Design principles of a microtubule polymerase
Figures

Polymerase activity requires at least two TOGs that need not be different, and a basic region; dimerization is not essential.
(A) Domain organization of Stu2, from N- to C-terminus. (B) All-yeast in vitro reconstitution assay. (left) Schematic of the TIRF assay. Axonemes seed unlabeled yeast microtubules in the presence of Stu2-eGFP. (right) Representative kymographs for wild-type Stu2 and for a monomeric Stu2 variant. Microtubule growth rates are determined by tracking the Stu2-eGFP spot on the microtubule end. (C) Inactivating TOG1 (blue box/points, TOG1* indicates the R200A mutation) or TOG2 (red box/points, TOG2* indicates the R519A mutation) weakens the polymerase activity of the dimer compared to the wild-type dimer (black). Smooth curves indicate a hyperbolic fit to the data. Black: each point represents n = 45 microtubules measured from three different chambers; red/blue: n = 20 microtubules. Error bars are SEM. (D) Eliminating dimerization reduces polymerase activity by sharply weakening end-binding affinity (green). End-binding affinity is restored by increasing the charge in the basic domain (purple). Data were fit as in C., and the WT data are repeated from that panel. Green: n = 45 microtubules measured from three different chambers; purple: n = 25 microtubules. Error bars are SEM. (E) Functional Stu2 variants containing only TOG2 (TOG2-TOG2, orange) or only TOG1 (TOG1-TOG1, pink). Smooth curves indicate a hyperbolic fit to the data. Data were fit as in C., and the WT data are repeated from that panel. Orange: n = 30 microtubules measured from two different chambers; pink: n = 20 microtubules measured. Error bars are SEM. See also Figure 1—figure supplement 1. (F) Summary statistics from the hyperbolic fits shown in C, D and E. See also Figure 1—figure supplement 1. The maximal activity decreases when the number of TOGs is reduced. Apparent end binding affinity (KM) does not depend strongly on the number of type of active TOGs. (G) Cartoon illustrating that polymerases with two tubulin-binding TOGs are about half as active as WT with its four TOGs.
-
Figure 1—source data 1
Numerical data associated with Figure 1.
- https://doi.org/10.7554/eLife.34574.004
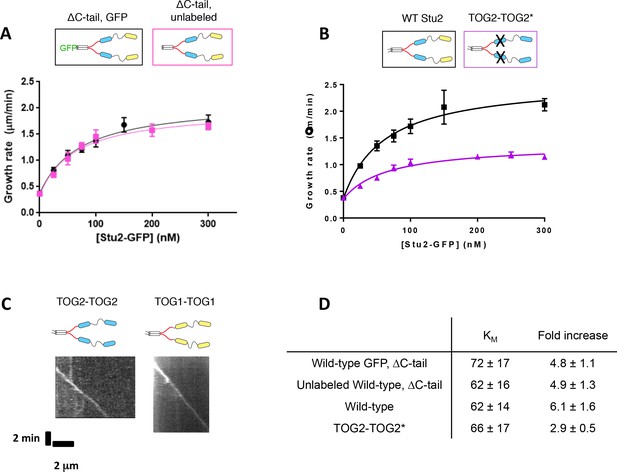
Additional data supporting the activity of various Stu2 constructs.
(A) A Stu2-eGFP variant lacking the C-terminal tail (ΔC-tail) exhibits similar polymerase activity with (black) or without (pink) a C-terminal eGFP tag. Smooth curves indicate a hyperbolic fit to the data. Black: N = 45 measured from three different chambers, pink: N = 25. Error bars show SEM. (B) Mutating the basic-proximal TOG2 in an all TOG2 polymerase (TOG2-TOG2*, purple) results in a slight decrease in polymerase activity. Black: N = 45 measured from three different chambers; purple: N = 20 measurements. Error bars show SEM. Smooth curves indicate a hyperbolic fit to the data. (C) Both TOG2-TOG2 polymerases (kymograph, left) and TOG1-TOG1 polymerases (kymograph, right) can tip-track growing microtubule ends. Representative kymographs for each sample contain 100 nM Stu2 mutant and 800 nM wild-type yeast tubulin. (D) Summary statistics of hyperbolic fits to the activity measurements shown in A and B.
-
Figure 1—figure supplement 1—source data 1
Numerical data associated with Figure 1—figure supplement 1.
- https://doi.org/10.7554/eLife.34574.005
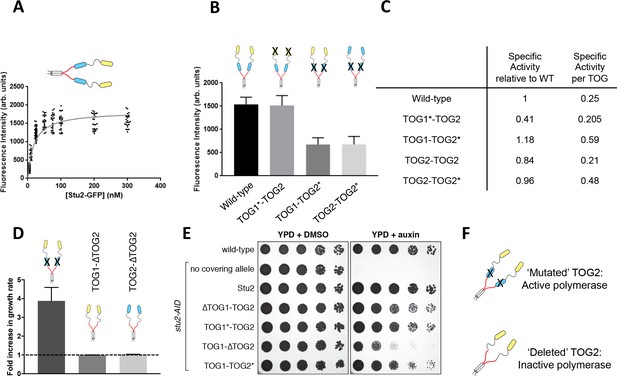
A unique functional role for the basic-proximal TOG domain.
(A) Fluorescence intensity of wild-type Stu2-eGFP dimers on the microtubule end. Intensities were determined as a function of Stu2 concentration by measuring the fluorescent ‘spot’ at the end of a growing microtubule. The data were fit with a hyperbolic function, yielding a half-maximal concentration of 19 nM, comparable to the half-maximal concentration for activity of 62 nM. N = 200 measured from two different chambers for 5 nM, N = 25 for 10, 50–300 nM, N = 50 measured from two different chambers for 25 nM (B) Interfering with tubulin binding by the basic-proximal TOG (TOG2*; R519A mutation) decreases the amount of Stu2 on the microtubule end, but interfering with tubulin binding by the N-terminal TOG (TOG1*; R200A mutation) does not detectably change the amount of Stu2 on the end. The reduced accumulation of TOG1-TOG2* cannot be ascribed to the loss of tubulin binding by TOG2, because TOG2-TOG2* also shows reduced accumulation of the microtubule end. Samples were analyzed with 200 nM Stu2 in the presence of 0.8 μM tubulin in dynamic assays. N = 50 for all. Error bars are SEM. (C) Specific activity of Stu2 variants relative to wild-type, on a per-polymerase (left) and per functional TOG (right) basis. Specific activity was obtained by dividing the values from the last column of Figure 1F by the relative amount of polymerase on the microtubule end (as determined in Figure 2B; one for WT and Stu2(TOG2-TOG2), 0.5 for Stu2(TOG1-TOG2*) and Stu2(TOG2-TOG2*)). Stu2 variants with the basic-proximal TOG inactivated show higher specificity. (D) Deleting the basic-proximal TOG domain (∆TOG2) abolishes polymerase activity and interaction with the microtubule lattice (see Figure 2—figure supplement 1), whether the N-terminal TOG domain is TOG1 or TOG2. Changes in polymerase activity may be attributable to the distance between the most basic-proximal TOG and the basic domain (see Figure 2—figure supplement 1D) The fold-increase in elongation rate is plotted for different polymerase variants. The dashed line indicates the rate of elongation for the ‘no Stu2’ control. N = 25 measurements for all; error bars are SEM. Samples contained 0.8 μM tubulin and 200 nM Stu2 variants. (E) Wild-type (SBY3), stu2-AID (SBY13772) and stu2-AID cells expressing various STU2-3V5 alleles from an ectopic locus (wildtype, SBY13901; TOG1∆, SBY13904; R200A, SBY13919; TOG2∆, SBY13907; R519A, SBY13925) were serially diluted (5-fold) and spotted on plates containing either DMSO or 500 μM auxin. Stu2 constructs deleted for TOG1 (∆TOG1-TOG2) or with TOG1 inactivated for tubulin binding (TOG1*-TOG2) display a mild defect in rescuing the loss of WT Stu2. A Stu2 construct deleted for TOG2 (TOG1-∆TOG2) shows a more severe loss of rescue activity. A Stu2 construct with TOG2 inactivated for tubulin binding (TOG1-TOG2*) yields full rescue activity. See also Figure 2—figure supplement 1. (F) Schematic cartoon summarizing that polymerase activity and Stu2 function requires a TOG domain adjacent to the basic region, even if that TOG is compromised for binding to unpolymerized tubulin.
-
Figure 2—source data 1
Numerical data associated with Figure 2.
- https://doi.org/10.7554/eLife.34574.008
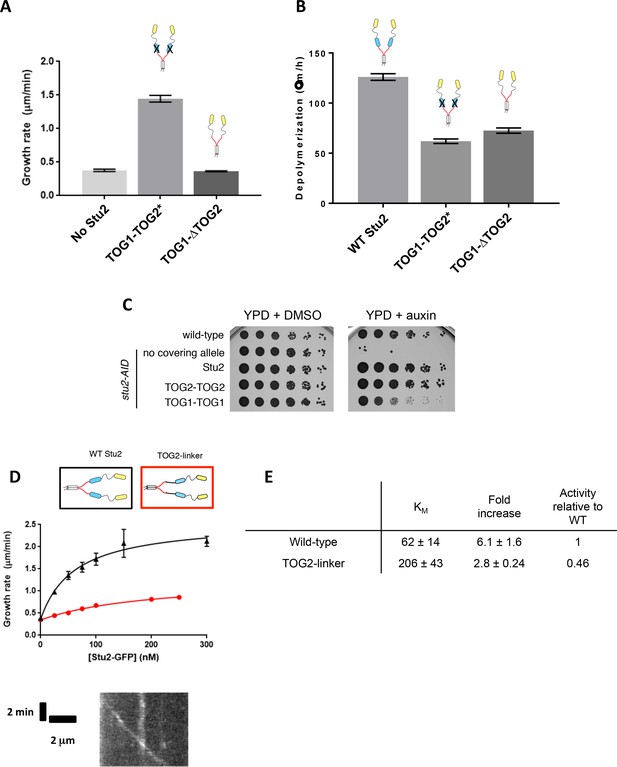
Effects of inactivating, deleting, or replacing the basic-proximal TOG domain.
(A) Deleting the TOG2 domain (Stu2(TOG1-ΔTOG2)) abolishes polymerase activity. Polymerase activity is retained when the TOG2 domain is inactivated by mutagenesis. Samples were analyzed with 200 nM Stu2 in the presence of 0.8 μM tubulin in dynamic assays. N = 25 end measurements for all; error bars are SEM. (B) Deleting the TOG2 domain (Stu2(TOG1-ΔTOG2)) does not compromise the tubulin binding activity of TOG1 as judged in a ‘depolymerase’ assay wherein TOG domains cause the depolymerization of stable microtubules. Samples were analyzed with 200 nM Stu2 in the presence unlabeled GTPγS stabilized MTs. N = 20 measurements for all. Error bars show SEM. (C) Wild-type (SBY3), stu2-AID (SBY13772) and stu2-AID cells expressing various STU2-3V5 alleles from an ectopic locus (wildtype, SBY13901; TOG1∆, SBY13904; TOG2-TOG2, SBY16931; TOG1-TOG1, SBY17595) were serially diluted (5-fold) and spotted on plates containing either DMSO or 500 μM auxin. A Stu2 variant in which TOG2 is replaced with a second copy of TOG1 (Stu2(TOG1-TOG1)) partially rescues the loss of endogenous Stu2 in a genetic rescue assay, but to a lesser degree than that of a Stu2(TOG2-TOG2) variant. (D) Inserting a flexible, 16 amino acid spacing linker between the end of TOG2 and the basic domain weakens the polymerase activity of Stu2 bud does not abolish end localization, shown in the representative kymograph (bottom). Black: N = 45 measured from three different chambers; red: N = 20 measurements. Error bars show SEM. Smooth curves indicate a hyperbolic fit to the data. (E) Summary statistics of hyperbolic fits to the activity measurements shown in D.
-
Figure 2—figure supplement 1—source data 1
Numerical data associated with Figure 2—figure supplement 1.
- https://doi.org/10.7554/eLife.34574.009
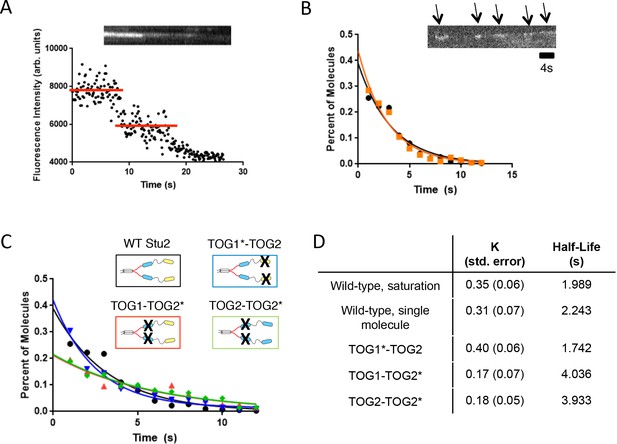
Inactivating tubulin binding by the basic-proximal TOG domain enhances processivity of Stu2.
(A) Example raw fluorescence intensity trace showing two-step photobleaching of a stationary, homodimeric Stu2-eGFP. (B) Lifetime distribution for single Stu2-eGFP molecules on the growing microtubule end under single molecule conditions (5 nM Stu2, black) or ‘spiked’ into a higher concentration reaction (200 nM Stu2 of which only 5 nM is labeled, orange). Inset kymograph illustrates instances of Stu2 ‘runs’ (arrows). Histogram summarizes n = 400 measurements (four independent trials at 1 μM tubulin) for 5 nM Stu2-eGFP and n = 200 measurements (two independent trials at 0.8 μM tubulin) made using 5 nM Stu2-eGFP with 195 nM Stu2-KCK unlabeled. Data, which are plotted as percent of total for comparative purposes, were fit with an exponential, yielding an average residence time of 2.2 s (see also D) for single molecule eGFP and 2.0 s (see also D) for the spike measurements. Scale bar is 4 s. (C) Lifetime histograms for Stu2 variants. Compromising tubulin binding by the basic-proximal TOG (TOG1-TOG2*, red trace) yields a roughly 2-fold increase in end residence time. Compromising the N-terminal TOG (TOG1*-TOG2, blue trace) does not increase end-residence time. TOG2-TOG2* (green trace) also shows an increase in end residence time. All samples contained 5 nM Stu2-eGFP variants and 1 μM unlabeled yeast tubulin. Two independent trials of n = 100 measurements for all mutants, yielding a total n = 200. Samples were fit with exponential as done in B to extract average residence times. (D) Tabulated summary of results from all exponential fits to residence time distributions in C.
-
Figure 3—source data 1
Numerical data associated with Figure 3.
- https://doi.org/10.7554/eLife.34574.011
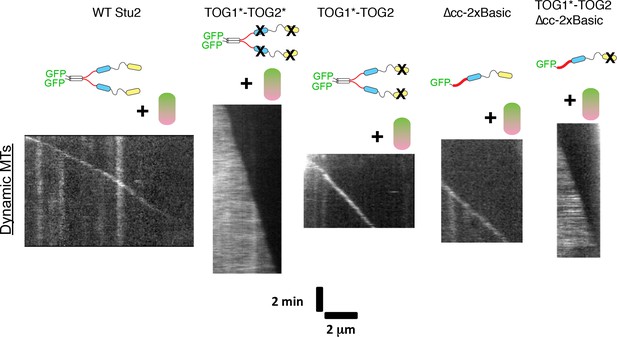
Polymerase elements required for preferential plus-end localization of Stu2.
Polymerase assays were carried out using wild-type tubulin (0.8 μM) and a panel of polymerase mutants (200 nM each). Representative kymographs are shown for Stu2 variants. Plus-end localization requires at least two tubulin-binding TOGs (WT, TOG1*-TOG2, ∆cc-2xBasic) and is not sensitive to how they are linked (compare TOG1*-TOG2 to ∆cc-2xBasic). Unexpectedly, polymerases with 0 (TOG1*-TOG2*) or one active TOGs (TOG1*-TOG2-∆cc-2xBasic) robustly coat the body of the microtubule. See also Figure 4—figure supplement 1.
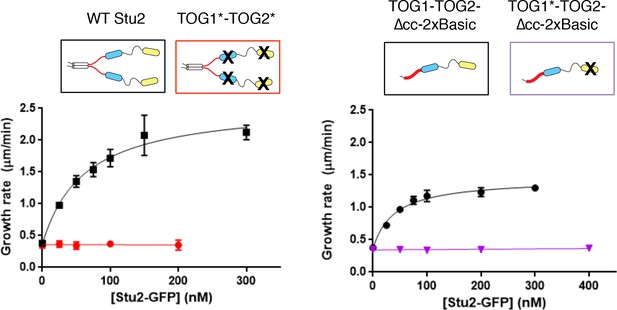
Additional data from assays using mutated TOGs.
‘Double dead’ Stu2 (TOG1*-TOG2*) (left) with both TOGs inactivated does not show polymerase activity (red). Data for wild-type Stu2 (black) are reproduced from Figure 1. Stu2(TOG1*-TOG2)-∆cc-2xBasic (right), which contains a single active TOG domain, does not show polymerase activity (purple). Data for Stu2(TOG1*-TOG2)-∆cc-2xBasic (black; this construct has two active TOGs) are reproduced from Figure 1. Black: N = 45 from three independent chambers; Red and Purple: N = 20 measurements. All error bars are SEM. Smooth curves indicate a hyperbolic fit to the data.
-
Figure 4—figure supplement 1—source data 1
Numerical data associated with Figure 4—figure supplement 1.
- https://doi.org/10.7554/eLife.34574.014

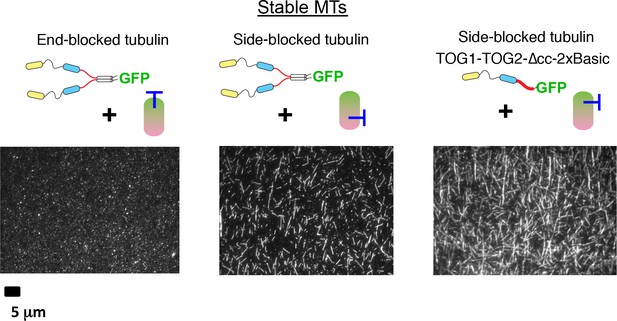
Unexpected antagonism between TOG: αβ-tubulin engagement and basic:lattice interactions.
(A) The structure of Stu2:TOG1 (slate) bound to yeast αβ-tubulin (α-tubulin in pink and β-tubulin in lime; PDB code 4FFB) is shown in cartoon representation. Red spheres indicate the approximate position of interface blocking mutations on the plus- (β:T175R,V179R) or minus-end (α:T350E) and of perturbing mutations on lateral interaction surfaces (α: H284A, β: F281A). All interface mutations are distant from the TOG-interacting surface. Orange spheres indicate the positions of the structured C-terminii of α- and β-tubulin that precede the charged ‘tails’. (B) Tubulin elements required to antagonize lattice binding by Stu2. Stu2:microtubule binding assays were monitored by TIRF and performed using stabilized microtubules as the substrate, 100 nM Stu2-eGFP, and 1 μM of tubulin mutants. Stu2 coats the stabilized microtubules when no unpolymerized tubulin in present (‘No tubulin’). Lattice-binding is substantially eliminated when wild-type tubulin is included as a competing binding partner (‘WT tubulin’). This tubulin-induced antagonism of lattice binding does not require the tubulin tails (‘∆tails tubulin’) or longitudinal contacts (‘end blocked tubulin’). Tubulin perturbed on the lateral interface does not effectively antagonize lattice binding by Stu2 (‘side blocked tubulin’), indicating that lateral contacts between TOG-bound tubulins are important. See also Figure 5—figure supplement 2. (C) Cartoon illustrating that lateral tubulin interactions (possibly transient) between TOG-bound tubulins antagonize interactions between the basic domain and the MT lattice. Tubulins bound to either the TOG1 or TOG2 domains are illustrated; other combinations of two tubulin-binding TOGs tested yield a similar result. (D) Control of lattice binding by unpolymerized tubulin is not an idiosyncratic property of Stu2. Zyg-9, a monomeric Stu2 family polymerase from C. elegans, also shows this tubulin-induced attenuation of microtubule lattice binding. 50 nM Zyg-9-mCherry was used with unlabeled GTPγS-stabilized yeast microtubules; 5 μM bovine tubulin was the competing binding partner.
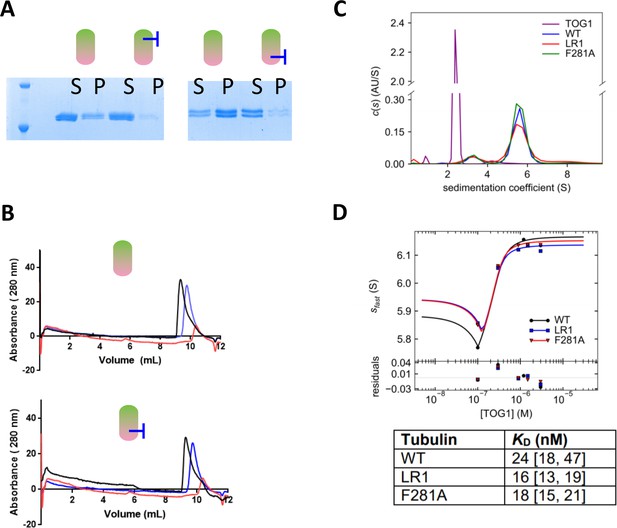
Assays characterizing the assembly and TOG-binding properties of tubulin with a mutated lateral interface.
(A) ‘Spin-down’ assays for microtubule assembly reveal by SDS-PAGE that mutations on the lateral interface of β-tubulin (left, F281A) or α-tubulin (right, H284A) reduce the extent of microtubule assembly compared to wild-type. S = supernatant, p=pellet. Assays were performed using 1 μM tubulin. (B) A gel filtration binding assay demonstrate that a lateral tubulin mutant, Tub1-H284A, binds a TOG domain comparably to wild-type tubulin. Left: Overlaid gel filtration traces of WT tubulin alone (blue), TOG1 alone (red), and the tubulin + TOG mix (black). The shift to earlier elution volumes indicates formation of a larger complex. Right: Overlaid gel filtration traces of α-H284A tubulin alone (blue), TOG1 alone (red, repeated from left panel), and the tubulin + TOG mix (black). A similar shift in elution indicating the presence of a TOG:tubulin complex was observed. All samples contain 1 μM tubulin variant and 1 µM TOG1. (C) Analysis of TOG1:αβ-tubulin interactions by sedimentation velocity. The plot shows c(s) distributions (color coded by sample) for each control sample: TOG1 alone (2 µM, purple), WT tubulin (0.3 µM, blue), ‘LR1’ plus-end blocked tubulin (β-tubulin: T175R, V179R) (0.3 µM, red) and ‘F281A’ (β-tubulin: F281A) (0.3 µM, green). (D) Isotherms for Stu2 TOG1 domain binding to WT tubulin (black), plus-end blocked tubulin ‘LR1’ (β-tubulin:T175R,V179R) (blue) and side-block tubulin ‘F281A’ (β-tubulin:F281A) (red). TOG1 binds with comparable affinity to all three tubulin variants including the two tubulin mutants (plus-end and side-block) and WT tubulin. Fitted binding affinities and 1 σ confidence intervals for fitted affinities (in parentheses) are shown in the table on the right.
Additional data from assays using mutated tubulins.
More experiments testing whether tubulin mutated on the lateral or longitudinal tubulin interfaces can antagonize interactions between the basic region and the microtubule. Results are consistent with Figure 5 in showing that lateral interactions between TOG-bound tubulins are important: lattice binding by Stu2 is attenuated in the presence of plus-end blocked tubulin (β-tubulin:T175R,V179R, left); lattice binding by dimeric or monomeric forms of Stu2 is not attenuated in the presence of a mutation on the lateral interface (α-tubulin:H284A, middle and right). Samples contain 100 nM Stu2-eGFP variant and 1 μM tubulin mutant; unlabeled, GTPγS-stabilized yeast microtubules were used as the MT binding substrate.

Mutation- or drug-induced perturbations that strength tubulin interactions result in loss of Stu2 polymerase activity and tip-tracking.
(A) Representative kymographs showing normal end tracking, with little lattice binding by Stu2 with wild-type yeast tubulin (left). End-tracking is lost, and lattice binding is enhanced, when β:T238A mutant tubulin was used (right; this mutant stabilizes tubulin:tubulin interactions). Loss of end-tracking and gain in lattice binding also occurred in the presence of wild-type tubulin plus the microtubule-stabilizing drug Epothilone-B (right). Kymographs contain 100 nM Stu2-eGFP and 500 nM yeast tubulin (wild-type or mutant as indicated). 4 μM Epothilone B was added to right panel. (B) Stu2 shows reduced polymerase activity in the presence of Epothilone-B or β:T238A mutant tubulin. Measurements were made with 500 nM tubulin (either wild-type or T238A) and 200 nM Stu2-eGFP. N = 25 measurements for all. All error bars are SEM. (C) Cartoon illustrating a ‘futile cycling’ to explain the gain in lattice binding associated with perturbations that strengthen tubulin:tubulin interactions. The preference of tubulin to be curved normally prevents release from TOGs away from the microtubule end. When tubulin:tubulin interactions are strengthened by drug- or mutation-induced perturbation, straightening becomes easier and Stu2 can prematurely release its TOG-bound tubulins away from the microtubule end.
-
Figure 6—source data 1
Numerical data associated with Figure 6.
- https://doi.org/10.7554/eLife.34574.019
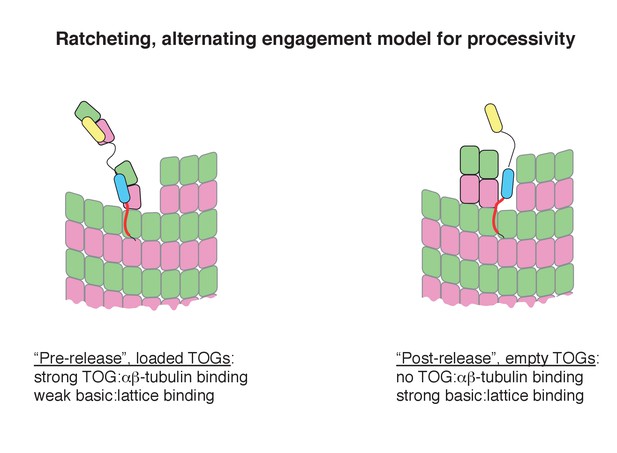
Speculative ‘ratcheting’ model for processivity.
The model was inspired by the fact that the TOG:tubulin engagement status regulates the lattice-binding activity of the basic region. We assume two limiting states for Stu2 on the microtubule end: a ‘loaded’ state wherein the basic region binds weakly to the lattice because at least two TOGs are engaged with tubulins, and an ‘empty’ state wherein the basic region binds strongly to the lattice because the TOGs have released their tubulins to the microtubule. In this ‘post-release’ state, strong interactions between the basic region and the lattice help retain the empty polymerase near the growing end where it is poised to capture ‘fresh’ unpolymerized tubulins for another round of activity. Capture and incorporation of fresh tubulins drives movement with the growing end.
Additional files
-
Supplementary file 1
Strains used in this study.
All strains are derivatives of SBY3 (W303).
- https://doi.org/10.7554/eLife.34574.021
-
Transparent reporting form
- https://doi.org/10.7554/eLife.34574.022





