Hepatic NF-kB-inducing kinase (NIK) suppresses mouse liver regeneration in acute and chronic liver diseases
Figures
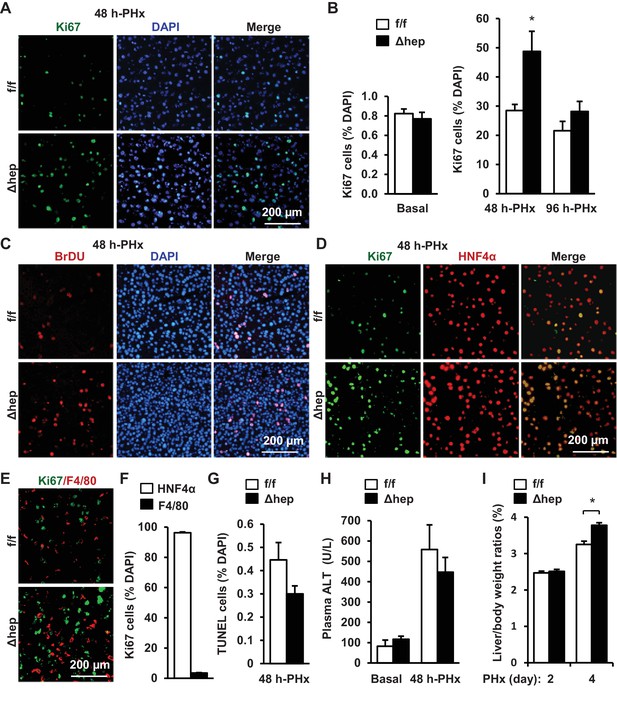
Hepatocyte-specific ablation of NIK accelerates reparative hepatocyte proliferation.
NIKf/f (n = 7) and NIKΔhep (n = 7) male mice (8 weeks) were subjected to PHx, and livers were harvested 48 hr or 96 hr later. (A) Representative immunostaining of liver sections (48 hr after PHx) with anti-Ki67. (B) Ki67+ cells were counted and normalized to total DAPI+ cells. (C) Representative immunostaining of liver sections (48 hr after PHx) with anti-BrdU antibodies. (D–E) Representative images of liver sections (48 hr after PHx) costained with anti-Ki67 and anti-HNF4α antibodies (D) or anti-Ki67 and anti-F4/80 antibodies (E). (F) Ki67+HNF4α+ and Ki67+F4/80+ cells were counted and normalized to total Ki67+ cells. (G) Liver cell death were assessed 48 hr after PHx using TUNEL reagents. (H) Plasma ALT levels. (I) Liver to body weight ratios (n = 8 per group). Data were statistically analyzed with two-tailed Student’s t test, and presented as mean ± SEM. *p<0.05.
-
Figure 1—source data 1
Hepatic NIK deficiency accelerates liver regeneration.
- https://doi.org/10.7554/eLife.34152.003
-
Figure 1—source data 2
PHx increases hepatocyte replications.
- https://doi.org/10.7554/eLife.34152.004
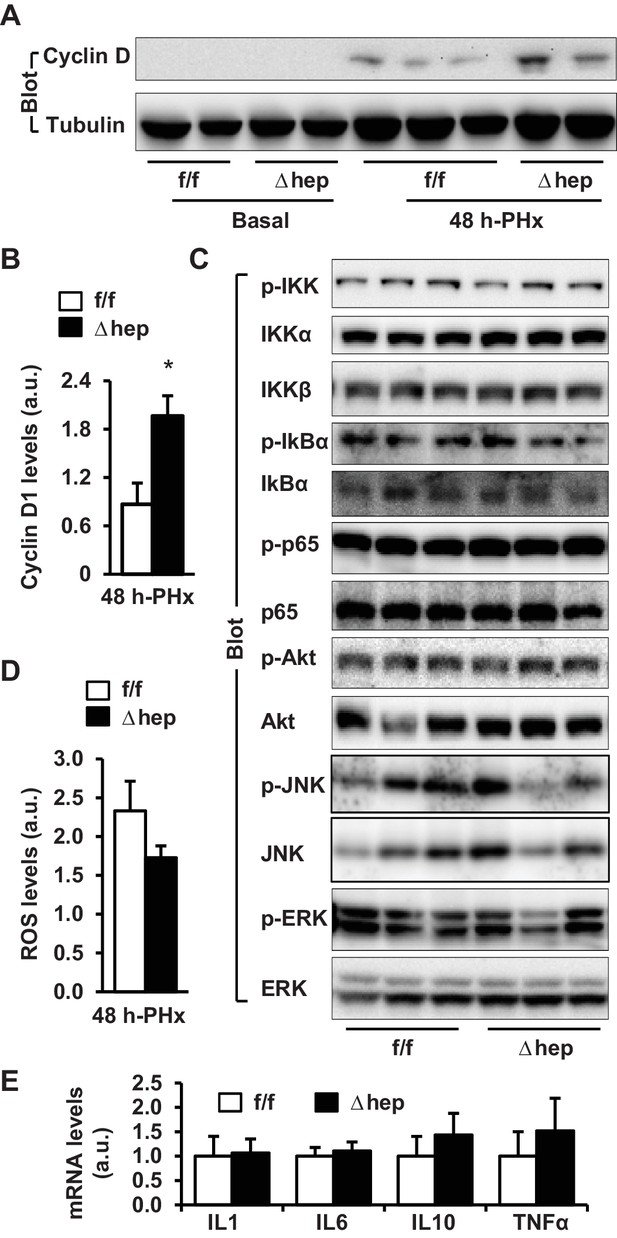
Hepatic NIK deficiency upregulates cyclin D1 without altering NF-kB1, Akt, and MAPK pathways in the liver.
NIKf/f and NIKΔhep male mice (8 weeks) were subjected to PHx. (A–B) Liver extracts were immunoblotted with anti-cyclin D1 antibody (48 hr after PHx). Cyclin D1 levels were quantified and normalized to α-tubulin levels (NIKf/f: n = 4, NIKΔhep: n = 4). (C) Liver extracts were immunoblotted with the indicated antibodies (4 hr after PHx). (D) Liver ROS levels 48 hr after PHx (normalized to liver weight). NIKf/f: n = 5, NIKΔhep: n = 6. (E) Liver cytokine expression was measured by qPCR and normalized to 36B4 expression (48 hr after PHx). NIKf/f: n = 5, NIKΔhep: n = 5. Data were statistically analyzed with two-tailed Student’s t test, and presented as mean ± SEM. *p<0.05.
-
Figure 2—source data 1
Hepatic NIK regulates hepatocyte cell cycle progression.
- https://doi.org/10.7554/eLife.34152.008
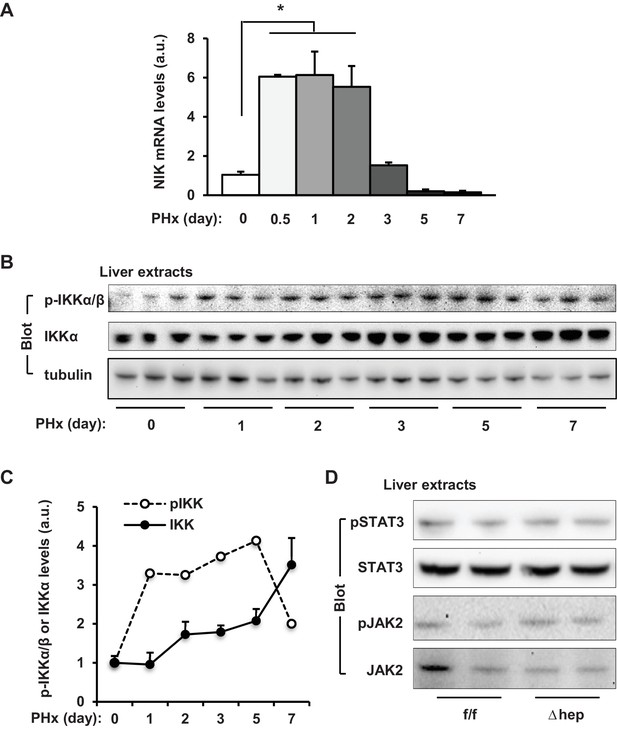
Effect of PHx on liver NIK pathway activation.
PHx was performed on C57BL/6 male mice. (A) Liver NIK mRNA abundance was measured by qPCR (normalized to 36B4 levels, n = 3 per group). (B–C) Liver extracts were immunoblotted with antibodies against phospho-IKKα/β, IKKα and α-tubulin. Liver phospho-IKKα/β (normalized to IKKα levels) and IKKα (normalize to α-tubulin levels) levels were measured on days 0–7 following PHx. (D) PHx was performed on NIKf/f and NIKΔhep male mice. Liver extracts were prepared from resected livers and blotted with antibodies against phospho-STAT3, STAT3, phospho-JAK2, and JAK2. Data were statistically analyzed with two-tailed Student’s t test, and presented as mean ± SEM. *p<0.05.
-
Figure 2—figure supplement 1—source data 1
PHx stimulates hepatic NIK expression.
- https://doi.org/10.7554/eLife.34152.007
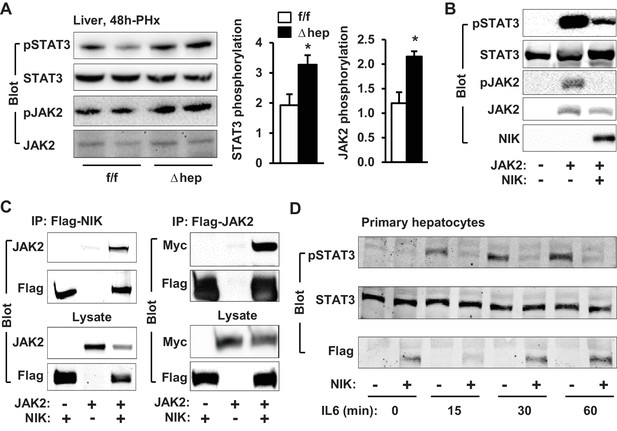
NIK inhibits the JAK2/STAT3 pathway.
(A) Liver extracts were prepared from NIKf/f and NIKΔhep males 4 hr after PHx and immunoblotted with anti-phospho-JAK2 and anti-phospho-STAT3 antibodies. Phosphorylation of JAK2 (pTyr1007/1008) and STAT3 (pTyr705) was normalized to total JAK2 and STAT3 levels, respectively. (B) STAT3 and JAK2 were coexpressed with or without NIK in HEK293 cells. Cell extracts were immunoblotted with the indicated antibodies. (C) NIK was coexpressed with JAK2 in HEK293 cells. Cell extracts were immunoprecipitated (IP) and immunoblotted with the indicated antibodies. (D) Mouse primary hepatocytes were transduced with NIK or β-gal adenoviral vectors and stimulated with IL6 (10 ng/ml). Cell extracts were immunoblotted with the indicated antibodies. Data were statistically analyzed with two-tailed Student’s t test, and presented as mean ± SEM. *p<0.05.
-
Figure 3—source data 1
NIK inhibits the JAK2/STAT3 pathway.
- https://doi.org/10.7554/eLife.34152.010
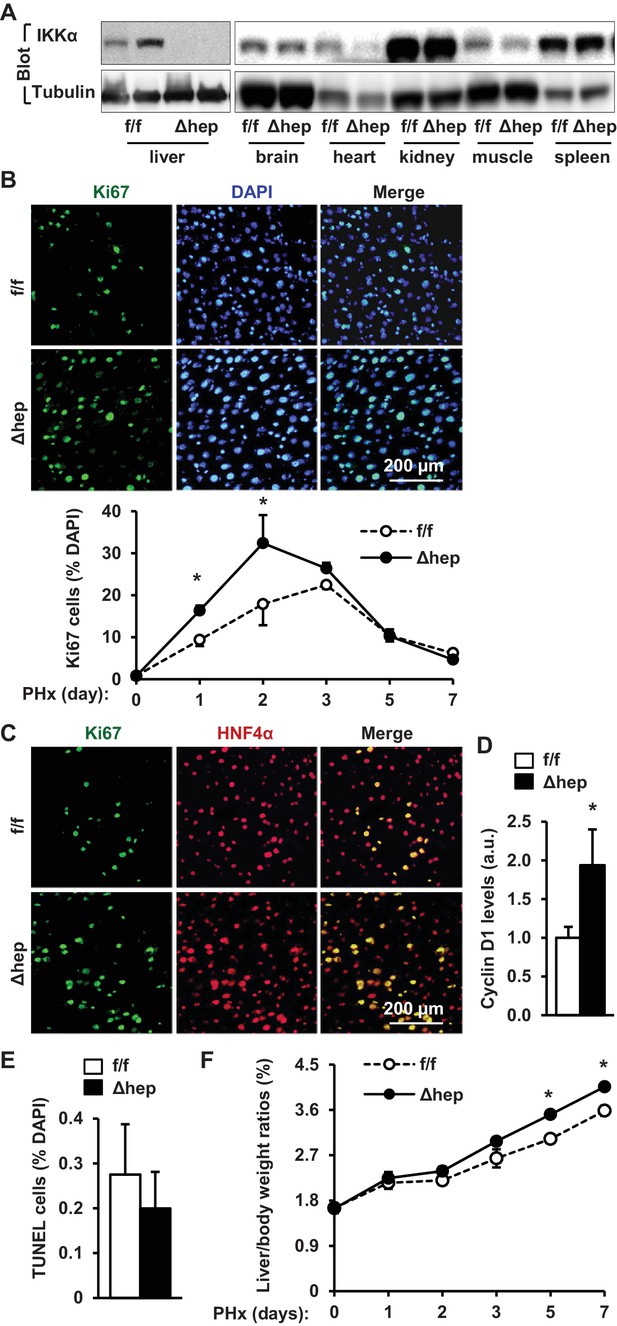
Ablation of hepatocyte IKKα accelerates hepatocyte reparative proliferation.
(A) Tissue extracts were immunoblotted with anti-IKKα or anti-α-tubulin antibodies. (B–F) IKKαf/f (n = 6) and IKKαΔhep (n = 6) male littermates were subjected to PHx, and livers were harvested 48 hr later. (B) Liver sections were immunostained with anti-Ki67 antibody, and Ki67+ cells were counted and normalized to total DAPI+ cells. Day 0 and 1: n = 4 per group; day 3: IKKαf/f: n = 6, IKKαΔhep: n = 8; day 5: IKKαf/f: n = 9, IKKαΔhep: n = 8; day 7: IKKαf/f: n = 6, IKKαΔhep: n = 5. (C) Representative images of liver sections costained with anti-Ki67 and anti-HNF4α antibodies. (D) Liver cyclin D1 was measured by immunoblotting (normalized to α-tubulin levels). (E) TUNEL-positive cells in liver sections. (F) Liver to body weight ratios. Day 0 and 1: n = 4 per group; day 3: IKKαf/f: n = 6, IKKαΔhep: n = 8; day 5: IKKαf/f: n = 9, IKKαΔhep: n = 8; day 7: IKKαf/f: n = 6, IKKαΔhep: n = 5. Data were statistically analyzed with two-tailed Student’s t test, and presented as mean ± SEM. *p<0.05.
-
Figure 4—source data 1
Hepatic IKKα regulates liver regeneration.
- https://doi.org/10.7554/eLife.34152.012
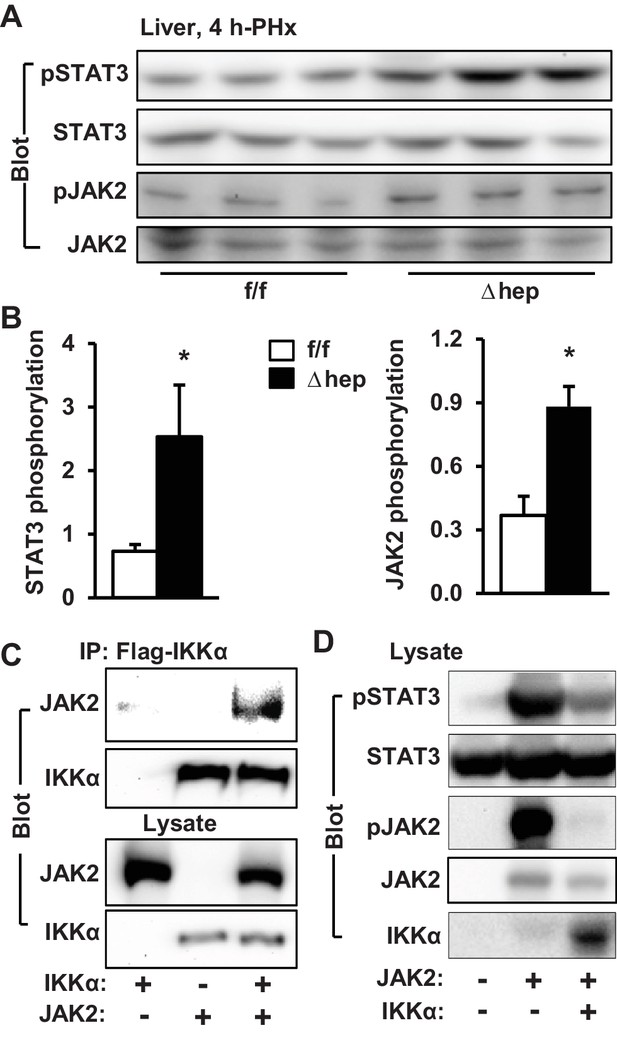
IKKα inhibits the JAK2/STAT3 pathway.
(A–B) Liver extracts were prepared 4 hr after PHx and immunoblotted with anti-phospho-JAK2 and anti-phospho-STAT3 antibodies. Phosphorylation of JAK2 (pTyr1007/1008) and STAT3 (pTyr705) was normalized to total JAK2 and STAT3 levels, respectively. IKKαf/f: n = 6, IKKαΔhep: n = 6. (C) IKKα and JAK2 were coexpressed in HEK293 cells. Cell extracts were immunoprecipitated (IP) and immunoblotted with the indicated antibodies. (D) STAT3 and JAK2 were coexpressed with IKKα in HEK293 cells. Cell extracts were immunoblotted with the indicated antibodies. Data were statistically analyzed with two-tailed Student’s t test, and presented as mean ± SEM. *p<0.05.
-
Figure 5—source data 1
IKKα regulates the JAK2/STAT3 pathway.
- https://doi.org/10.7554/eLife.34152.017
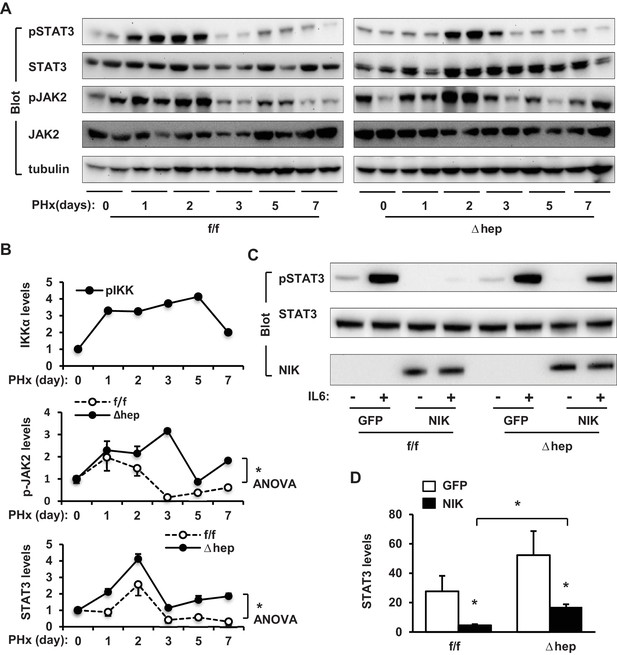
The effect of PHx on activation of liver IKKα and JAK2/STAT3 pathways.
IKKαf/f and IKKαΔhep male mice were subjected to PHx, and livers were harvested on days 0–7. (A) Liver extracts were immunoblotted with the indicated antibodies. (B) Phosphorylation of JAK2 (normalized to total JAK2 levels) and STAT3 (normalized to total STAT3) were analyzed using ANOVA (n = 3 per group). IKKα phosphorylation (Figure 2—figure supplement 1C) was replotted here. (C–D) Primary hepatocytes were transduced with GFP or NIK adenoviral vectors and stimulated with IL6 (10 ng/ml) for 15 min. Cell extracts were immunoblotted with the indicated antibodies. Phosphorylation of STAT3 were normalized to total STAT3 (n = 3 per group). Data were statistically analyzed with two-tailed Student’s t test, and presented as mean ± SEM. *p<0.05.
-
Figure 5—figure supplement 1—source data 1
PHx regulates the hepatic JAK2/STAT3 pathway.
- https://doi.org/10.7554/eLife.34152.015
-
Figure 5—figure supplement 1—source data 2
Regulation of the JAK2/STAT3 pathway by NIK/IKKα pathways.
- https://doi.org/10.7554/eLife.34152.016
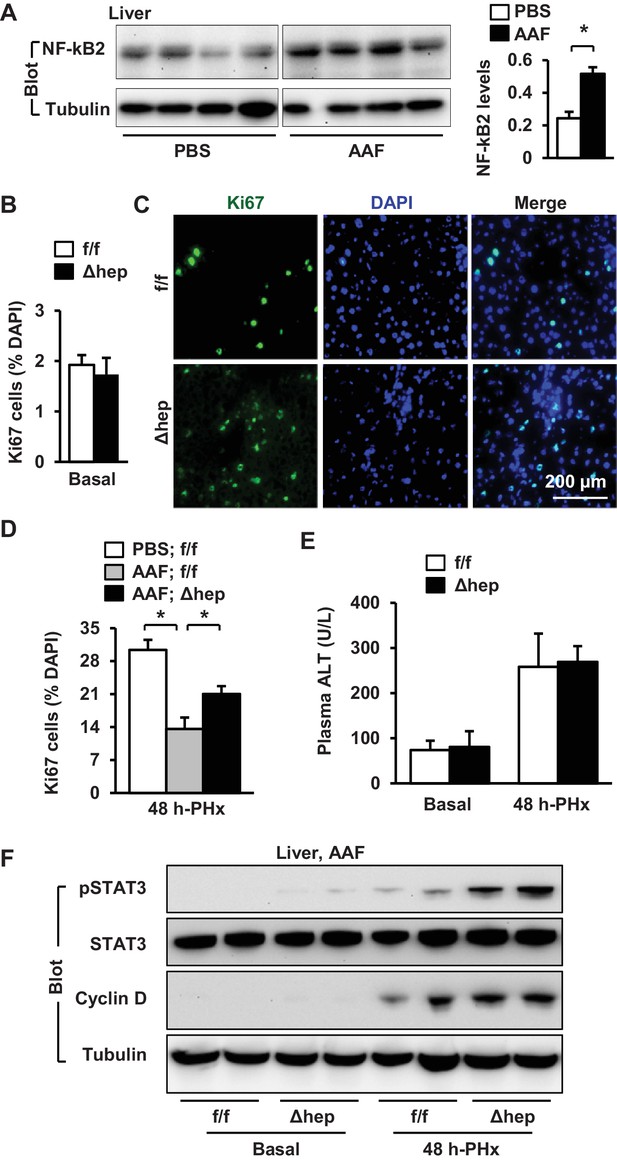
Ablation of hepatocyte NIK reverses AAF-induced impairment in hepatocyte reparative proliferation.
(A) C57BL/6 males (8 weeks) were treated with PBS or AAF (10 mg/kg body weight, gavage) daily for 10 days. NF-kB2 p52 in liver extracts was immunoblotted with anti-NF-kB2 antibody (normalized to α-tubulin levels). PBS: n = 4, AAF: n = 4. (B–G) NIKf/f and NIKΔhep males were treated with PBS or AAF (10 mg/kg body weight) for 10 days and then subjected to PHx. Livers were harvested 48 hr later. (B) Baseline Ki67+ cell number in resected liver sections obtained from PHx. NIKf/f: n = 5, NIKΔhep: n = 4. (C) Representative immunostaining of liver sections (AAF-treated) with anti-Ki67 antibody. (D) Ki67+ cell number in liver sections (normalized to DAPI+ cells). PBS;NIKf/f: n = 3, AAF;NIKf/f: n = 5, AAF;NIKΔhep: n = 5. (E) Plasma ALT levels. NIKf/f: n = 3, NIKΔhep: n = 4. (F) Liver extracts were immunoblotted with the indicated antibodies. Data were statistically analyzed with two-tailed Student’s t test, and presented as mean ± SEM. *p<0.05.
-
Figure 6—source data 1
Hepatic NIK regulates hepatocyte proliferation in AAF-treated mice.
- https://doi.org/10.7554/eLife.34152.022
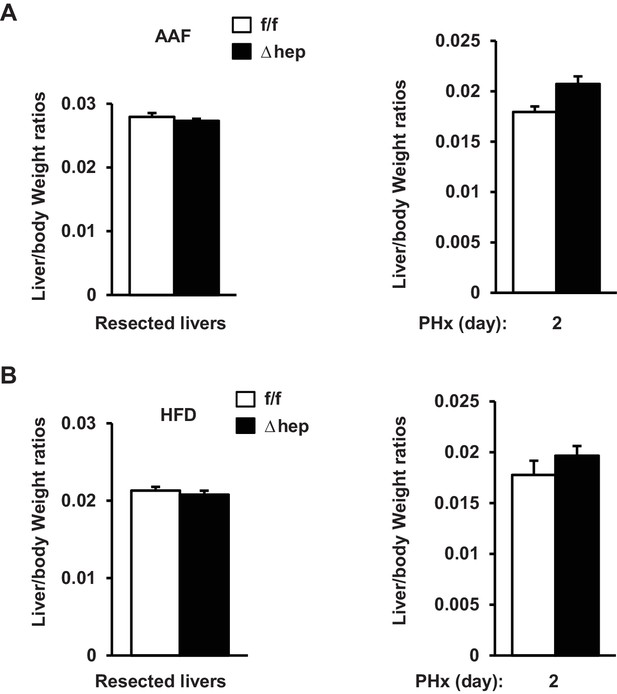
Hepatic NIK inhibits reparative hepatocyte proliferation.
(A) NIKf/f and NIKΔhep males were treated with PBS or AAF (10 mg/kg body weight) for 10 days and then subjected to PHx. Resected liver weight and liver weight 2 days post-PHx were normalized to body weight. Resected: n = 3 per group; PHx: n = 5 per group. (B) NIKf/f and NIKΔhep males were fed a HFD for 10 weeks followed by PHx. Resected liver weight and liver weight 2 days post-PHx were normalized to body weight. Resected: n = 5 per group; PHx: n = 5 per group. Data were statistically analyzed with two-tailed Student’s t test, and presented as mean ± SEM.
-
Figure 6—figure supplement 1—source data 1
Baseline hepatocyte proliferation in AAF-treated mice.
- https://doi.org/10.7554/eLife.34152.020
-
Figure 6—figure supplement 1—source data 2
Baseline hepatocyte proliferation in HFD-fed mice.
- https://doi.org/10.7554/eLife.34152.021
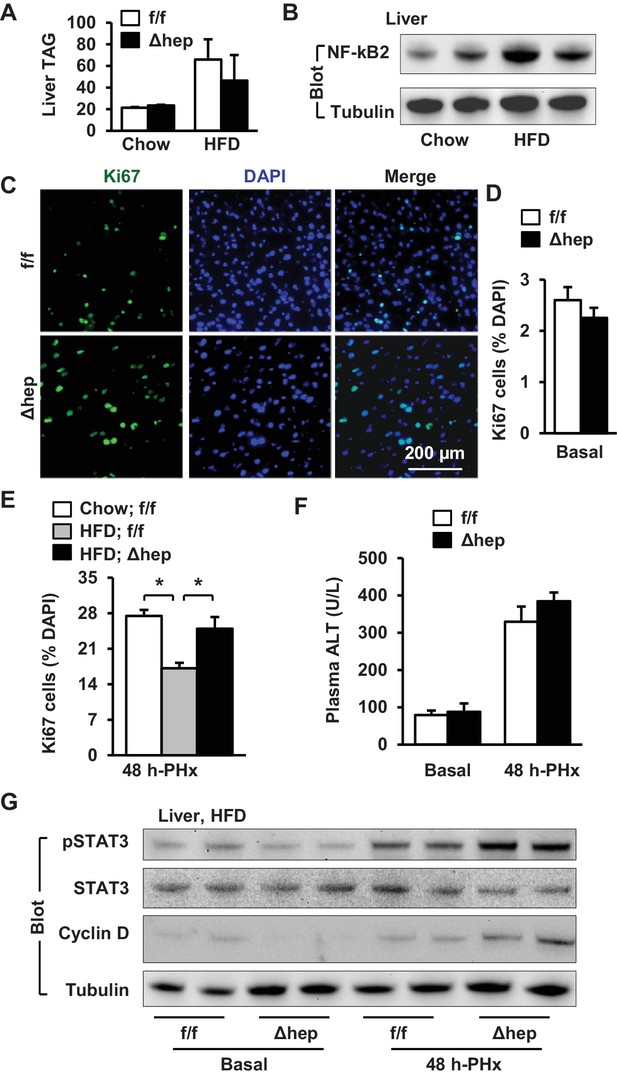
Hepatic NIK deficiency corrects impaired hepatocyte reparative proliferation in mice with NAFLD.
(A–B) C57BL/6 males (8 weeks) were fed a normal chow diet (n = 5) or a HFD (n = 5) for 10 weeks. (A) Liver TAG levels (normalized to liver weight). (B) NF-kB2 p52 in liver extracts was immunoblotted with anti-NF-kB2 antibody (normalized to α-tubulin levels). (C–H) NIKf/f and NIKΔhep males were fed a HFD for 10 weeks followed by PHx, and livers were harvested 48 hr after PHx. (C) Representative immunostaining of liver sections with anti-Ki67 antibody. (D) Baseline Ki67+ cell number in resected liver sections obtained from PHx. NIKf/f: n = 4, NIKΔhep: n = 4. (E) Liver Ki67+ cell number (normalized to DAPI+ cells). Chow;NIKf/f: n = 3, HFD;NIKf/f: n = 5, HFD; NIKΔhep: n = 4. (F) Plasma ALT levels. NIKf/f: n = 3, NIKΔhep: n = 4. (G) Liver extracts were immunoblotted with the indicated antibodies. Data were statistically analyzed with two-tailed Student’s t test, and presented as mean ± SEM. *p<0.05.
-
Figure 7—source data 1
Hepatic NIK regulates hepatocyte proliferation in HFD-fed mice.
- https://doi.org/10.7554/eLife.34152.024
Tables
| Reagent type | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Ki67 | Vector lab | VP-RM04 | 1:100 |
| Antibody | NIK | Abcam | ab191591 | 1:2000 |
| Antibody | IKK beta | Cell Signaling Technology | 8943 | 1:5000 |
| Antibody | IKK alpha | Cell Signaling Technology | 2682 | 1:5000 |
| Antibody | p-IKKa/b | Cell Signaling Technology | 2697 | 1:5000 |
| Antibody | STAT3 | Santa Cruz | sc-8019 | 1:1000 |
| Antibody | p-STAT3 | Cell Signaling Technology | 9145 | 1:5000 |
| Antibody | JAK2 | Cell Signaling Technology | 3230 | 1:5000 |
| Antibody | p-JAK2 1007/1008 | Cell Signaling Technology | 3776 | 1:5000 |
| Antibody | Myc | Santa Cruz | sc-40 | 1:1000 |
| Antibody | Flag | Sigma | F1804 | 1:5000 |
| Antibody | p85 | Home-raised | N/A | 1:5000 |
| Antibody | α-tubulin | Santa Cruz | sc-5286 | 1:1000 |
| Antibody | JNK | Cell Signaling Technology | 9258 | 1:5000 |
| Antibody | p-JNK | Cell Signaling Technology | 4668 | 1:5000 |
| Antibody | ERK1/2 | Cell Signaling Technology | 9102 | 1:5000 |
| Antibody | p-ERK1/2 | Cell Signaling Technology | 4370 | 1:5000 |
| Antibody | NF-kB2 | Cell Signaling Technology | 4882 | 1:5000 |
| Antibody | p65 | Cell Signaling Technology | 8242 | 1:5000 |
| Antibody | p-p65 | Cell Signaling Technology | 3033 | 1:5000 |
| Antibody | IkB alpha | Cell Signaling Technology | 4812 | 1:5000 |
| Antibody | p-IkB alpha | Cell Signaling Technology | 9246 | 1:5000 |
| Antibody | AKT | Cell Signaling Technology | 4091 | 1:5000 |
| Antibody | p-Akt | Cell Signaling Technology | 4060 | 1:5000 |
| Antibody | Cyclin D1 | Cell Signaling Technology | 2978 | 1:5000 |
| Antibody | F4/80 | eBioscience | 14–4801 | 1:100 |
| Antibody | HNF4 alpha | Santa Cruz | sc-8987 | 1:100 |
| Antibody | CK8 | Developmental Studies Hybridoma Bank | Troma I | 1:100 |
| Antibody | BrDU | Cell Signaling Technology | 5292 | 1:100 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.34152.025





