Synthetic single domain antibodies for the conformational trapping of membrane proteins
Figures
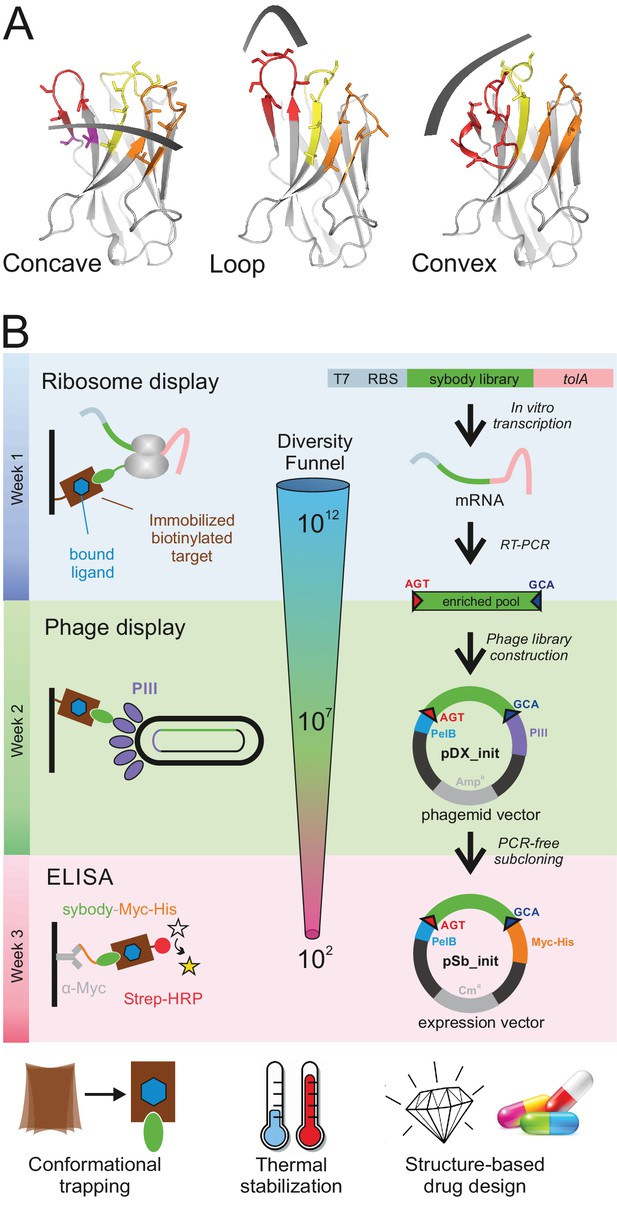
Selection of sybodies against membrane proteins within three weeks.
(A) Three synthetic libraries exhibiting highly variable randomized surfaces (concave, loop and convex) each harboring a diversity of 9 × 1012 were designed based on thermostabilized nanobody frameworks. CDR1, CDR2 and CDR3 are colored in yellow, orange and red, respectively. (B) The in vitro selection platform is built as a selection cascade, starting with 1012 sybodies displayed on ribosomes for pre-enrichment, followed by a focused phage display library of 107 clones and binder identification by ELISA (typically 96 clones). The platform builds on fragment exchange (FX) cloning using Type IIS restriction sites encoded on the phage display (pDX_init) and expression vector (pSb_init) backbones, which generate AGT and GCA sticky ends for PCR-free subcloning. Key elements for reliable selections against membrane proteins are the shape variability of the sybody libraries, exceptionally high experimental diversities using ribosome display and the change of display system during the selection process.
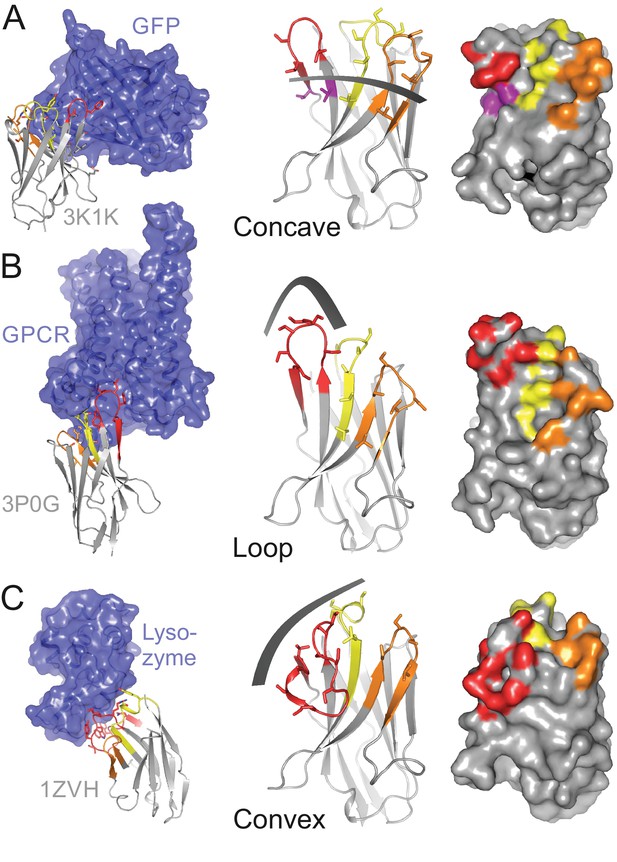
Variable sybody scaffolds based on three camelid nanobodies.
CDR1, CDR2 and CDR3 are colored in yellow, orange and red, respectively. In the left panel, crystal structures of camlid nanobodies in complex with GFP (PDB: 3K1K) (A), a GPCR (PDB: 3P0G) (B) and Lysozyme (PDB: 1ZVH) (C) are shown, which served as starting point to delineate scaffolds for randomization. Nanobody residues contacting the target proteins are depicted as sticks. The target proteins are colored in blue. In the middle panel, homology models of three framework nanobodies are shown as cartoons and randomized residues (defined as serines and threonines in these examples) are highlighted as sticks. The three sybody libraries exhibit a concave (A), loop (B) or convex (C) binding surface, respectively. The right panel shows the randomized surface of the three libraries with the side chains of the randomized positions highlighted in color. Note that the concave library contains randomized residues outside of the CDR regions, which are colored in purple.
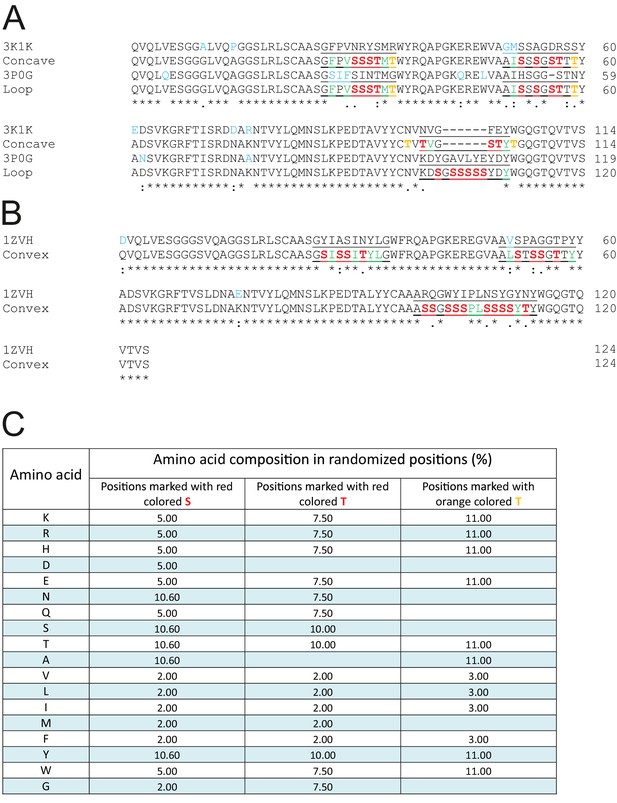
Framework sequences and randomized positions.
(A and B) Sequences of the framework sybodies are aligned with the sequences of their natural precursors. The frameworks of the concave and the loop library are identical (A) while the convex library has its own scaffold (B). Residues of the natural precursor nanobodies differing from the framework sequence are marked in blue. The three CDR regions are underlined. Invariant CDR residues contributing to the hydrophobic core of the respective scaffold are marked in green. Note that the differently shaped libraries exhibit alternative sets of invariant CDR residues that precisely match the corresponding scaffolds. This harmonization is a critical and unprecedented feature of our synthetic nanobdy libraries, as it allows for the first time to include variable CDR lengths without the risk of scaffold destabilization. Randomized residues are highlighted as red S (for which randomization mixture one was used), as red T (mix 2) and orange T (mix 3). (C) Amino acid composition of randomized positions obtained by three different trinucleotide randomization mixtures. The rationale behind the three randomization mixtures is provided in the main text.
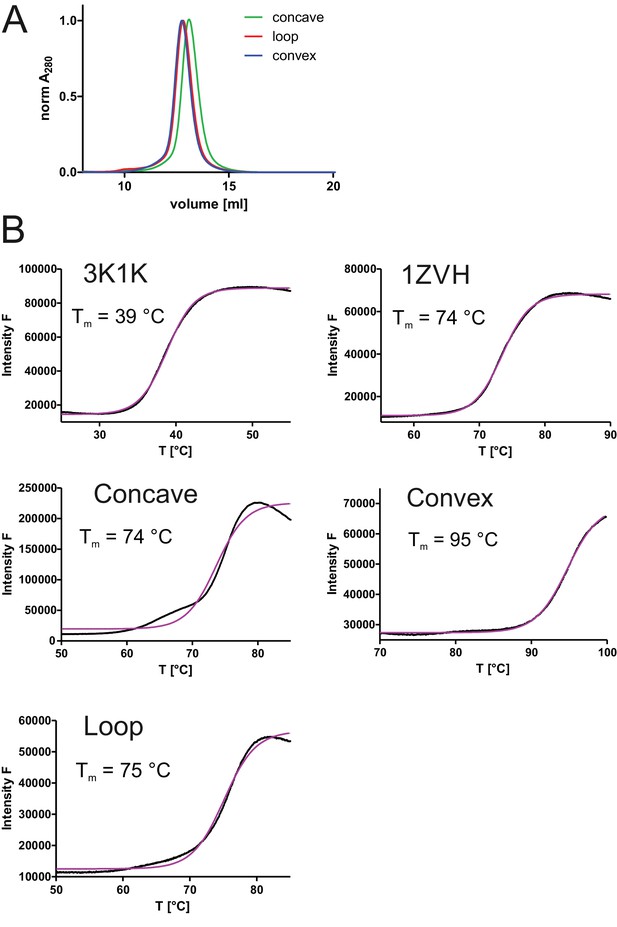
Biophysical characterization of sybodies.
Three framework sybodies representing the concave, the loop and the convex library and containing serines and threonines in the randomized positions were generated by gene synthesis (sequences provided in Figure 1—figure supplement 2). (A) SEC analysis of periplasmatically expressed concave, loop and convex framework sybodies using a Superdex 75 300/10 GL column. (B) Determination of melting temperature (Tm) of framework sybodies and their natural precursors 3K1K and 1ZVH using dye SYPRO Orange (ThermoFluor). Representative data of two technical replicates are shown.
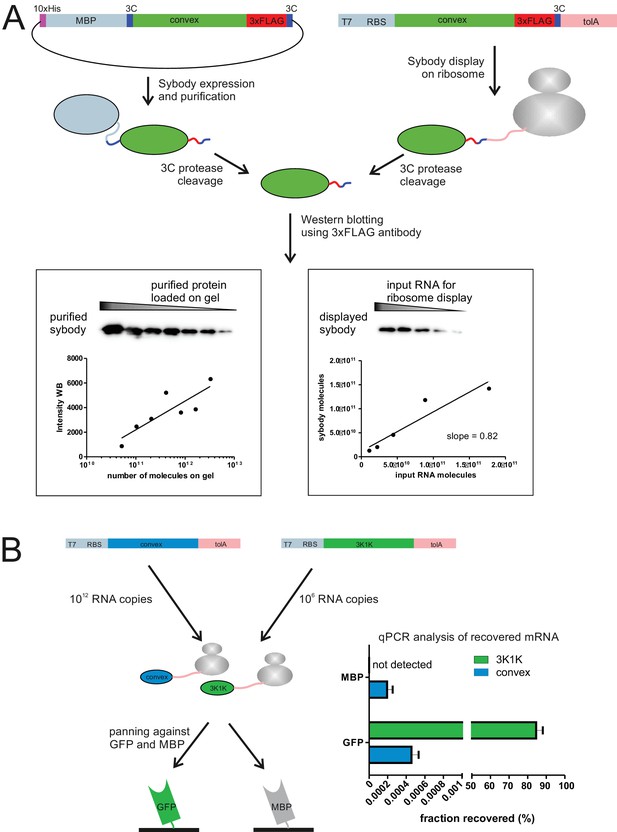
Ribosome display of single domain antibodies.
(A) The non-randomized convex sybody was either purified containing a C-terminal 3x-FLAG tag or displayed on ribosomes containing the same tag using the commercial kit PUREfrexSS (GeneFrontier). 3C protease cleavage was used to liberate the displayed sybody from the ribosomal complex. Western blotting analysis using anti-3x-FLAG antibody and purified sybody as standard revealed a display efficiency of 82% of input mRNA for ribosome display. (B) 106 mRNA molecules encoding the GFP-specific 3K1K nanobody were displayed on ribosomes using PUREfrexSS together with 1012 mRNA molecules encoding the non-randomized convex sybody. The ribosomal complexes were pulled down using either biotinylated GFP or MBP immobilized on magnetic beads. The mRNA of isolated ribosomal complexes was isolated, reverse transcribed and the resulting cDNA was analyzed by qPCR performing technical triplicates. This analysis revealed that 84.6 ± 3.5% (error corresponds to standard deviation) of the input 3K1K mRNA was retrieved on GFP-coated beads, while virtually no background binding of the non-randomized convex sybody nor 3K1K binding to MBP was observed.
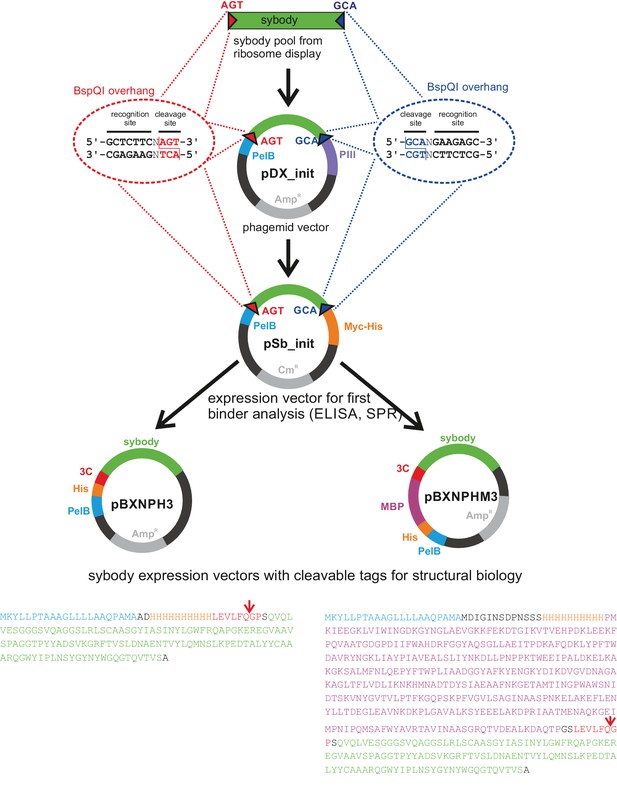
FX cloning vector series for phage display and purification of sybodies and nanobodies.
Sybody pools from ribosome display (or nanobodies from immunized camelids) are amplified with primers containing restriction sites of Type IIS enzyme BspQI (isoschizomer of SapI) to generate AGT and GCA overhangs. BspQI restriction sites generating the same overhangs were introduced into the backbones of vector pDX_init for phage display and pSb_init for periplasmatic expression and attachment of Myc- and His-tag. Note that in pDX_init and pSb_init the BspQI restriction sites are part of the sybody open reading frame. Finally, sybodies/nanobodies are sub-cloned from pSb_init to the destiny vectors pBXNPH3 or pBXNPHM3 for periplasmic expression. Tag-less sybodies/nanobodies for structural biology purposes can be obtained by 3C protease cleavage. Importantly, the vector series permits for PCR-free subcloning once the sybodies have been inserted into phage display vector pDX_init. The vectors were made available through Addgene (for Addgene IDs, see Table 3).
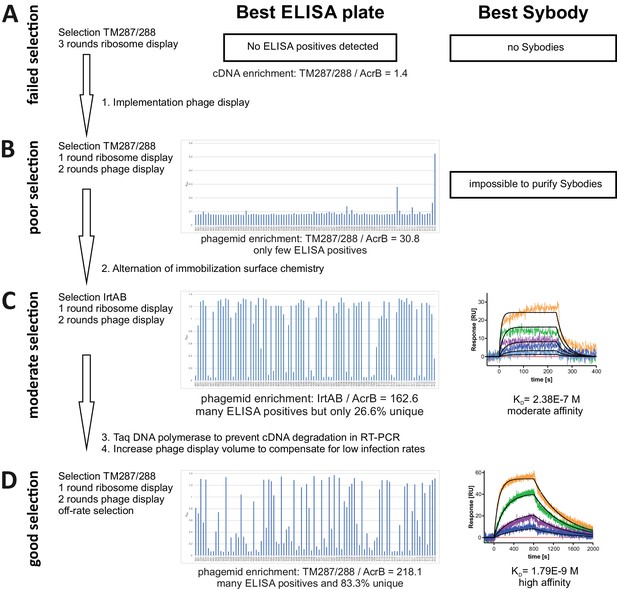
Improvement of the sybody selection procedure.
(A) Three rounds of ribosome display using the same type of magnetic beads for target immobilization (Dynabeads Myone Streptavidin T1) failed to generate sybodies against ABC transporter TM287/288. Pool enrichment against TM287/288 compared to negative control AcrB was poor. No positive ELISA hits were identified. (B) Sybody selections against TM287/288 were performed applying one round of ribosome display followed by two rounds of phage display using Dynabeads Myone Streptavidin T1 for target immobilization. The pool was enriched approximately 30 fold and a few positive ELISA hits were found. Purification of identified sybodies failed. (C) Sybody selections against ABC transporter IrtAB, a homologue of TM287/288 sharing a sequence identity of 27%, was performed as in (B), but using different immobilization chemistries (Dynabeads Myone Streptavidin T1 for ribosome display, Maxisorp microtiter plates for the first phage display round and Dynabeads Myone Streptavidin C1 for the second phage display round) to suppress accumulation of background binders. Strong enrichment was observed and a high number of positive ELISA hits were identified. Only 27% of positive ELISA hits were unique sybodies with moderate affinities. (D) Final optimized sybody selection protocol as described in the materials and methods section. Diversity bottlenecks were removed by using Taq DNA polymerase for cDNA amplification and increasing the working volume of the first phage display round. An off-rate selection step was introduced in the second phage display round. Enrichment and number of ELISA hits was similar to the selection shown in (C). The number of unique ELISA hits increased to 83% and high affinity binders were obtained. The binders obtained in (D) against TM287/288 are described in detail in main Figures 3 and 4.
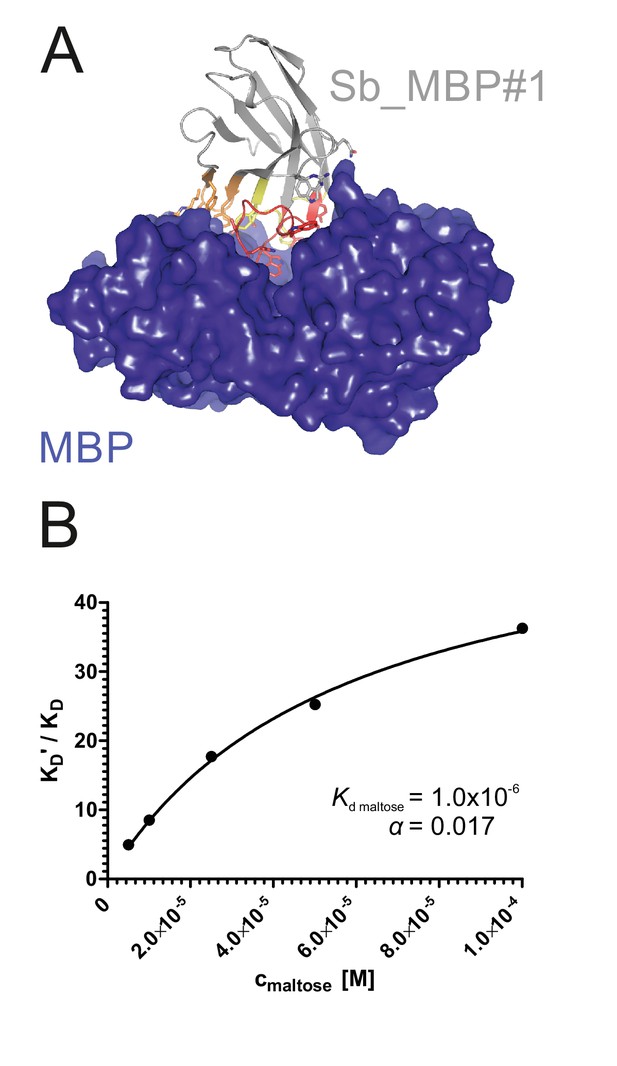
Structural and biochemical characterization of convex sybody Sb_MBP#1.
(A) Crystal structure of the Sb_MBP#1/MBP complex. MBP is shown as blue surface, the convex sybody Sb_MBP#1 is shown as grey cartoon with CDRs 1–3 colored in yellow, orange and red, respectively. Sybody residues mediating contacts to MBP are shown as sticks. (B) Maltose and sybody Sb_MBP#1 compete for binding to MBP. In the depicted Schild analysis, the sybody affinity ratios determined in the presence (KD’) and absence (KD) of maltose is plotted against the maltose concentration. The binding affinity for maltose KD,maltose was determined as 1.0 µM. The allosteric constant α amounts to 0.017, that is the ratio KD’/KD saturates at a value of 58.
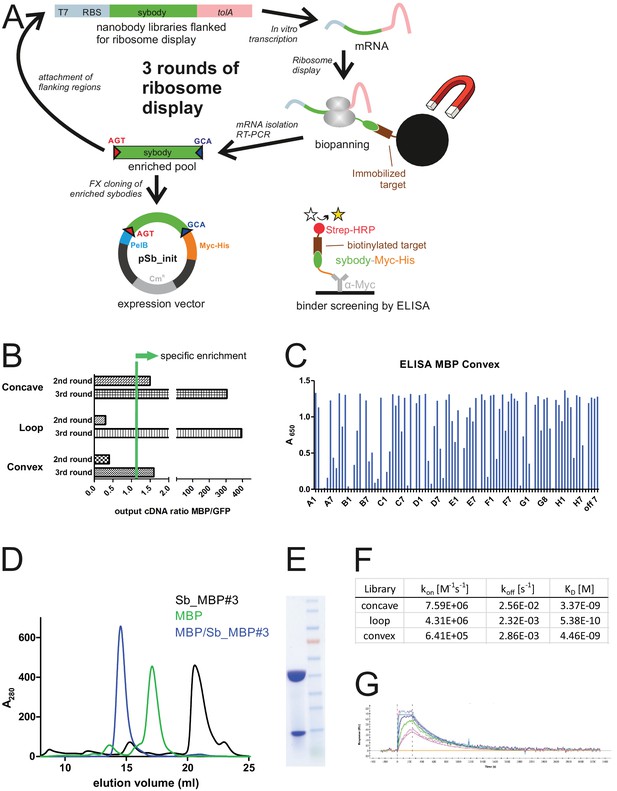
Sybody selections against MBP.
(A) Sybodies were selected against MBP using three rounds of ribosome display and MBP immobilized on magnetic beads. Sybodies were expressed in pSb_init and analyzed by ELISA. (B) Binder enrichment was monitored using qPCR by comparing the cDNA output after panning against the target MBP versus the control protein GFP. (C) ELISA analysis of convex pool after selection round 3. MBP-specific DARPin off7 was used as positive control (Binz et al., 2004). (D) SEC analysis of sybody Sb_MBP#3 alone and in complex with MBP using a Superdex 200 300/10 GL column. (E) SDS-PAGE analysis of Sb_MBP#3/MBP complex after SEC. (F) KD, kon and koff values of the highest affinity sybodies obtained from the concave, loop and convex library, as measured by SPR. (G) SPR traces of the loop sybody exhibiting an affinity of 0.5 nM.
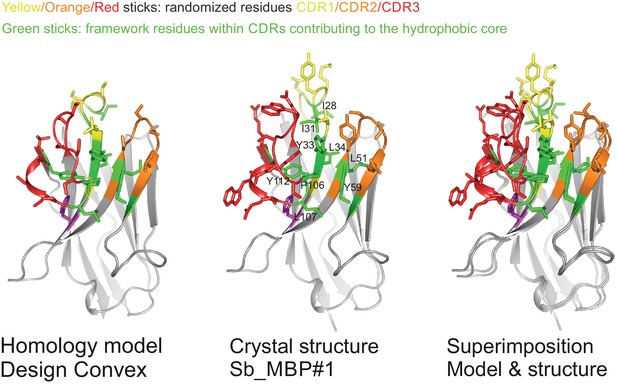
Validation of sybody library design.
Comparison of homology model of non-randomized convex sybody based on the coordinates of 1ZVH with the structure of selected convex sybody Sb_MBP#1 (determined in complex with MBP). CDR residues contributing to the hydrophobic core are highlighted as green sticks, randomized residues as sticks colored in yellow, orange and red for CDR1, CDR2 and CDR3, respectively.
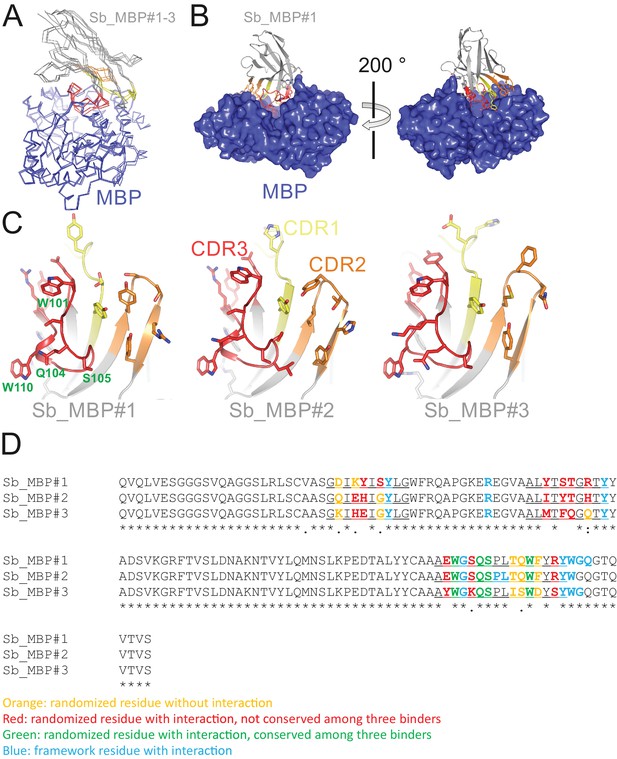
Detailed analysis of sybody-MBP complex structures.
(A) Structures of MBP (blue) in complex with Sb_MBP#1–3 (grey with CDR1, CDR2 and CDR3 in yellow, orange and red, respectively). The coordinates of MBP were used to perform superimposition. (B) Interaction of Sb_MBP#1 with MBP, shown along the MBP cleft from both sides. Sybody residues contacting MBP (distance ≤4 Å) are shown as sticks. (C) Detailed view of interacting residues of sybodies Sb_MBP#1–3. In the left panel, four randomized residues of CDR3 which are invariant among the three binders are labeled. (D) Sequence alignment of Sb_MBP#1–3. The CDR regions are underlined.
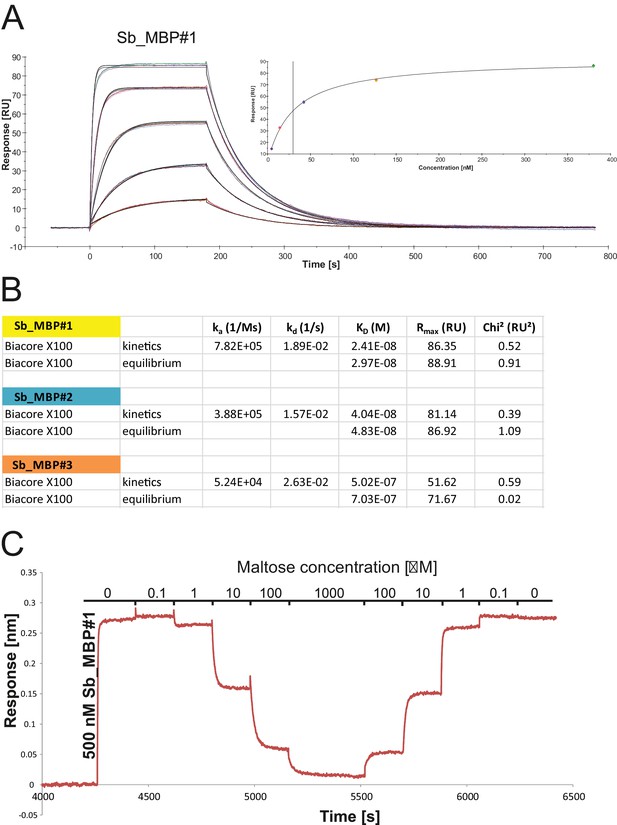
Biophysical analysis of sybody-MBP interactions.
(A) SPR analysis of interaction between sybody Sb_MBP#1 (analyte) and biotinylated MBP (ligand), determined as technical triplicates for each analyte concentration. Concentrations of Sb_MBP#1: 0, 4.7, 14.1, 42.2, 126.7, 380 nM. Data were fitted with a 1:1 binding model to obtain kon, koff and KD, kinetics. Inset shows binding equilibrium data to determine KD, equilibrium. Sybodies Sb_MBP#2 and Sb_MBP#3 were analyzed accordingly. (B) Data table summarizing the values obtained from SPR analysis shown in (A). (C) Displacement of 500 nM Sb_MBP#1 bound to immobilized MBP by addition of increasing maltose concentrations was monitored using the Octet RED96 System.
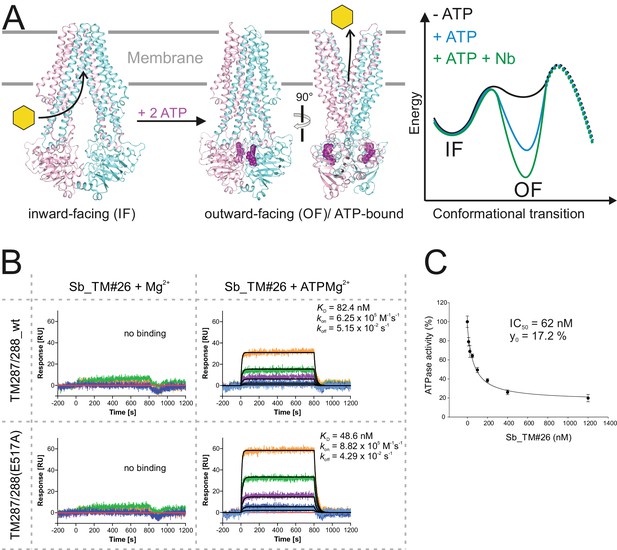
Conformational trapping of ABC transporter TM287/288.
(A) In the absence of nucleotides, ABC transporter TM287/288 adopts its inward-facing (IF) state and captures substrates from the cytoplasm. ATP binding is required to achieve a partial population of the outward-facing (OF) state, which allows for substrate exit to the cell exterior. Sybodies were selected in the presence of ATP against the transporter mutant TM287/288(E517A), which is incapable of ATP hydrolysis and predominantly populates the OF state in this condition. (B) SPR analysis of loop sybody Sb_TM#26 in the presence and absence of ATP using wildtype TM287/288 and TM287/288(E517A) as ligands. Concentrations of Sb_TM#26: 0, 1, 3, 9, 27, 81 nM. (C) ATPase activities of wildtype TM287/288 at increasing concentrations of Sb_TM#26. Error bars report the standard deviation of technical triplicates. IC50 corresponds to the sybody concentration required for half-maximal inhibition and y0 to the residual ATPase activity at saturating sybody concentrations.
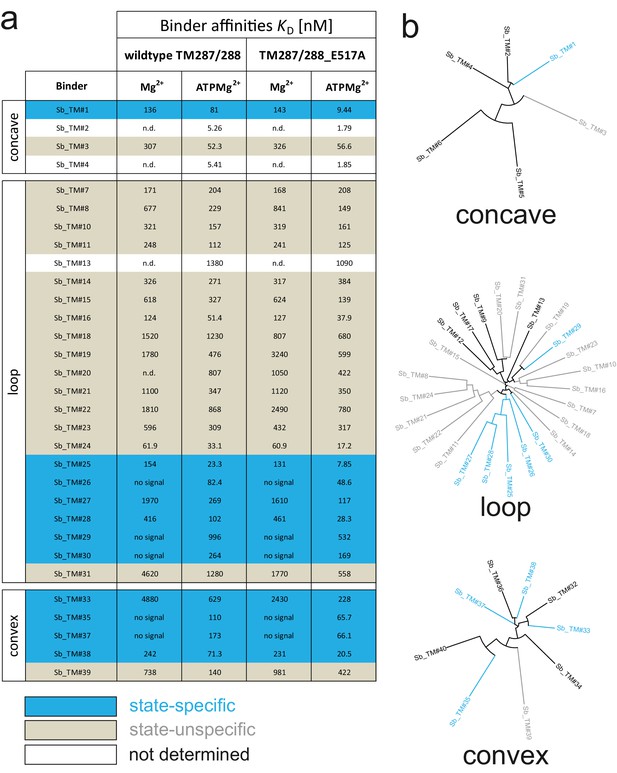
Analysis of sybodies raised against ABC transporter TM287/288.
(A) Binding affinities of 31 sybodies belonging to the concave, loop and convex library were determined by kinetic SPR measurements using the ProteOn XPR36 Protein Interaction Array System in the presence and absence of ATP and using wildtype TM287/288 and the ATPase-deficient mutant TM287/288(E517A) as ligands. Binders which exhibit an affinity increase of at least ten-fold against TM287/288(E517A) in the presence of ATP were defined as state-specific and are marked in blue. (B) Phylogenetic trees of sybodies specific against TM287/288 as determined by ELISA. Note that some of the sybodies were not analyzed by SPR either due to low yields during purification or poor SPR data.

Sequence alignment of concave sybodies raised against TM287/288.
https://doi.org/10.7554/eLife.34317.020
Sequence alignment of loop sybodies raised against TM287/288.
https://doi.org/10.7554/eLife.34317.021
Sequence alignment of convex sybodies raised against TM287/288.
https://doi.org/10.7554/eLife.34317.022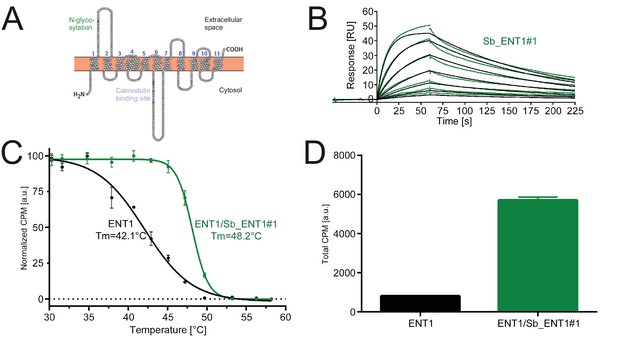
Conformation-specific binding of Sb_ENT1#1 to the inhibition state of human ENT1.
(A) Snake plot of human ENT1. (B) SPR analysis of Sb_ENT1#1 binding to biotinylated ENT1 revealing a KD of 40 nM. (C) Scintillation proximity assay thermal shift (SPA-TS) analysis of human ENT1 in the presence and absence of Sb_ENT1#1 using [3H]-NBTI inhibitor. Error bars correspond to standard deviations of technical triplicates. Sb_ENT1#1 stabilizes an inhibited conformation as evidenced by a shift of the apparent melting temperature (Tm) by 6.1°C and (D) a 7-fold increase of the absolute SPA signal measured at 30.1°C.
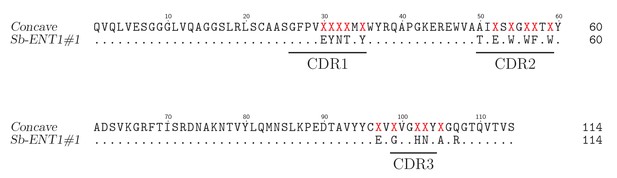
Sequence of Sb_ENT1#1.
https://doi.org/10.7554/eLife.34317.024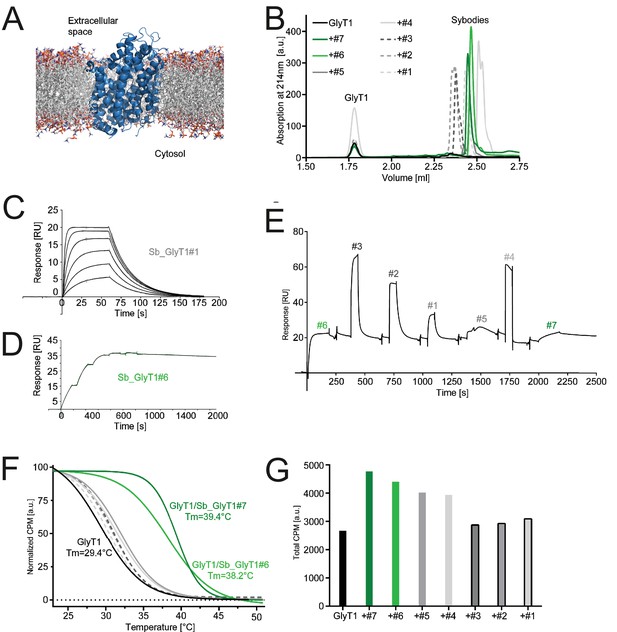
Inhibition-state specific sybodies against human GlyT1.
(A) Schematic of a GlyT1 homolog (PDP ID: 4M48) embedded in a lipid bilayer, illustrating the limited number of surface-accessible epitopes. (B) RP8-HPLC analysis of sybody-GlyT complexes previously separated by SEC. (C, D) SPR analysis of Sb_GlyT1#1 (KD = 307 nM) and Sb_GlyT1#6 (KD = 494 pM). Due to a slow off-rate, SPR analysis of Sb_GlyT1#6 was performed in a single cycle measurement. (E) SPR analysis reveals binding of Sb_GlyT1#1–4 to the GlyT1/Sb_GlyT1#6 complex, indicating the presence of two binding epitopes. Sb_GlyT1#5 and Sb_GlyT1#7 compete for binding with Sb_GlyT1#6. (F) SPA-TS analysis of Sb_GlyT1#1–7 using [3H]-Org24598 reuptake inhibitor. Shifts of the melting temperature (Tm) are highest for Sb_GlyT1#6 and Sb_GlyT1#7 with values of 8.8 and 10°C, respectively, and correlate well with (G) increased absolute SPA signals measured at 19°C.

Sequence alignment of sybodies raised against GlyT1.
https://doi.org/10.7554/eLife.34317.026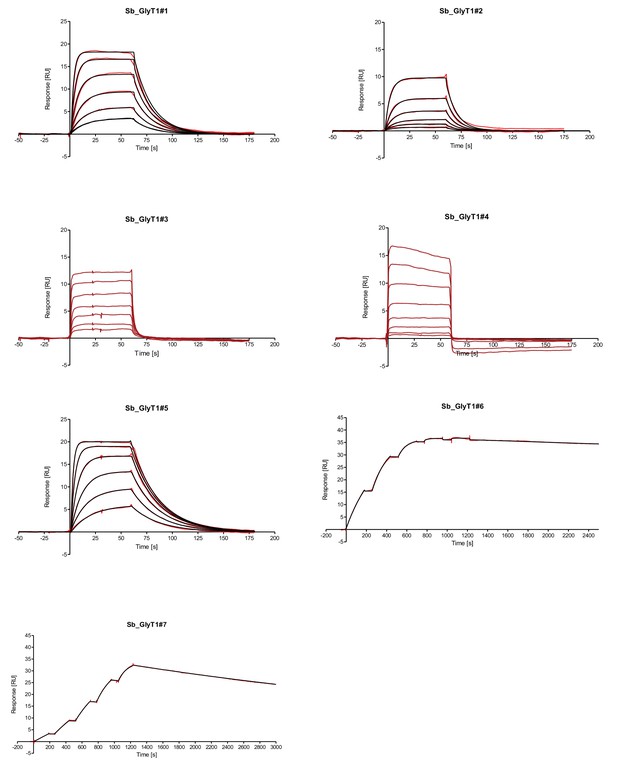
SPR analysis of sybodies raised against ENT1 and GlyT1.
Data were fitted using a 1:1 binding model. Representative data of replicates measured on two different SPR chips are shown.
Tables
Features of the three sybody libraries.
https://doi.org/10.7554/eLife.34317.009| Library | Template PDB entry/target | Binding interface in template | Length of CDR3 | Number of randomized residues in library | Theoretical diversity of library |
|---|---|---|---|---|---|
| concave | 3K1K/GFP | 672 Å2 | 6 aa | 15 | 8.3 × 1017 |
| loop | 3P0G/GPCR | 901 Å2 | 12 aa | 16 | 4.3 × 1019 |
| convex | 1ZVH/Lysozyme | 533 Å2 | 16 aa | 18 | 2.8 × 1022 |
DNA sequences of non-randomized sybodies and flanking regions for ribosome display
https://doi.org/10.7554/eLife.34317.010| Framework sequence concave | CAGGTTCAGCTGGTTGAGAGCGGTGGTGGCCTGGTCCAAGCTGGCGGTTCGCTGCGTCTGAGCTGCGCCGCAAGCGGTTT CCCGGTGAGCAGCAGCACGATGACCTGGTATCGTCAGGCACCGGGCAAAGAACGTGAGTGGGTCGCGGCGATTTCCAGCT CTGGTAGCACCACGACCTACGCAGATTCTGTTAAGGGCCGCTTTACCATCAGCCGCGACAACGCGAAGAATACGGTCTAT TTGCAGATGAATAGCCTGAAACCGGAAGATACCGCGGTTTACTACTGTACCGTGACCGTGGGTAGCACGTACACGGGCCA AGGTACCCAAGTGACTGTGAGC |
| Framework sequence loop | CAGGTTCAGCTGGTTGAGAGCGGTGGTGGCCTGGTCCAAGCTGGCGGTTCGCTGCGTCTGAGCTGCGCCGCAAGCGGTTT CCCGGTGAGCAGCAGCACGATGACCTGGTATCGTCAGGCACCGGGCAAAGAACGTGAGTGGGTCGCGGCGATTTCCAGCT CTGGTAGCACCACGACCTACGCAGATTCTGTTAAGGGCCGCTTTACCATCAGCCGCGACAACGCGAAGAATACGGTCTAT TTGCAGATGAATAGCCTGAAACCGGAAGATACCGCGGTTTACTACTGTAACGTGAAAGACAGCGGTAGCTCCAGCAGCTC CTACGACTATTGGGGCCAAGGTACCCAAGTGACTGTGAGC |
| Framework sequence convex | CAAGTCCAGCTGGTGGAATCGGGTGGTGGTAGCGTCCAGGCGGGTGGTAGCCTGCGTCTGAGCTGTGCGGCTAGCGGCTC TATTTCCAGCATCACGTACCTGGGCTGGTTTCGCCAGGCACCGGGCAAAGAGCGTGAGGGCGTCGCAGCGCTGAGCACCA GCTCCGGTACCACCTACTACGCGGACAGCGTTAAGGGTCGTTTCACGGTGAGCCTGGACAACGCCAAGAATACCGTGTAT CTGCAAATGAACAGCTTGAAACCGGAAGATACTGCTTTGTATTACTGCGCGGCAGCCAGCAGCGGCTCCAGCAGCCCGCT GTCTAGCAGCAGCTATACGTACTGGGGTCAGGGCACCCAAGTTACCGTTTCT |
| 5’ flank ribosome display | TAATACGACTCACTATAGGGAGACCACAACGGTTTCCCTCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATATCC ATGGGTAGT |
| 3’ flank ribosome display | GCAAAGCTTTATATGGCCTCGGGGGCCGAATTCGGATCTGGTGGCCAGAAGCAAGCTGAAGAGGCGGCAGCGAAAGCGGC GGCAGATGCTAAAGCGAAGGCCGAAGCAGATGCTAAAGCTGCGGAAGAAGCAGCGAAGAAAGCGGCTGCAGACGCAAAGA AAAAAGCAGAAGCAGAAGCCGCCAAAGCCGCAGCCGAAGCGCAGAAAAAAGCCGAGGCAGCCGCTGCGGCACTGAAGAAG AAAGCGGAAGCGGCAGAAGCAGCTGCAGCTGAAGCAAGAAAGAAAGCGGCAACTGAAGCTGCTGAAAAAGCCAAAGCAGA AGCTGAGAAGAAAGCGGCTGCTGAAAAGGCTGCAGCTGATAAGAAAGCGGCAGCAGAGAAAGCTGCAGCCGACAAAAAAG CAGCAGAAAAAGCGGCTGCTGAAAAGGCAGCAGCTGATAAGAAAGCAGCGGCAGAAAAAGCCGCCGCAGACAAAAAAGCG GCAGCGGCAAAAGCTGCAGCTGAAAAAGCCGCTGCAGCAAAAGCGGCCGCAGAGGCAGATGATATTTTCGGTGAGCTAAG CTCTGGTAAGAATGCACCGAAAACGGGGGGAGGGGCGAAAGGGAACAATGCTTCGCCTGCCGGGAGTGGTAATACTAAAA ACAATGGCGCATCAGGGGCCGATATCAATAACTATGCCGGGCAGATTAAATCTGCTATCGAAAGTAAGTTCTATGACGCA TCGTCCTATGCAGGCAAAACCTGTACGCTGCGCATAAAACTGGCACCCGATGGTATGTTACTGGATATCAAACCTGAAGG TGGCGATCCCGCACTTTGTCAGGCTGCGTTGGCAGCAGCTAAACTTGCGAAGATCCCGAAACCACCAAGCCAGGCAGTAT ATGAAGTGTTCAAAAACGCGCCATTGGACTTCAAACCGTAG |
FX cloning vectors for phage display and sybody production
https://doi.org/10.7554/eLife.34317.016| Vector name | Description | Resistance marker | Addgene ID |
|---|---|---|---|
| pDX_init | E.coli entry and expression vector for FX cloning system, N-terminal PelB signal sequence and C-terminal fusion to PIII for phage display using M13 phages. Nanobodies and sybodies are inserted and excised using SapI or BspQI. | Amp | #110101 |
| pSb_init | E.coli entry and expression vector for FX cloning system, N-terminal PelB signal sequence and C-terminal Myc- and 6xHis-tag. Nanobodies and sybodies are inserted and excised using SapI or BspQI. | Cm | #110100 |
| pBXNPH3 | E. coli expression vector for FX cloning system, N-terminal PelB signal sequence followed by 10xHisTag and 3C cleavage site. Nanobodies and sybodies are inserted using SapI or BspQI. | Amp | #110098 |
| pBXNPHM3 | E.coli expression vector for FX cloning system, N-terminal PelB signal sequence followed by 10xHisTag, maltose binding protein and 3C cleavage site | Amp | #110099 |
| SB_concave | pBXNPHM3 containing non-randomized framework sybody of the concave library | Amp | #110102 |
| SB_loop | pBXNPHM3 containing non-randomized framework sybody of the loop library | Amp | #110103 |
| SB_convex | pBXNPHM3 containing non-randomized framework sybody of the convex library | Amp | #110104 |
Data collection and refinement statistics
https://doi.org/10.7554/eLife.34317.017| Sb_MBP#1 (PDB: 5M13) | Sb_MBP#2 (PDB: 5M14) | Sb_MBP#3 (PDB: 5M15) | |
|---|---|---|---|
| Data Collection | |||
| Space group | P212121 (19) | P212121 (19) | P212121 (19) |
| Cell dimensions | |||
| a, b, c (Å) | 58.298 82.789 102.583 | 57.890 57.950 281.540 | 57.030 57.780 286.530 |
| α, β, γ (°) | 90.00 90.00 90.00 | 90.00 90.00 90.00 | 90.00 90.00 90.00 |
| Resolution (Å) | 50–1.37 | 50–1.6 | 50–1.9 |
| Rmeas (%) 1) | 6.5 (60.9) | 5.9 (124) | 7.8 (146.6) |
| I/σI | 15.26 (3.47) | 16.98 (1.82) | 21.44 (2.17) |
| CC1/2 (%) | 99.9 (86.3) | 99.9 (68.5) | 100 (60.5) |
| Completeness (%) | 99.4 (97.7) | 100 (100) | 100 (100) |
| Redundancy | 6.1 | 6.5 | 12.9 |
| Refinement | |||
| Resolution (Å) | 50–1.37 | 50–1.6 | 50–1.9 |
| No. reflections (work/test) | 102618/5131 | 126118/6307 | 75931/3797 |
| Rwork/Rfree (%) | 16.82/18.60 | 19.04/21.56 | 20.92/25.70 |
| No. atoms | |||
| Protein | 3873 | 7640 | 7619 |
| Water | 694 | 1040 | 422 |
| B-factor (Å2) | |||
| Total | 20.1 | 34.4 | 50.1 |
| R.m.s deviations | |||
| Bond lengths (Å) | 0.005 | 0.003 | 0.003 |
| Bond angles (°) | 0.750 | 0.591 | 0.623 |
-
1) Values in parentheses are for the last resolution shell
Characterization of sybodies raised against ENT1 and GlyT1.
https://doi.org/10.7554/eLife.34317.028| Sybody | kon [M−1S−1] | koff [s−1] | KD [M] (kinetics) | KD [M] (equilibrium) | ΔTm(SPA-TS) [°C] | SPA signal (fold increase) |
|---|---|---|---|---|---|---|
| Sb ENT1#1 | 1.86E+05 | 7.44E-03 | 4.00E-08 | 6.1 | 7 | |
| Sb_GlyT1#1 | 1.88E+05 | 5.77E-02 | 3.07E-07 | 0.9 | 1.2 | |
| Sb_GlyT1#2* | 3.68E+04 | 9.28E-02 | 2.52E-06 | 1.5 | 1.1 | |
| Sb_GlyT1#3** | 1.54E-07 | 1.7 | 1.1 | |||
| Sb_GlyT1#4** | 4.761E-07 | 2.1 | 1.5 | |||
| Sb_GlyT1#5 | 4.54E+05 | 3.72E-02 | 8.19E-08 | 2.6 | 1.5 | |
| Sb_GlyT1#6*** | 1.00E+05 | 4.99E-05 | 4.94E-10 | 8.8 | 1.6 | |
| Sb GlyT1#7*** | 2.01E+04 | 1.85E-04 | 9.18E-09 | 10 | 1.8 |
List of Primers
https://doi.org/10.7554/eLife.34317.029| Primers for library assembly (triplets designated 111, 222 and 333 correspond to the trinucleotide mixtures 1–3 for randomization; all primers in 5’ to 3’ orientation) | |
|---|---|
| CDR1_a_b | GCA AGC GGT TTC CCG GTG 111 111 111 222 ATG 333 TGG TAT CGT CAG GCA CCG G |
| CDR1_c | C TGT GCG GCT AGC GGC 111 ATT 111 111 ATC 222 TAC CTG GGC TGG TTT CGC C |
| CDR2_a_b | GA AGA CCT GTC GCG GCG ATT 111 AGC 111 GGT 111 222 ACG 333 TAC GCA GAT TCT GTT AAG GGC CG |
| CDR2_c | CGA AGA CCT GCA GCG CTG 111 ACC 111 111 GGT 222 ACC TAC TAC GCG GAC AGC G |
| CDR3_a | GA AGA CCT GCG GTT TAC TAC TGT 333 GTG 222 GTG GGT 111 222 TAC 333 GGC CAA GGT ACC CAA GTG AC |
| CDR3_b | CGC GAA GAC CTC GTG AAA GAC 111 GGT 111 111 111 111 111 TAC GAC TAT TGG GGC CAA GGT ACC CAA GTG AC |
| CDR3_c | GAA GAC CTC TGC GCG GCA GCC 111 111 GGC 111 111 111 CCG CTG 111 111 111 111 TAT 222 TAC TGG GGT CAG GGC ACC CAA GTT ACC GTT TCT |
| FW1_a_b_for | CAG GTT CAG CTG GTT GAG AGC |
| FW1_a_b_rev | CAC CGG GAA ACC GCT TGC |
| FW1_c_for | CAA GTC CAG CTG GTG GAA TCG |
| FW1_c_rev | GCC GCT AGC CGC ACA G |
| FW2_a_b_rev | ATG CAT GGT CTC ACG ACC CAC TCA CGT TCT TTG CCC GGT GCC TGA CGA TAC CA |
| FW2_c_rev | ATG CAT GGT CTC ACT GCG ACG CCC TCA CGC TCT TTG CCC GGT GCC TGG CGA AAC CAG CCC AGG |
| FW3_a_b_for | CGC AGA TTC TGT TAA GGG CCG |
| FW3_c_for | ACC TAC TAC GCG GAC AGC G |
| FW4_a_b_rev | GCT CAC AGT CAC TTG GGT ACC TTG GCC |
| FW4_c_rev | AGA AAC GGT AAC TTG GGT GCC CTG |
| Link1_a_b_for | ATG CAT GAA GAC CTG TCG CGG CG |
| Link1_a_b_rev | ATG CAT GGT CTC ACG ACC CAC |
| Link1_c_for | TAT ATC GAA GAC CTG CAG CGC TG |
| Link1_c_rev | ATG CAT GGT CTC ACT GCG ACG |
| Link2_a_for | TAT ATC GAA GAC CTG CGG TTT ACT ACT G |
| Link2_a_rev | ATG CAT GGT CTC ACC GCG GTA TCT TCC GGT TTC |
| Link2_b_for | ATG CAT GGT CTC ACC GCG GTA TCT TCC GGT TTC |
| Link2_b_rev | ATG CAT GGT CTC ACA CGT TAC AGT AGT AAA CCG CGG |
| Link2_c_for | ATA TAT GAA GAC CTC TGC GCG GC |
| Link2_c_rev | ATG CAT GGT CTC AGC AGT AAT ACA AAG CAG TAT CTT CCG G |
| Primers for vector construction | |
| pBXNPH3_#1 | CAG CAG TCC GGC AGC AGC GGT CGG CAG CAG GTA TTT CAT GGT TAA TTC CTC CTG TTA GCC |
| pBXNPH3_#2 | CTC CTC GCT GCC CAG CCT GCA ATG GCC GCA GAT CAC CAT CAT CAT CAC CAT CAT CAT CAT CAT TTA |
| pBXNPHM3_#1 | ATA TAT GCG GCC GCC ATA GTG ACT GGA TAT GTT G |
| pBXNPHM3_#2 | CAT GGT TAA TTC CTC CTG TTA GCC CAA AAA |
| pBXNPHM3_#3 | AAA TAC CTG CTG CCG ACC GCT GCT GCT GGT |
| pBXNPHM3_#4 | ATA TAT GCG GCC GCA TTA GGC ACC CCA GGC TTT A |
| pBXNPH3_blunt_for | CTC ATG ACC AAA ATC CCT TAA CGT GAG |
| pBXNPH3_EcoRI_rev | ATA TAT GAA TTC ATG GGG AGA CCC CAC ACT AC |
| pDX_init_#1 | ATA TAT GCT CTT CAA GCG GAA GAG AGC CCA ATA CGC AAA CCG |
| pDX_init_#2 | CGT TAG TAA ATG AAT TTT CTG TAT GAG GTT TTG |
| pDX_init_#3 | GAA CCT GAA GCC CAG TAC CCG TAC |
| pDX_init_#4 | CGT ACG GGT ACT GGG CTT CAG GTT |
| pDX_init_#5 | TAT AAC TTG AAG AGC CGG CTG CCA TGG CCG GCT GGG CC |
| pDX_init_#6 | TAT AGC AGG AAG AGC TCA CCA CCA TCA CCA TCA CGA ACC TG |
| pDX_init_#7 | TAT AGC TCT TCA AGT CTG CCC ACA TAT ACC TGC CGT TC |
| pDX_init_#8 | TAT AGC TCT TCC TGC AGA CAC GTG TCA CGT GAG GCC |
| Cm_EcoRI_for | GCT CAT GAA TTC CCC GCG CG |
| Cm_blunt_rev | GTG CAA TGT AAC ATC AGA GAT TTT GAG ACA C |
| Nb_init_for | ATG CAG GAA GAG CTG GCG AAC AAA AAC TCA TCT CAG AAG AGG ATC TG |
| Nb_init_rev | ATA CTT GAA GAG CCG GCC ATT GCA GGC TGG GCA G |
| RD_FX_pRDV_for | ATA TAT GCT CTT CTG CAA AGC TTT ATA TGG CCT CGG GGG C |
| RD_FX_pRDV_rev1 | TAT ATA GCT CTT CAA CTA CCC ATG GAT ATA TCT CCT TCT TAA AGT TAA AC |
| pRDV_SL_for | AGA CCA CAA CGG TTT CCC TCT AGA AAT AAT TTT GTT TAA CTT TAA G |
| pRDV_SL_rev | CCC TAT AGT GAG TCG TAT TAA TTT CGA TGG |
| GS-Linker_FW | GGC GGT GGC GGT AGT AGA AGA GCG AGC TGC AGA CTG |
| GS-Linker_RV | GCC GGA ACC ACT TGG ACC TTG AAA CAA AAC TTC TAA ATG ATG |
| Primers for target amplification | |
| GFP_FX_FW | TAT AGC TCT TCT AGT CAA TTC AGC AAA GGA GAA GAA CTT TTC |
| GFP_FX_RV | TAT AGC TCT TCT TGC TGC ACT AGT TTT GTA GAG CTC ATC C |
| MBP_FX_FW | ATA TAT GCT CTT CTA GTA AAA TCG AAG AAG GTA AAC TGG TAA TCT GG |
| MBP_FX_RV | TAT ATA GCT CTT CAT GCG CTA CCC GGA GTC TGC GC |
| IrtAB_FX_FW | ATA TAT GCT CTT CTA GTC TTC GTG GAC TGG GTG CCC GCG ACC AT |
| IrtAB_FX_RV | TAT ATA GCT CTT CAT GCC CGT GCC GTC GAC CCG ATC GCC CAC TC |
| Primers for ribosome and phage display | |
| Med_FX_for | ATA TGC TCT TCT AGT CAG GTT CAG CTG GTT GAG AGC G |
| Med_FX_rev | TAT AGC TCT TCA TGC GCT CAC AGT CAC TTG GGT ACC |
| Long_FX_for | ATA TGC TCT TCT AGT CAA GTC CAG CTG GTG GAA TCG |
| Long_FX_rev | TAT AGC TCT TCA TGC AGA AAC GGT AAC TTG GGT GCC C |
| RT_Primer | CTT CAG TTG CCG CTT TCT TTC TTG |
| Medium_ORF_for | AGT CAG GTT CAG CTG GTT GAG AGC G |
| Medium_ORF_rev | TGC GCT CAC AGT CAC TTG GGT ACC |
| Long_ORF_for | AGT CAA GTC CAG CTG GTG GAA TCG |
| Long_ORF_rev | TGC AGA AAC GGT AAC TTG GGT GCC C |
| 5'_flank _for | CGA AAT TAA TAC GAC TCA CTA TAG GGA GAC |
| tolAk_rev | CCG CAC ACC AGT AAG GTG TGC GGT TTC AGT TGC CGC TTT CTT TCT |
| tolAk_2 | CCG CAC ACC AGT AAG GTG TGC |
| 5'_flank _rev | TAT AGC TCT TCA ACT ACC CAT GGA TAT ATC TCC |
| 3’_flank_for | TAT AGC TCT TCT GCA AAG CTT TAT ATG GCC TC |
| Medium_ORF_5'_rev | CGC TCT CAA CCA GCT GAA CCT GAC T |
| Long_ORF_5'_rev | CGA TTC CAC CAG CTG GAC TTG ACT |
| Medium_ORF_3’_for | GGT ACC CAA GTG ACT GTG AGC GCA |
| Long_ORF_3'_for | GGG CAC CCA AGT TAC CGT TTC TGC A |
| Primers for qPCR | |
| qPCR_RD_5’_for | GGG AGA CCA CAA CGG TTT CCC |
| qPCR_ RD_S and M_5’_rev | CAC CGG GAA ACC GCT TGC GGC |
| qPCR_ RD_L_5’_rev | GCC GCT AGC CGC ACA GCT C |
| qPCR_ RD_tolA_3’_for | GCC GAA TTC GGA TCT GGT GGC |
| qPCR_ RD_tolA_3’_rev | CTG CTT CTT CCG CAG CTT TAG C |
| qPCR_PD_pDX_for | GAC GTT CCG GAC TAC GGT TCC |
| qPCR_PD_pDX_rev | CAC AGA CAG CCC TCA TAG TTA GC |
| qPCR_3K1K_for | AGT GCC GGT GAT CGT AGC AG |
| qPCR_3K1K_rev | CCC AAT ATT CAA AGC CCA CGT T |
Primers and megaprimers used to assembly the sybody libraries
https://doi.org/10.7554/eLife.34317.030| Library | CDR | Randomized primer | Megaprimer | Assembly primer | Outer primers |
|---|---|---|---|---|---|
| concave | CDR1 | CDR1_a_b | Megaprimer 1 | FW2_a_b_rev | FW1_a_b_for/Link1_a_b_rev |
| CDR2 | CDR2_a_b | Megaprimer 2 | – | Link1_a_b_for/Link2_a_rev | |
| CDR3 | CDR3_a | – | – | Link2_a_for/FW4_a_b_rev | |
| loop | CDR1 | CDR1_a_b | Megaprimer 1 | FW2_a_b_rev | FW1_a_b_for/Link1_a_b_rev |
| CDR2 | CDR2_a_b | Megaprimer 3 | – | Link1_a_b_for/Link2_b_rev | |
| CDR3 | CDR3_b | – | – | Link2_b_for/FW4_a_b_rev | |
| convex | CDR1 | CDR1_c | Megaprimer 4 | FW2_c_rev | FW1_c_for/Link1_c_rev |
| CDR2 | CDR2_c | Megaprimer 5 | – | Link1_c_for/Link2_c_rev | |
| CDR3 | CDR3_c | – | – | Link2_c_for/FW4_c_rev |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.34317.031





