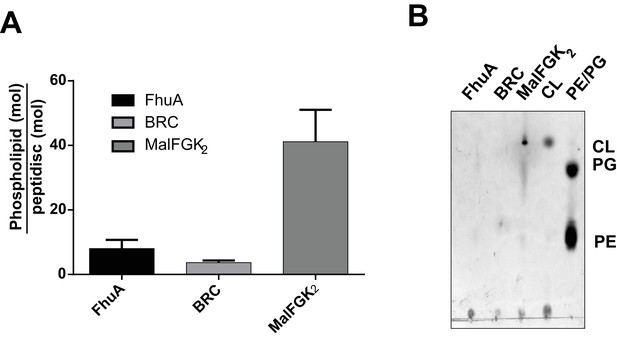
Quantification of phospholipids trapped in peptidiscs.
(A) Calculated number of phospholipids per peptidisc. Phospholipid content was determined by Malachite green assay after acid digestion of lipid extracts. Error bars represent standard deviation derived from three separate measurements. B) TLC analysis of lipid extracts obtained from 10 µg MalFGK2 peptidisc, 10 µg FhuA peptidiscs and 20 µg BRC peptidiscs, as well as pure lipid standards Cardiolipin (CL), 1,2-dioleoyl-sn-glycero-3-phosphoglycerol (PG), and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (PE).
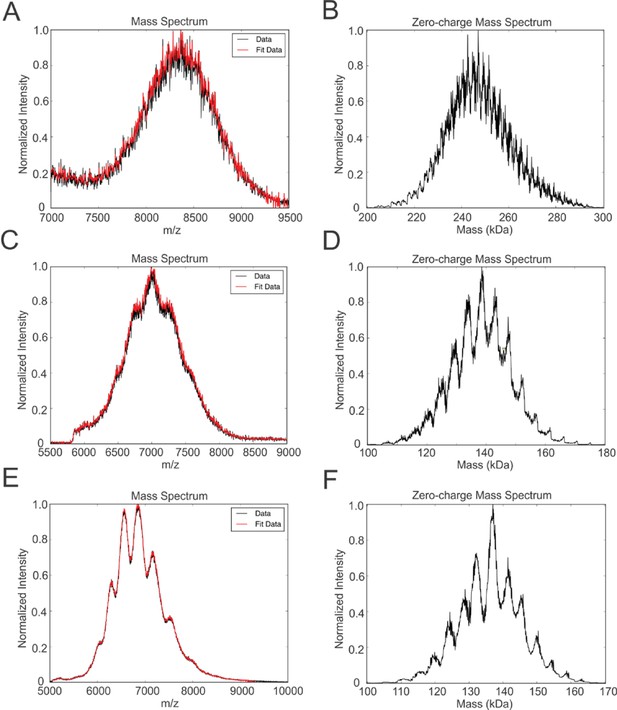
Native mass spectrometry of intact peptidiscs.
Panels A, C, E are mass spectra acquired in positive ion mode for aqueous ammonium acetate solutions (100 mM, pH 7, 22°C) of MalFGK2-NSPr, BRC-NSPr and FhuA-NSPr, respectively. Panels B, D, and F are deconvoluted mass spectra of the peptidiscs shown in A, C, and E, respectively.
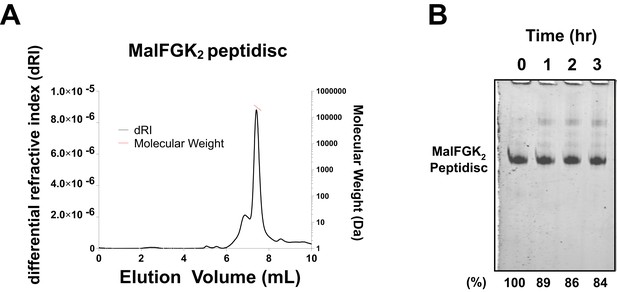
Stability of MalFGK2 peptidisc.
(A) Multi-angle light scattering analysis of MalFGK2 reconstituted in peptidisc. MalFGK2-NSPr (100 µg) was left for 3 days at 4°C before analysis by SEC-MALS. Protein sample was injected and protein concentration tracked through differential refractive interferometry (dRI, black trace). Molecular weight was calculated for the fractions corresponding to the peak of MalFGK2-NSP (red trace). (B) Structural stability of MalFGK2-NSPr. The MalFGK2 peptidisc was incubated at 30°C in Buffer A for the indicated time, then analyzed by BN-PAGE.
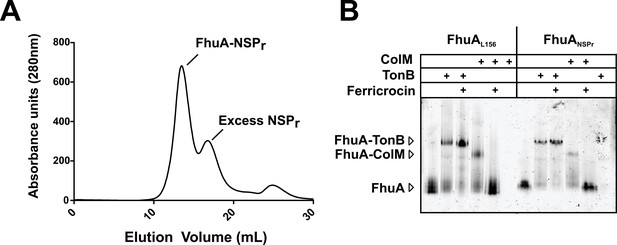
Binding activity of FhuA in nanodiscs and peptidiscs.
(A) Typical SEC profile of FhuA reconstituted in peptidisc (FhuA-NSPr) using an ‘on-column’ reconstitution protocol as described in Materials and methods. (B) The FhuA transporter reconstituted in nanodiscs (FhuA-MSPL156) or peptidiscs (FhuA-NSPr) was incubated with the C-terminal TonB23-329 fragment (2 µg) or with colicin M (5 µg), with or without ferricrocin as indicated. Samples were analyzed by CN-PAGE and Coomassie-blue staining of the gel.
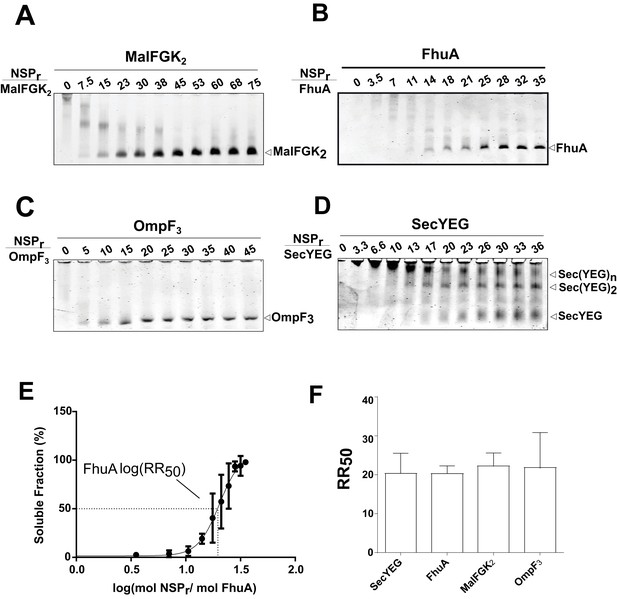
Express ‘in-gel’ method for determining optimal reconstitution ratio.
(A) NSP and MalFGK2 were mixed at the indicated molar ratio for 2 min in Buffer A containing a low amount of detergent (~0.008% DDM) before loading onto CN-PAGE. Peptidisc reconstitution occurs during migration in the detergent-free gel environment. The same experiment was performed with. (B) FhuA,. (C) OmpF3,. (D) SecYEG. (E) Reconstitution efficiency of FhuA as a function of the NSP concentration. The protein band FhuA-NSP in B) was quantified with Image J and the data plotted as log (mol NSP/mol FhuA). The data were fitted with a Boltzmann sigmoidal function to generate a curve describing the reconstitution efficiency and the half-maximal reconstitution ratio (RR50). (F) The RR50 was determined for other target proteins as in (E). Error bars represent the standard deviation from three separate reconstitution experiments.
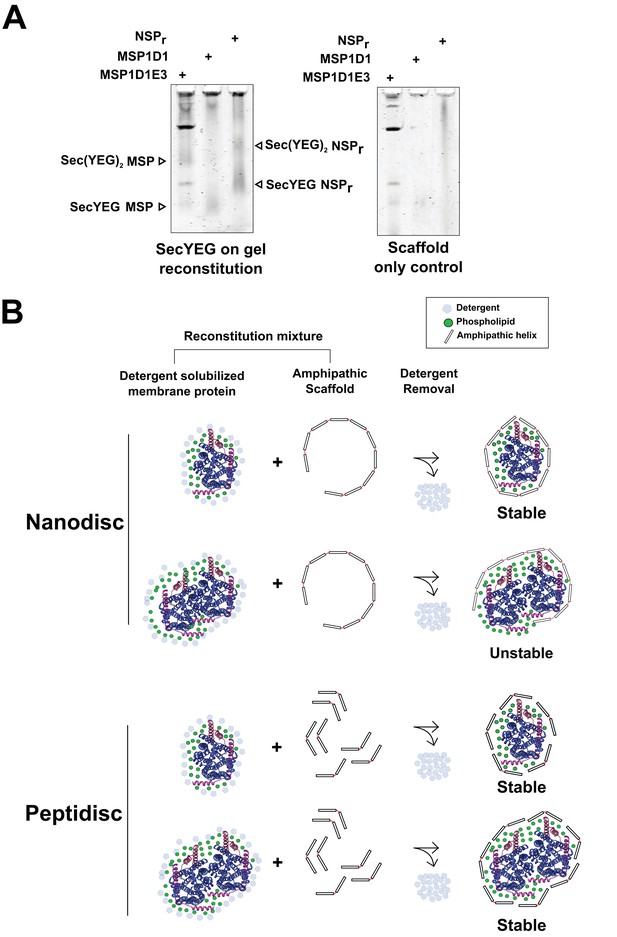
Capture of SecYEG monomer and dimer in peptidisc and nanodisc.
(A) Left panel; the SecYEG complex (2.5 µg) was incubated with the indicated scaffolds proteins (1.25 µg each) in Buffer A + 0.02% DDM. The sample was diluted threefold in detergent-free Buffer A and immediately analyzed by CN-PAGE. Right panel; the experiment on left panel was performed without addition of SecYEG. (B) Illustration representing possible reconstitution products of the SecYEGn complex into MSP1D1 and NSPr. The subunits SecY, SecE, and SecG are represented as blue, pink, and magenta, respectively.
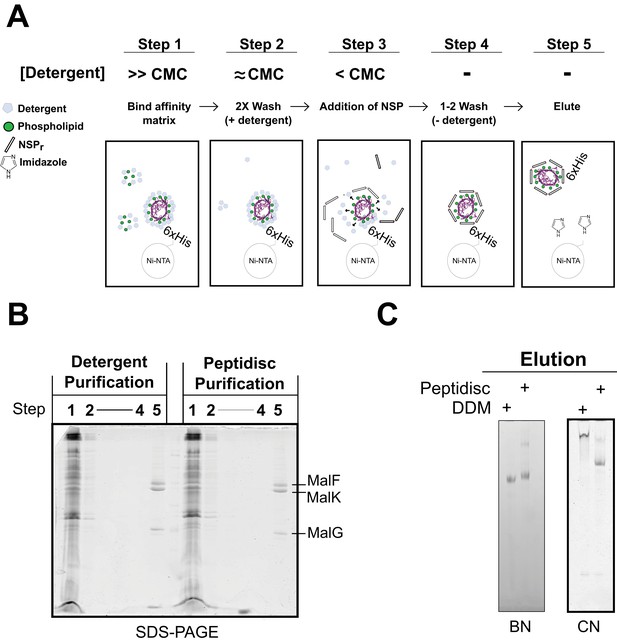
Direct ‘on-beads’ reconstitution during membrane protein purification.
(A) Principle of the ‘on-beads’ reconstitution. Step 1: the tagged protein is extracted from the membrane with excess of detergent buffer (>CMC) and incubated with the affinity resin. Step 2: The beads are washed twice with the detergent buffer near its critical micelle concentration (~CMC). Step 3: The beads are incubated with buffer containing excess NSP and limited amount of detergent (<CMC). Step 4: The beads are washed in detergent-free buffer to remove unbound NSP and residual detergent. Step 5: The protein captured in peptidiscs is eluted from the column in detergent-free solution. (B) SDS-PAGE and. (C) Native-PAGE analysis of the his-tagged MalGFK2 complex purified following conventional detergent method and ‘on-beads’ peptidisc detergent-free method.
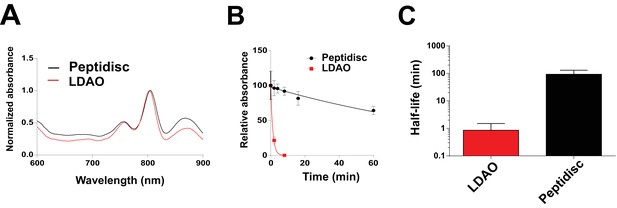
Thermostability of the BRC complex in peptidiscs.
(A) Absorbance scans of the BRC (1 µM) in detergent solution (0.03% LDAO, red trace) and in peptidisc (black trace). Scans were normalized to the value measured at 803 nm (the absorbance peak of the accessory bacteriochlorophylls). (B) Decrease in absorbance of the BRC at 803 nm after incubation at 65°C for the indicated time. (C) Calculated half-life of the BRC in peptidisc and LDAO at 65°C. The data in B) were fit with an exponential decay function to determine the corresponding half-life. Error bars represent the standard deviation from three separate experiments.
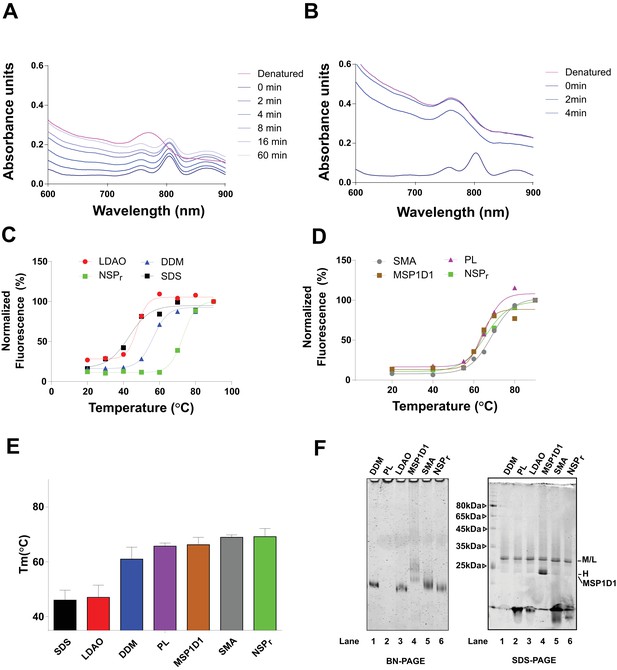
Effect of peptidisc on BRC stability.
(A) Absorbance scans of the BRC complex (1 µM) in peptidisc after incubation at 65°C for up to 1 hr (blue traces). Incubation at 90°C leads to full release of the bacteriochlorophyll pigment (magenta trace). (B) Absorbance scans of the BRC (1 µM) after incubation at 65°C in 0.03% LDAO for up to 4 min. (C) Fluorescence of the BRC in peptidisc (green trace), 0.1% LDAO (red trace), 0.02% DDM (blue trace), and 0.1% SDS (black trace). The BRC (1 µM) was incubated for 5 min at the indicated temperature before fluorescence was measured (700 nm; excitation at 680 nm). (D) The experiment repeated as in C), with BRC reconstituted into MSP1D1 (1:2 BRC:MSP1D1 molar ratio, brown trace), SMA (0.1%, grey trace), Proteoliposomes (1:1600:400 BRC:DOPC:DOPG), and peptidiscs (green trace). Fluorescence were normalized to 100% after denaturation for 5 min at 90 degrees. (E) Data were fitted using a Boltzmann sigmoidal function to calculate the melting temperature (Tm). (F) Analysis of reconstituted BRC fractions on BN-PAGE (left panel) and SDS-PAGE (right panel).
Tables
Calculated and observed molecular weight and scaffold stoichiometry of peptidiscs
https://doi.org/10.7554/eLife.34085.009| Peptidisc | Molecular weight measured by ESI-MS (kDa) | Measured NSPr stoichiometry (NSPr/disc) | Measured lipid stoichiometry (Lipid/Disc) | Calculated molecular weight* (kDa) |
|---|---|---|---|---|
| MalFGK2-NSPr | 247 ± 24 | 10 (±2): 1 | 41 (±10): 1 | 251 ± 12 |
| BRC-NSPr | 138 ± 18 | 9 (±1): 1 | 4 (±1): 1 | 138 ± 5 |
| FhuA-NSPr | 137 ± 18 | 10 (±2): 1 | 8 (±3): 1 | 131 ± 9 |
-
*The formula for the calculated molecular weight is as follows: MWpeptidisc = MW(protein)+n(MWNSPr)+m(MWLipid); where n is the measured NSPr stoichiometry, m is the measured lipid stoichiometry, MWLipid = 0.8 kDa, MWNSPr = 4.5 kDa, and MWprotein = 173 kDa, 80 kDa, and 94 kDa for MalFGK2, FhuA, and BRC, respectively. For NSPr and Lipid stoichiometry, the standard deviation is derived from three separate measurements.
Molecular weight, diameter, and scaffold stoichiometry of MalFGK2 reconstituted in peptidisc.
https://doi.org/10.7554/eLife.34085.010| Measured molecular weight (kDa)* | Measured diameter † | Calculated stoichiometry (scaffold/disc)‡ | |
|---|---|---|---|
| MalFGK2 Peptidisc | 250 ± 17 | 11.7 ± 1.4 | 12 (±2):1 |
-
*Molecular weight calculated from SEC-MALS data (Fig. S1). The standard error is derived from three independent SEC-MALS experiments. †Diameter of MalFGK2-peptidiscs determined by negative stain electron microscopy Figure 1B, assuming a perfectly circular shape. ‡Stoichiometry (n), based on the measured diameter of the particles, was calculated with the following formula: π(ddisc-2dα-helix) = (n/2)LNSPr; where dα-helix represents the diameter of an alpha-helix (0.5 nm), ddisc represents the measured disc diameter, and LNSPr represents length of the NSPr peptide.
Additional files
-
Supplementary file 1
Amphipathic scaffolds used in this study.
The length of the scaffold proteins was calculated by multiplying the number of amino acids (aa) by 1.5 Å, which is the rise given by an amino acid structured in an alpha-helix. For the MSPs scaffolds, the number of amino acids was from the TEV cleavage site (ENYLFQ//GXXX) to the C-terminus of the proteins.
- https://doi.org/10.7554/eLife.34085.017
-
Supplementary file 2
Native gel buffer recipes.
- https://doi.org/10.7554/eLife.34085.018
-
Supplementary file 3
Protein molecular weights.
- https://doi.org/10.7554/eLife.34085.019
-
Transparent reporting form
- https://doi.org/10.7554/eLife.34085.020





