Mitochondria-specific photoactivation to monitor local sphingosine metabolism and function
Figures
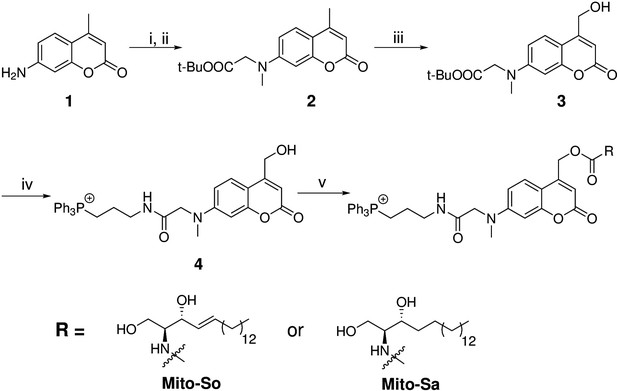
Synthesis of Mito-caged sphingosine (Mito-So) and sphinganine (Mito-Sa).
Conditions: (i), tert-butyl bromoacetate, DIPEA, NaI, ACN, reflux, 80%; (ii) MeI, NaH, 0, 88%; (iii) SeO2, xylene, reflux; NaBH4, MeOH, 58%; (iv) TFA/DCM, 0; (3-Aminopropyl)triphenylphosphonium bromide, HBTU, DIPEA, DCM, 25%; (v) Bis-(4-nitrophenyl)carbonate, sphingosine or sphinganine, DIPEA, DMF, 60, 39–50%.
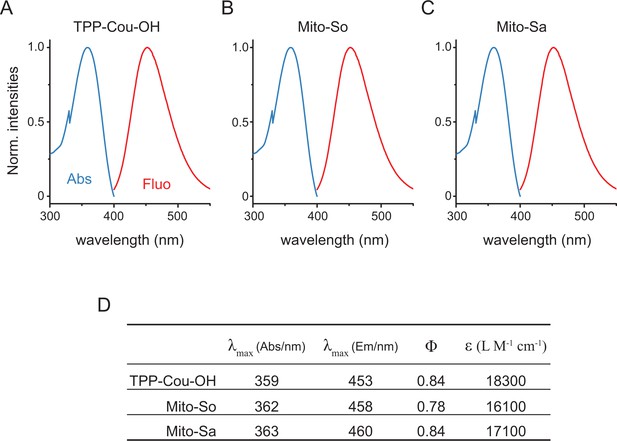
Photophysical properties of mitochondria-specific caged molecules.
(A, B, C) Normalized absorption (in blue) and emission (in red) spectra of the caged molecules; (D) List of maximal absorption, emission, quantum yield, extinction coefficient of the caged molecules. All measurements were acquired in ethanol solution; quantum yields were calculated using coumarin 1 (=0.73 in ethanol) as a reference compound (Jones et al., 1985).
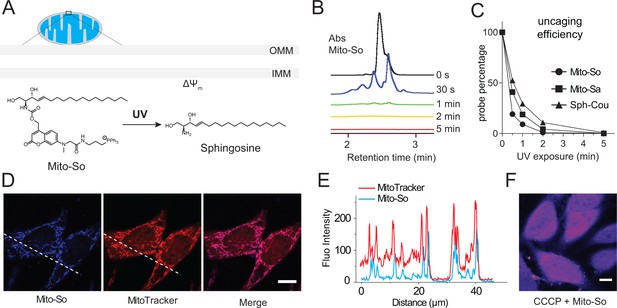
Uncaging efficiency and localization of mitochondria-targeted caged sphingosine.
(A) Illustration of mitochondria-targeted photoactivation of sphingosine. (B) LC-MS spectral of Mito-So absorbance after exposing Mito-So (100 μM) solution (10% DMSO in water) under UV light for indicated time. (C) Uncaging efficiency of Mito-So, Mito-Sa, and Sph-Cou. All probes were treated using procedures described in (B) and quantified by the corresponding absorbance. (D) Representative fluorescence images of Hela cells stained with Mito-So (5 μM) and MitoTracker (50 nM). (E) Intensity profiles of the white dotted line (D) in Mito-So and MitoTracker channel, respectively. (F) Representative fluorescence images of Mito-So staining in the presence of CCCP (10 μM). Scale bar: 10.
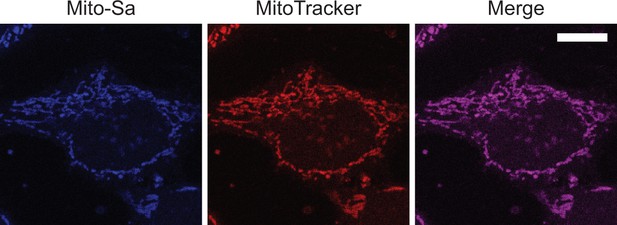
Fluorescence images Mito-Sa staining in living cells.
Representative fluorescence images of live Hela cells stained with Mito-So (5) and MitoTracker (50) following procedures described in the materials and methods. Scale bar: 10.
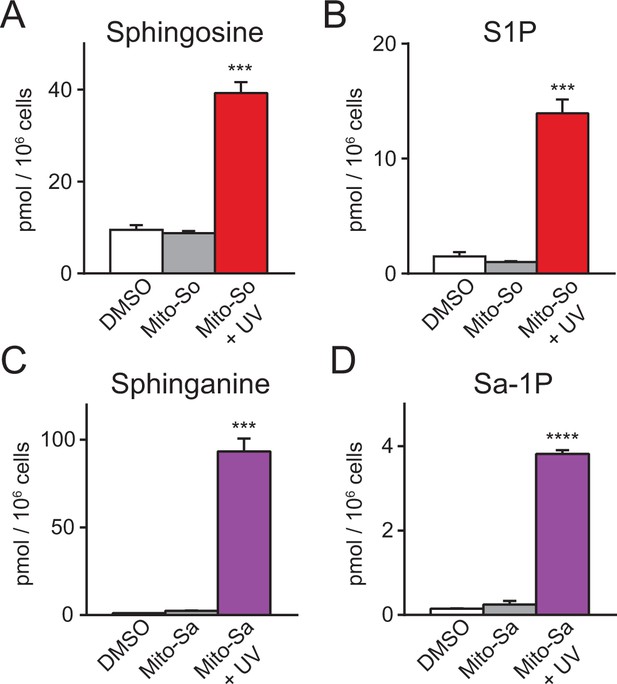
Sphingolipid analysis of mito-caged probes in Hela cells.
Fully confluent cells in 60 mm culture dishes were incubated with the caged probes (2 μM) for 15 min, washed, irradiated under 350–450 nm UV light for 2 min on ice. Cells were collected immediately after UV irradiation. Lipids were extracted and measured by LC-MS. Values were normalized with respect to the amount of C17 internal standards and cell numbers. Data represents the average of three independent experiments. Error bars represent SEM.
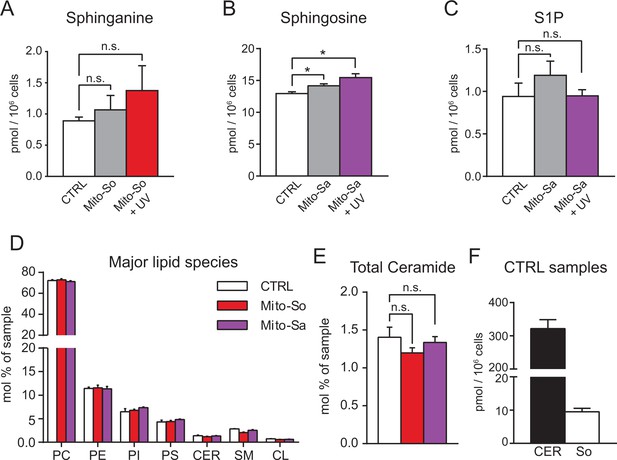
Lipid analysis of mito-caged probes in Hela cells.
Sphinganine 1-phosphate (Sa-1P) levels were not plotted due to the low signals that cannot be separated from background. (A, B, C, F) Values were normalized with respect to the amount of C17 internal standards and cell numbers. (D, E) Values were normalized with respect to the amount of internal standards and were plotted as percentage of all lipid signals. (F) Total ceramides and sphingosine levels in DMSO controls; of note, the two lipid classes were extracted and detected using different protocols. Nevertheless, the data here provides an estimation and direct comparison of endogenous ceramides and sphingosine. Data represents the average of three independent experiments. Error bars represent SEM. *p<0.05, n.s., not significant, student's t-test.
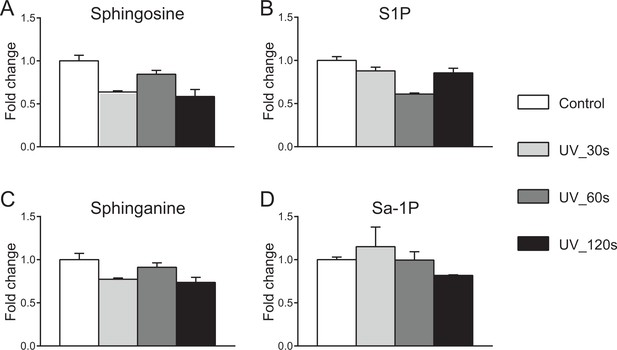
The effects of UV irradiation on sphingolipid levels.
Cells in 60 mm culture dishes were irradiated under 350–450 nm UV light on ice for indicated time period. Cells were collected immediately after UV irradiation. Extracted lipids were derivatized by AQC, and measured by LC-MS/MS. Values were normalized with respect to the amount of C17 internal standards and cell numbers. Data represents the average of three independent experiments. Error bars represent SEM.
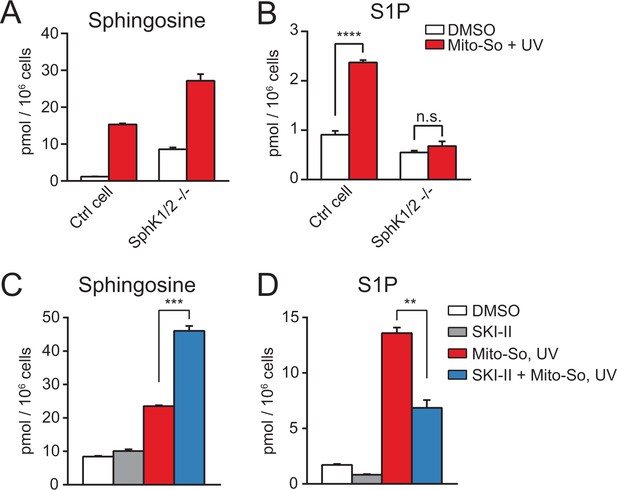
Sphingolipid analysis after Mito-So uncaging in sphingosine kinase double knockout Hela MZ cells (A, B) or inhibitor treated Hela cells (C, D).
Values were normalized with respect to the amount of C17 internal standards and cell numbers. Data represents the average of three independent experiments. Error bars represent SEM. **p<0.01, ***p<0.001, ****p<0.0001, n.s., not significant, student's t-test.
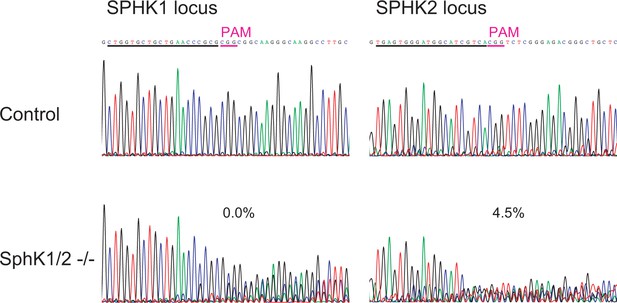
Analysis of mutation rates in knockout cells.
Each target locus was analyzed by PCR direct sequencing. High rates of aberrant sequences are observed around the estimated cutting site, at 3 bp from the NGG protospacer adjacent motifs (PAMs), only in the intended mutant cells. Guide RNA target sequences are underlined. The indicated values are the percentage of signals equivalent to control samples that were calculated by TIDE (tracking of indels by decomposition) analysis.
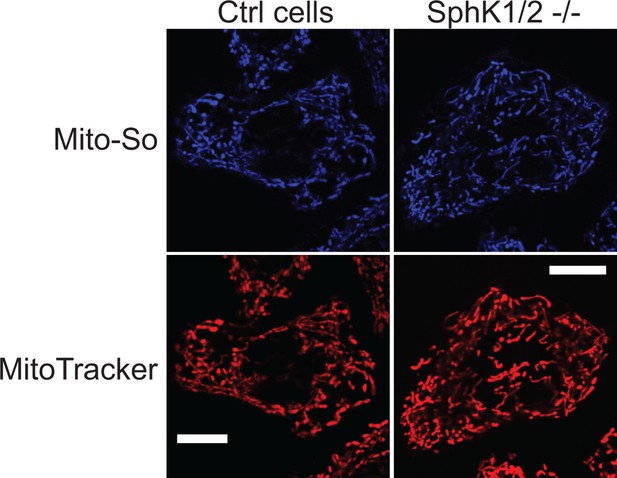
Fluorescence images of Mito-So in SphK KO cells.
Representative fluorescence images of sphingosine kinase knockout cells stained with Mito-So (5 μM) and MitoTracker (50 nM) following procedures described in the materials and methods. Scale bar: 10 μm.
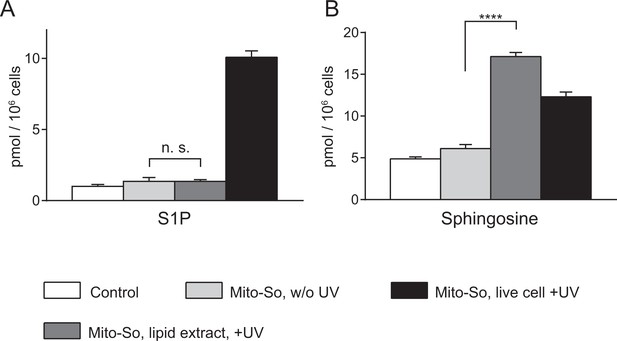
Mito-So was not phosphorylated in Hela cells in its caged form.
Cells were incubated with DMSO or caged probes (2 μM) for 15 min. From left to right, cells were collected only for lipid extraction (control; Mito-So, w/o UV); or cells were collected for extraction, dried, re-suspended in 200 water and irradiated for 10 min on ice (Mito-So, lipid extract,+UV); or cells were irradiated for 2 min on ice before lipid extraction (Mito-So, live-cell +UV). Extracted lipids were derivatized by AQC, and measured by LC-MS/MS. Values were normalized with respect to the amount of C17 internal standards and cell numbers. Data represents the average of three independent experiments. Error bars represent SEM. ****p<0.0001, n.s., not significant, student's t-test.
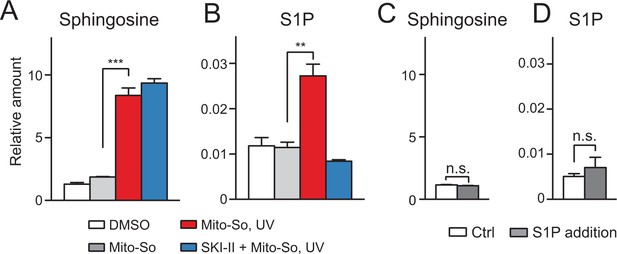
Sphingolipid analysis after Mito-So uncaging in purified mitochondria from mouse liver.
(A, B) Mitochondria were incubated with Mito-So (10 μM) for 10 min, ±SKI II (50 μM), irradiated under UV light for 2 min on ice, incubated for 15 min at 37°C prior to pelleting by centrifugation and lipid extraction. (C, D) C18 sphingosine 1-phosphate (10 pmol, approx. 10 times of the control) was added to the mitochondria, following the same experimental and extraction procedures. Values were normalized with respect to the amount of C17 internal standards and cell numbers. Data represents the average of three independent experiments. Error bars represent SEM. **p<0.01, ***p<0.001, ****p<0.0001, n.s., not significant, student's t-test.
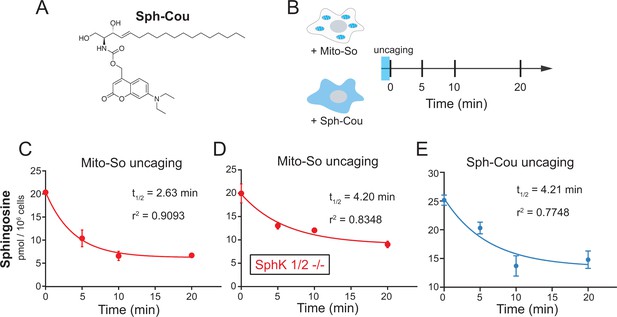
Real-time metabolism of mito-caged sphingosine and globally-caged sphingosine in living cells.
(A) Chemical structure of Sph-Cou. (B) Illustration of uncaging followed by real-time detection. Cells were first treated with caged probes for 15 min, washed, irradiated by UV light for 2 min on ice, incubated at 37°C, and then collected for lipid extraction at different time points. (C, D , E) Changes of sphingosine levels over time after uncaging. The decay curves were generated by fitting into the one phase decay functions. Values were normalized with respect to the amount of C17 internal standards and cell numbers. Data represent the average of three independent experiments. Error bars represent SEM.
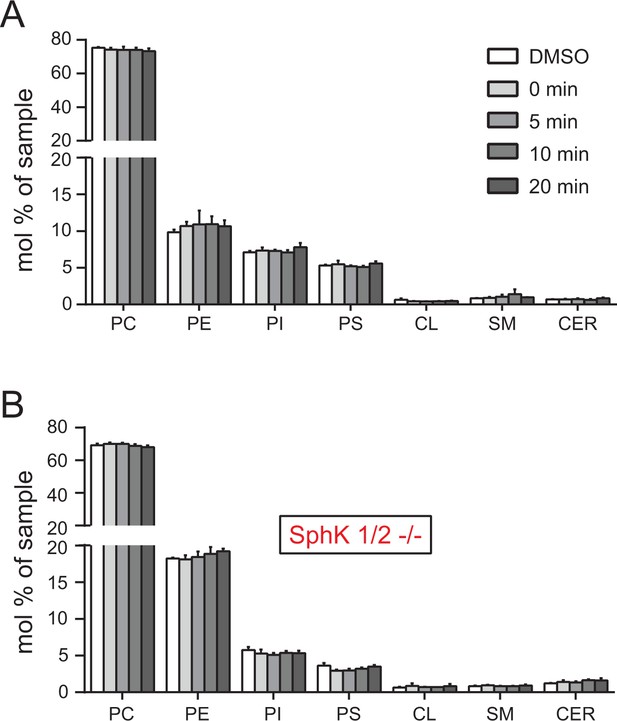
Total lipid analysis of mito-caged probes in Hela cells and SphK dKO cells.
No significant differences due to the different conditions were found. Values were normalized with respect to the amount of internal standards and were plotted as percentage of all lipid signals. Data represents the average of three independent experiments. Error bars represent SEM.
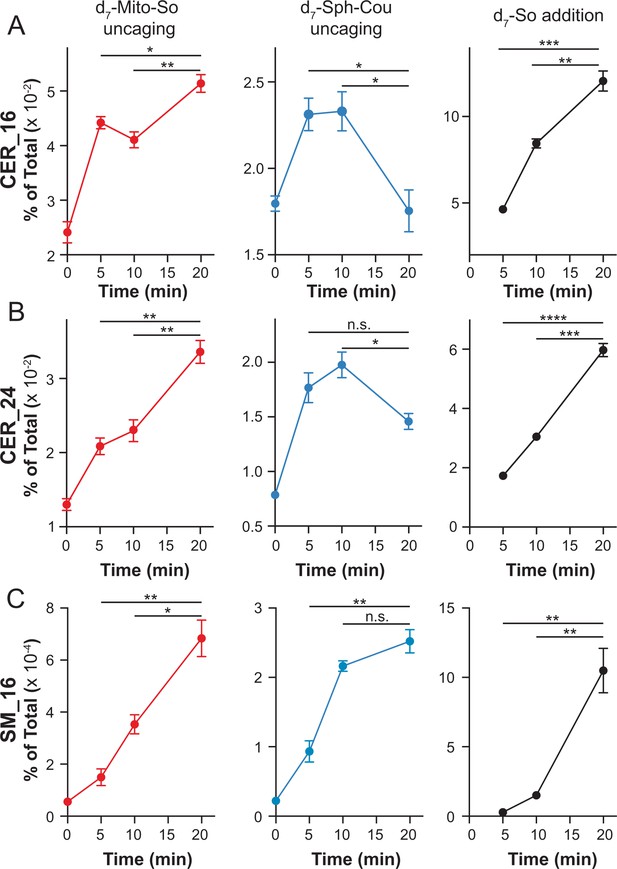
Real-time detection of d7-sphingolipids in living Hela cells after uncaging of d7-Mito-So, d7-Sph-Cou or direct addition of d7-sphingosine.
Values were normalized with respect to the amount of internal standards and were plotted as percentage of all lipid signals. Data represent the average of three independent experiments. Error bars represent SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, n.s., not significant, students' t-test.
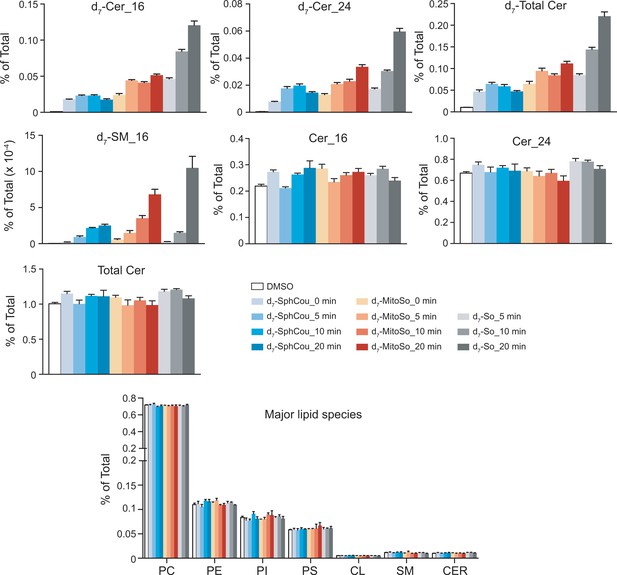
Total lipid profiles after releasing sphingosine in different subcellular compartments.
Values were normalized with respect to the amount of internal standards and were plotted as percentage of all lipid signals. Data represent the average of three independent experiments. Error bars represent SEM.
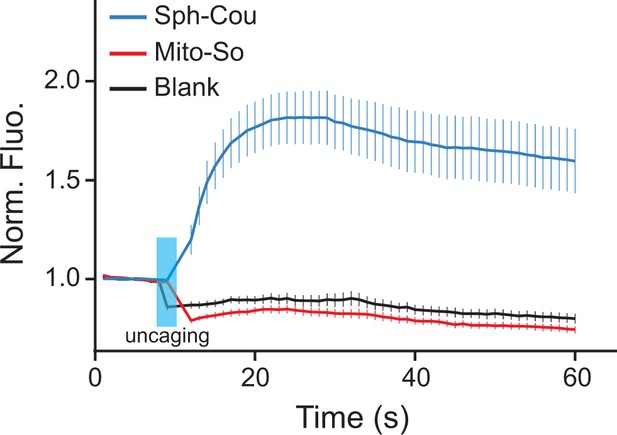
Calcium responses after photo-releasing sphingosine from caged precursors.
(A) Mean traces of normalized fluorescence intensity after uncaging of Mito-So, Sph-Cou, or blank. Hela cells were loaded with Fluo-4 AM (5 μM), together with Sph-Cou (5 μM) or Mito-So (5 μM) prior to UV illumination. Cells were irradiated for 4 s by a 405 nm laser at 37°C. Error bars represent SEM. n > 10.
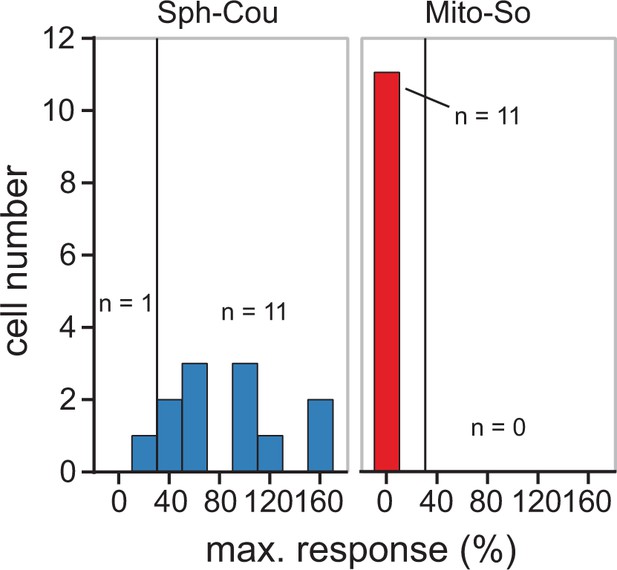
Histogram distribution of maximal calcium responses compared to the baseline in each cell, with the threshold set at 20% increase (black vertical line).
https://doi.org/10.7554/eLife.34555.020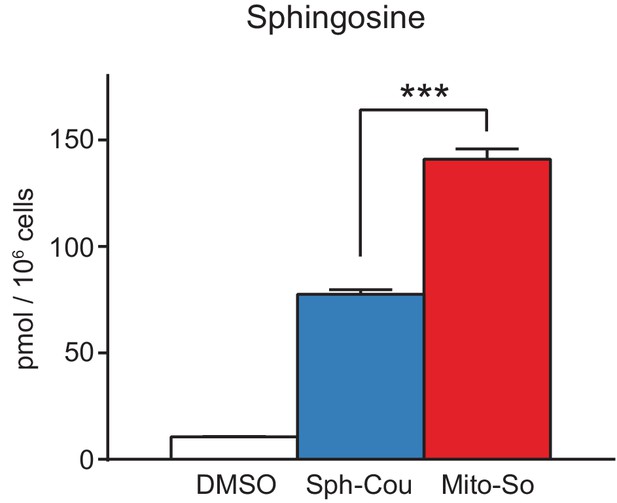
Comparison of cellular uptake between Sph-Cou and Mito-So.
Cells were incubated with DMSO, Sph-Cou or Mito-So (5 μM) for 15 min prior to lipid extraction. Extracted lipids were re-suspended in 200 μL water and illuminated for 10 min on ice. Samples were derivatized by AQC and measured by LC-MS/MS. Values were normalized with respect to the amount of C17 internal standards and cell numbers. Data represents the average of three independent experiments. Error bars represent SEM. ***p<0.001, student's t-test.
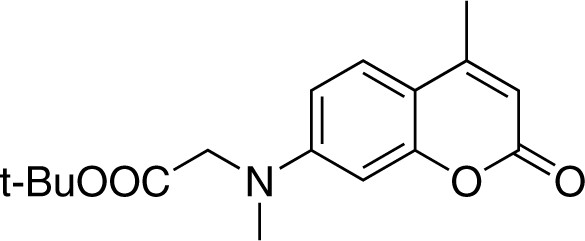
Structure for 7-[(tert-butoxycarbonylmethyl)-methylamino]-4-methylcoumarin.
https://doi.org/10.7554/eLife.34555.022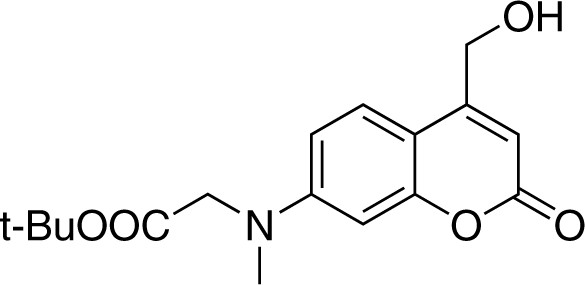
Strucutre for 7-[(tert-butoxycarbonylmethyl)-methylamino]-4-(hydroxymethyl)coumarin.
https://doi.org/10.7554/eLife.34555.023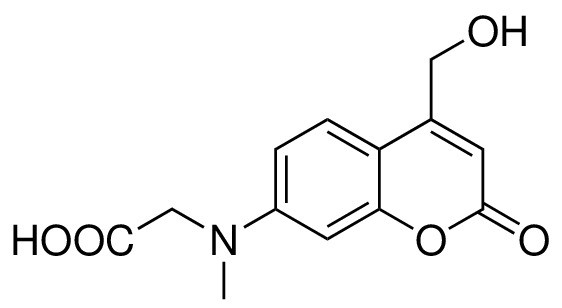
Structure for 7- [(carboxymethyl)-methylamino]-4-(hydroxymethyl)coumarin.
https://doi.org/10.7554/eLife.34555.024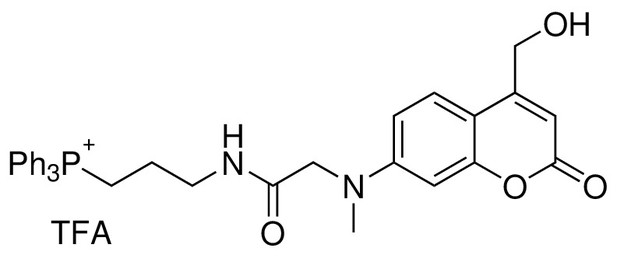
Structure for TPP-Cou-OH.
https://doi.org/10.7554/eLife.34555.025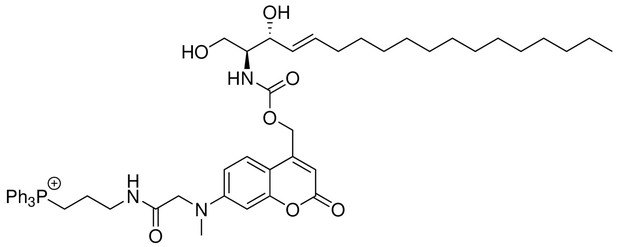
Structure for Mito-So.
https://doi.org/10.7554/eLife.34555.026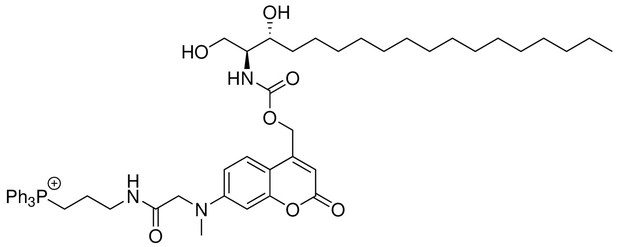
Structure for Mito-Sa.
https://doi.org/10.7554/eLife.34555.027Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.34555.028





