Mild myelin disruption elicits early alteration in behavior and proliferation in the subventricular zone
Figures
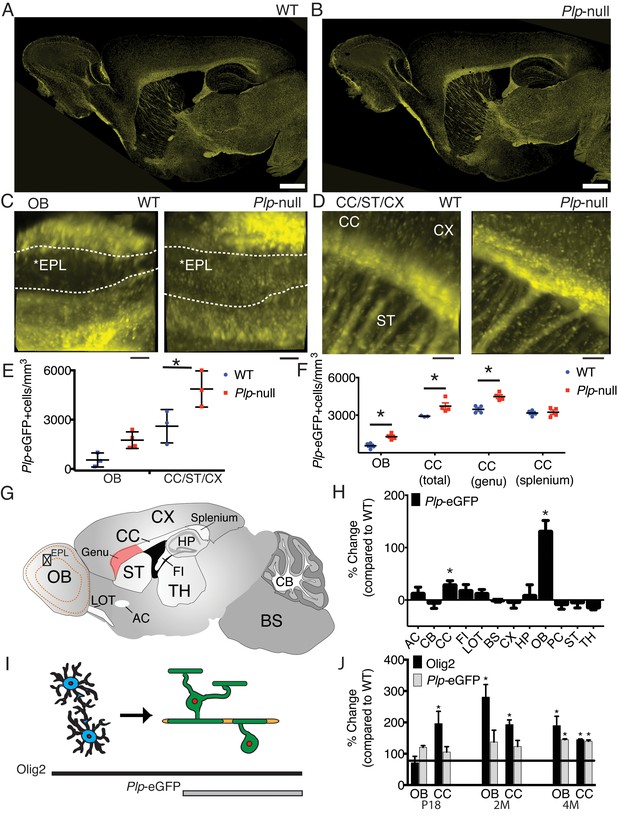
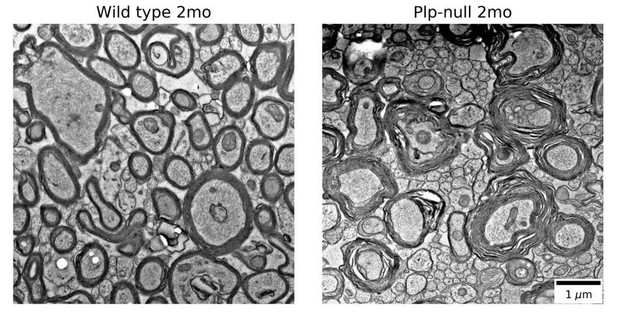
Region-specific increase in oligodendrocyte density in Plp1-null mice.
(A-B) Confocal image of Plp1-eGFP expression in 6M WT and Plp1-null. Bar = 1 mm. C and D. Plp1-eGFP imaged with cleared tissue digital scanned light-sheet microscopy (C-DSLM) in cleared brain tissue. (C) Plp1-eGFP in cleared WT and Plp1-null OB. EPL is outlined. Bar = 50 μm. (D) Plp1-eGFP in cleared cortex. Bar = 100 μm. Image plane extracted from 1.3 × 1.3 × 6 mm axial image stack. Acquired using 10 μm exciting light-sheet, 10x/0.28 detection objective with 0.65 μm in-plane and 1 μm axial spacing. (E) Cell density in cleared tissue imaged with C-DSLM was increased in the CC of the 6M Plp1-null (F(1,9) = 15, 2-way ANOVA; p<0.01). (F) MATLAB quantification of confocal images reveals an increase in Plp1-eGFP cells in the OB (1283 ± 122 vs 558 ± 112 cells/mm3; p<0.01) and CC (genu: 4483 ± 155 vs 3462 ± 152 cells/mm3, p<0.01) of Plp1-null mice n = 4/genotype. (G) Brain regions (Table 1). (H) Percent change in cell number in 6M Plp1-null vs. WT. (I) Olig2 Plp1-eGFP in the oligodendrocyte lineage. (J) Olig2+ cells were increased in P18 CC (195 ± 39% of WT), 2M OB (280 ± 40%) and CC (192 ± 15%), and 4M OB (189 ± 30%) and CC (144 ± 2%) in Plp1-null mice. Plp1-eGFP cells were also increased in 4M OB (145 ± 3%) and CC (140 ± 4%) in Plp1-null mice. WT and Plp1-null samples compared using repeated measures ANOVA. *p<0.01.
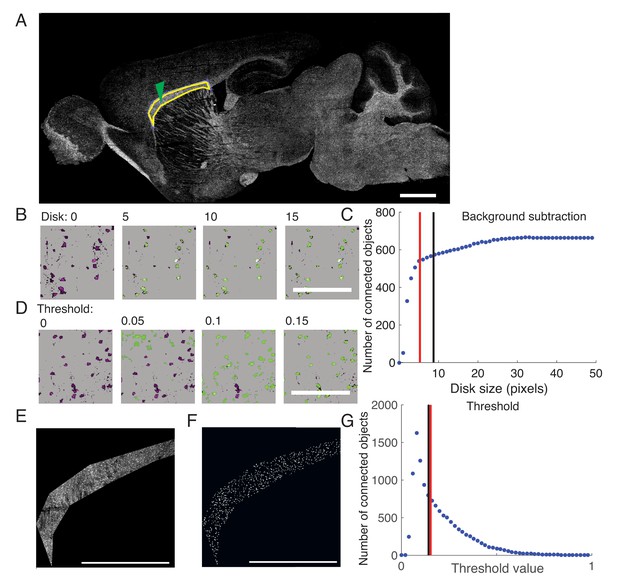
Semi-automatic quantification of the density of labeled oligodendrocytes in tissue sections implemented in MATLAB.
(A) Tiled image of sagittal 30 μm brain sections imaged at 25x with a confocal microscope. Yellow indicates the ROI (genu of the CC). Green arrow points to the location of the images in (B,D) Scale bar = 1 mm. (B) Background subtraction is determined by the average pixel value in a disk with a user-defined size. Representative images showing the impact of the connected objects identified (green) compared to original image (magenta). Scalebar = 100 μm. (C) Small disk size reduces the number of objects identified. Red line = average disk size used for Plp1-null. Black line = average disk size for WT. (D) Threshold value is determined by a blind user by comparing connected objects identified (green) to original image (magenta). Scale bar = 100 μm. (E) Original image of ROI. Scalebar = 1 mm. (F) Final image of connected objects. Scale bar = 1 mm. (G) Threshold values become increasingly restrictive with greater value. Red line = average threshold used for Plp1-null. Black line = average threshold for WT.
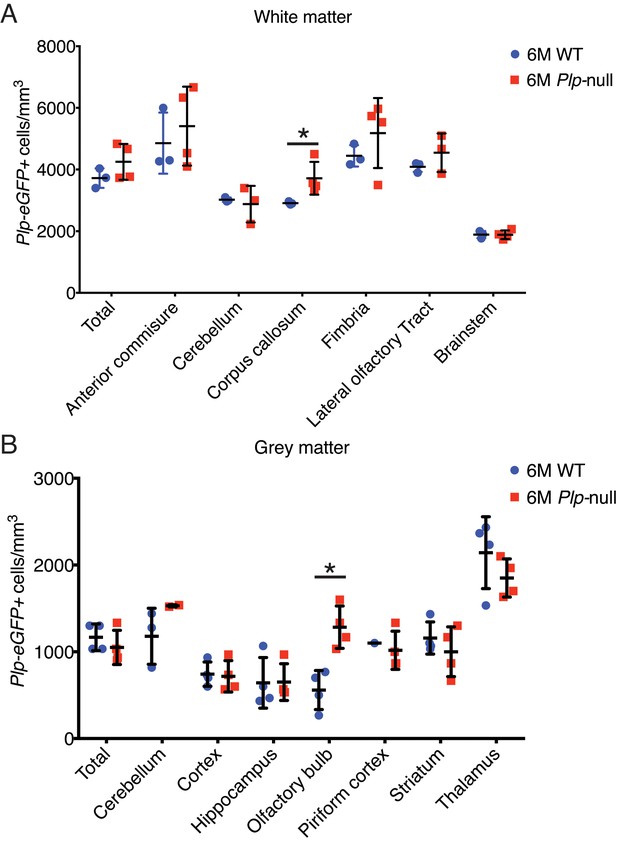
6M old Plp1-null mice exhibit regional differences in oligodendrocyte density.
(A) Oligodendrocyte density in white matter regions demonstrated a region-specific increase in the corpus callosum. (B) A region-specific increase was also observed in the olfactory bulb. (n = 4 mice/genotype, two-way ANOVA, *p<0.01).
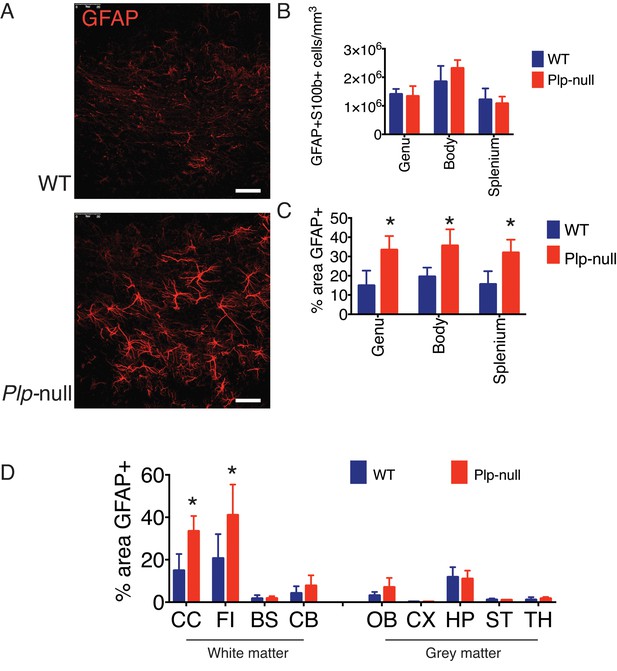
Region-specific astrogliosis was observed in white matter of 2M- old Plp1-null mice.
(A) GFAP immunoreactivity in WT and Plp1-null mice in the CC. Scale bar = 20 μm. (B) No alteration in the density of astrocytes was observed in the CC of Plp1-null mice as quantified by cells with both S100b and GFAP immunoreactivity. (C) Increased percent area covered by GFAP immunoreactivity was observed throughout the regions of the CC. (D) The CC and fimbria (FI) exhibited increased percent area covered by GFAP in Plp1-null mice. (n = 3 mice/genotype, two-way ANOVA, *=p < 0.01). Data is represented as mean ±S.E.M.
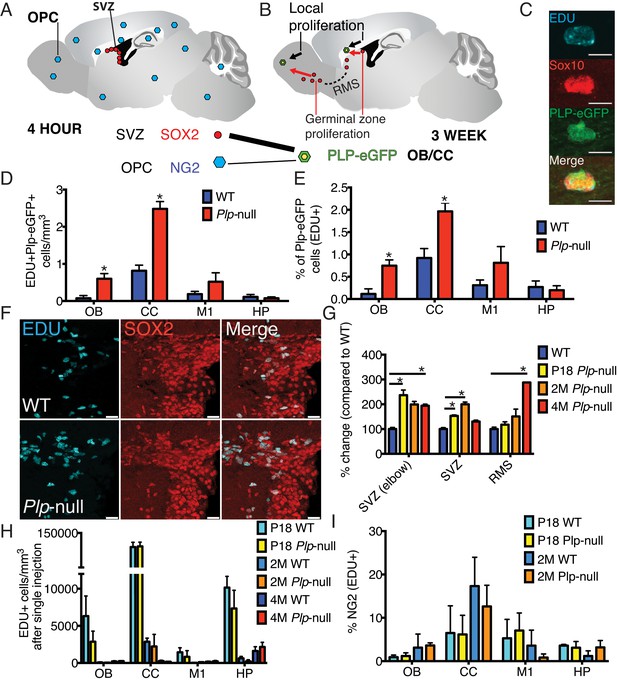
New oligodendrocytes in 4M Plp1-null mice.
(A) Location of proliferating NG2+ OPCs throughout the brain, or SOX2+ progenitors in the SVZ 4 hr after a single EdU injection. (B) EdU-labeled cells at 4 hr give rise to EdU + Plp1-eGFP + oligodendrocytes 3 weeks after injection. (C) New oligodendrocytes identified using Sox10 and Plp1-eGFP. Bar = 10 μm. (D-E) Increase in density of EdU + Plp1-eGFP + oligodendrocytes (D) and percent of Plp1-eGFP+ cells expressing EdU (E) in Plp1-null and WT mice after 3 weeks (*p<0.01, n = 3/genotype). (F) Sox2 and EDU cells in the SVZ in WT and Plp1-null samples. Bar = 25 μm. (G) % change (Plp1-null to WT) of SOX2+ progenitors that were EdU+ after a single EdU injection in SVZ or RMS. Compared by repeated measures ANOVA. *p<0.01. (H) No differential density of EdU + cells was observed in the OB, CC, M1 or HP after a single injection. (I) The % of NG2 +cells labeled by single EdU injection was comparable at P18 or 2M.
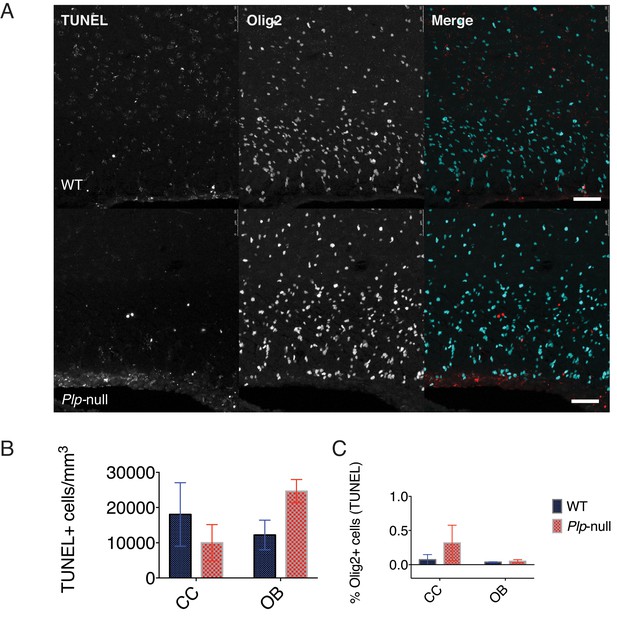
No alteration was found in oligodendrocyte cell death assayed with TUNEL in Plp1-null mice.
(A) Image of TUNEL and Olig2 expression in the CC of 2M WT and Plp1-null mice. Scale bar = 50 μm.(B) No change in the density of TUNEL+ cells in Plp1-null mice. (C) No alteration in the percentage of Olig2+ cells that were TUNEL+. (n = 3 mice/genotype, two-way ANOVA, p>0.05 for all comparisons). Data is represented as mean ±S.E.M.
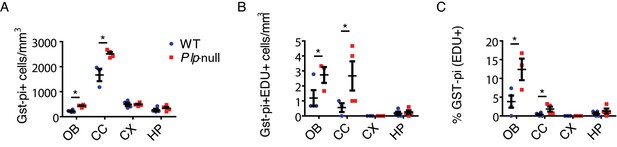
Density of mature oligodendrocytes (Gst-pi+).
(A) The density of mature oligodendrocytes (Gst-pi+) is significantly increased in 4-month Plp1-null mice in the OB (424 ± 26 vs 222 ± 26, p<0.01, RM-ANOVA) and CC (2512 ± 75 vs 1669 ± 246, p<0.01, RM-ANOVA). The number of EDU + Gst-pi+oligodendrocytes is also increased in the Plp1-null OB (2.7 ± 0.5 vs 1.2 ± 0.5, p<0.01, RM-ANOVA) and CC (2.7 ± 1 vs 0.6 ± 0.3, p<0.01, RM-ANOVA). (C) The % Gst-pi +cells that are EDU + is increased in the OB (12 ± 2.7 vs 3.8 ± 1.6) and CC (1.8 ± 0.7 vs 0.4 ± 0.23) of Plp1-null mice. N = 4 mice/genotype.
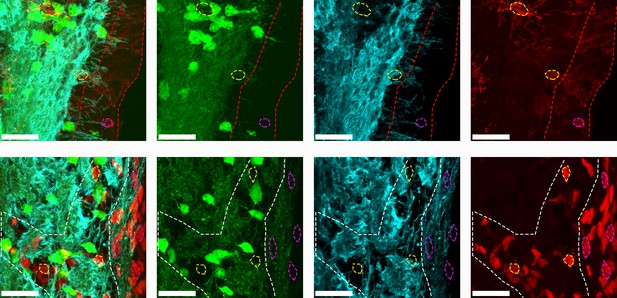
PLP1 expression observed by Plp1-eGFP transgene expression (green) and PLP1 staining (blue) indicates that myelinated fibers are located near and possibly within the SVZ (red line).
PLP1 is located in myelinated processes as shown by the overlap with Plp1-eGFP processes and is difficult to colocalize with cell bodies. Plp1-eGFP was not observed in the cell bodies of NG2+ cells (top panel) in the parachyma (yellow outline) or SVZ (pink outline). Plp1-eGFP was also not observed in the Sox2+ progenitors in the elbow (white line, yellow outline) or SVZ (pink outline).
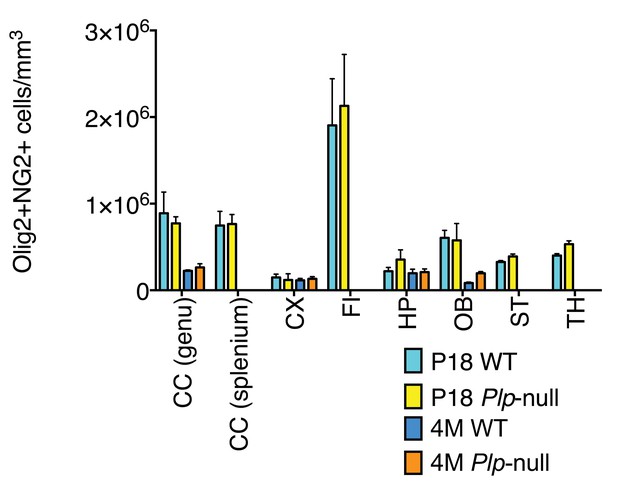
No alteration in NG2 +OPC density in Plp1-null mice.
(A) Density of cells that co-expressed Olig2 and NG2 did not differ between WT and Plp1-null at P18 or 4 months in any region analyzed. (n = 3 mice/genotype for P18, n = 4 mice/genotype for 4 months, two-way ANOVA). Data is represented as mean ±S.E.M.
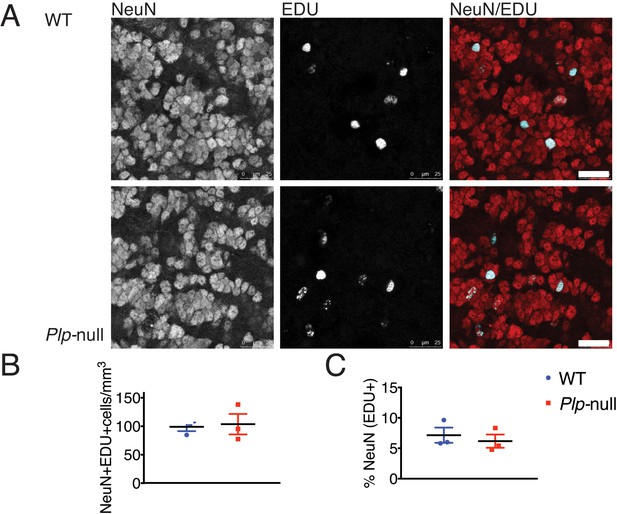
Neurogenesis in Plp1 -null mice was unaltered.
(A) NeuN+ EDU+ cells in WT and Plp1-null mice. Scale bar = 25 μm. (B) Density of NeuN+ EDU+ cells in the OB in WT and Plp1-null mice 3 weeks after EDU injection (n = 3 mice/genotype). (C) % NeuN+ cells (EDU+) in OB in WT and Plp1-null mice 3 weeks after EDU injection (n = 3 mice/genotype).
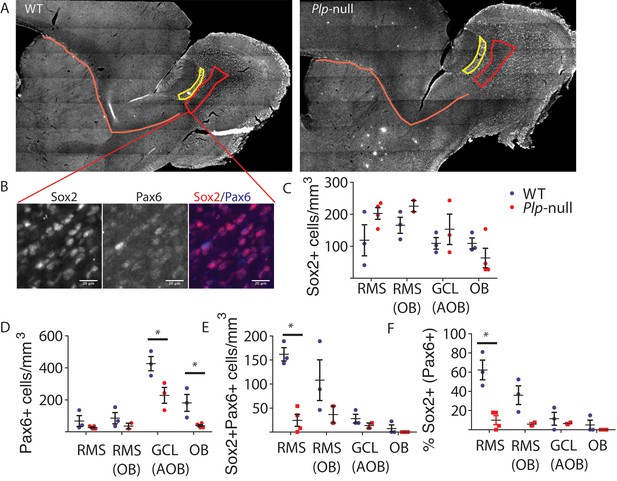
Pax6 expression in the RMS.
(A) Pax6 staining in the RMS (orange line), the RMS entry into the OB (red box), and the GCL of the AOB (yellow box). (B) Colocalization of Sox2 and Pax6 in the RMS. Scale bar = 20 um. (C) The number of Sox2+ cells was not significantly altered in 4 m Plp1-null mice (p>0.05, RM-ANOVA). (D) The number of Pax6+ cells was significantly reduced in the granule cell layer of the OB (38 ± 6.8 vs 181 ± 53) and GCL of the AOB (227 ± 50 vs 427 ± 44, p<0.01, RM-ANOVA). (E) The density of Sox2+ Pax6+ cells was significantly reduced in the RMS (0 vs 7.5 ± 7.5, p<0.01, RM-ANOVA). (F) The % of Sox2 +cells that expressed Pax6 was reduced in the RMS (0 vs 5 ± 5, p<0.01, RM-ANOVA). N = 3 WT and 4 Plp1-null mice. RMS = rostal migratory stream, OB = main olfactory bulb, GCL = granule cell layer, AOB = accessory olfactory bulb.
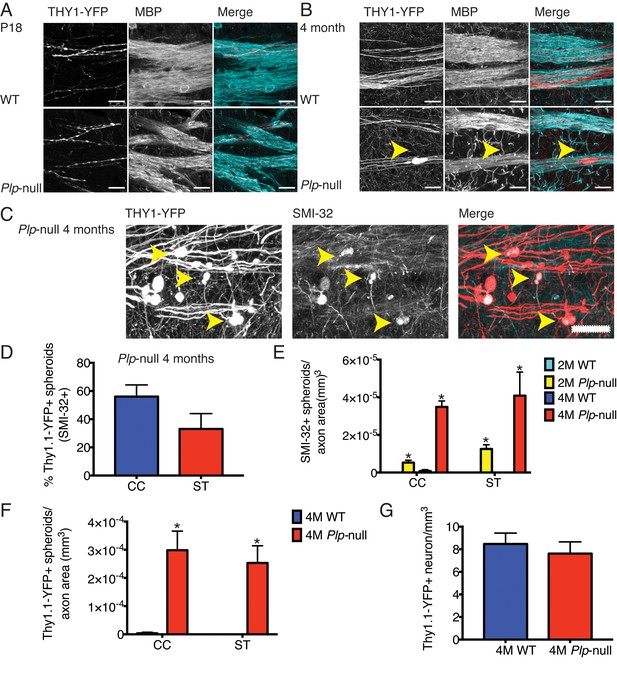
Cortical axon disruption in Plp1-null mice.
(A) Thy1-YFP-positive axons were myelinated in P18 striatum and were surrounded by myelin basic protein (MBP) +processes. At P18, the developing Thy1-YFP axons had varicosities in both WT and Plp1-null mice. Scale bar = 20 μm. (B) At 4 months, a significant number of spheroids were visible in Plp1-null mice in the striatum. Scale bar = 20 μm. (C) SMI-32 was observed in Thy1-YFP+ spheroids in 4M Plp1-null mice. Yellow arrowhead indicates location of spheroid. Scale bar = 20 μm. (D) Percentage of Thy1-YFP+ spheroids that were SMI-32+ in 4M Plp1-null mice in CC (56 ± 8%) and striatum (ST) (33 ± 11%). (E) Density of SMI-32+ spheroids increased in 2M (0 ± 0 versus 5.0 × 10−5 ± 1.0 × 10−5) and 4M Plp1-null mice (7.4 × 10−7 ± 7.4 × 10−7 versus 3.0 × 10−4 ± 3.0 × 10−6). (F) A significant increase in the number of spheroids per axon area was observed in 4M Thy1-YFP + Plp1 null mice. (G) No change in the density of Thy1-YFP+ layer V neurons was observed. n = 3 mice/genotype. WT and Plp1-null were compared with a repeated measures ANOVA. *p<0.01.
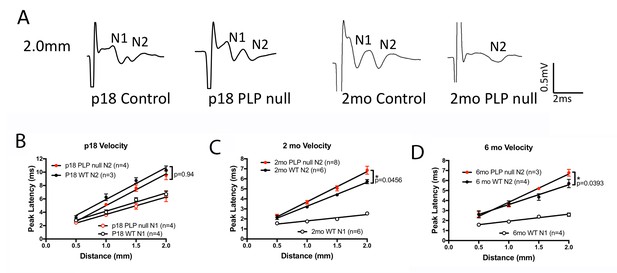
The compound action potential (CAP) for the corpus callosum differs between Plp1-null mice and controls.
(A) Representative traces of callosal CAP recordings elicited by electrical stimulation 2.0 mm away. Fast (N1) and slow (N2) peaks are labeled. The N1 peak is missing for the 2M Plp1-null trace. (B-D) Peak latency vs. distance data for 18D (B), 2 M (C) and 6 M (D) mice. Conduction velocity: N1 ~1.5 m/s. p values are shown for analysis of covariance.
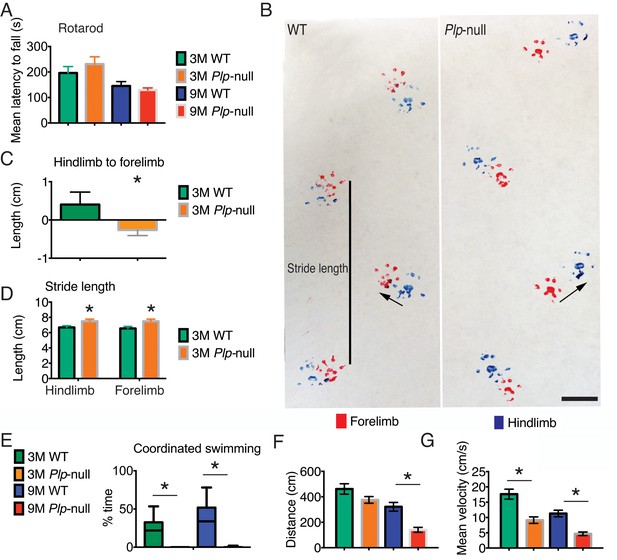
Motor deficits in Plp1-null mice.
(A) Rotarod performance was not altered in 3 and 9M Plp1-null mice. Mean latency to fall (average of three trials, two-way ANOVA, p=0.64, n = 10/ genotype). (B) Gait pattern of 3M Plp1-null and WT siblings. 3M Plp1-null mice overstep their hind limbs as indicated by the arrows. Bar = 1 cm. (C) The directionality from hindlimb to forelimb was reversed in 3M Plp1-null mice (0.4 ± 0.1 vs −0.3 ± 0.03 cm; unpaired t-test, n = 10/ genotype, *p<0.01). (D) Plp1-null mice display a greater stride length (forelimb: 7.2 ± 0.2 vs 6.3 ± 0.1 cm; hindlimb: 7.0 ± 0.2 vs 6.3 ± 0.1 cm; unpaired t-test, n = 10/ genotype, *p<0.01). (E) Coordinated swimming pattern was never observed in Plp1-null mice. Swimming patterns were quantified (Materials and methods) and Plp1-null mice scored 0 (3M: 26 ± 7.6 vs 0 ± 0, 9M: 35 ± 6.2 vs 0 ± 0; two-way ANOVA, n = 10/genotype, *p<0.01). (F-G) 9M Plp1-null mice (G: distance traveled = 140 ± 19 cm; F: mean velocity = 4.7 ± 0.6) performed worse than WT (G: distance traveled = 321 ± 34 cm; F: mean velocity = 11 ± 1.1; two-way ANOVA, n = 10 mice/genotype, *p<0.01). (G) 3M Plp1-null displayed slower mean velocity (F: 9.1 ± 1.1 vs 18 ± 1.6 cm/s; two-way ANOVA, n = 10 mice/genotype, *p<0.01). Means ± s.e.m (A,C,D,F,G) or whisker plot showing min-max (E).
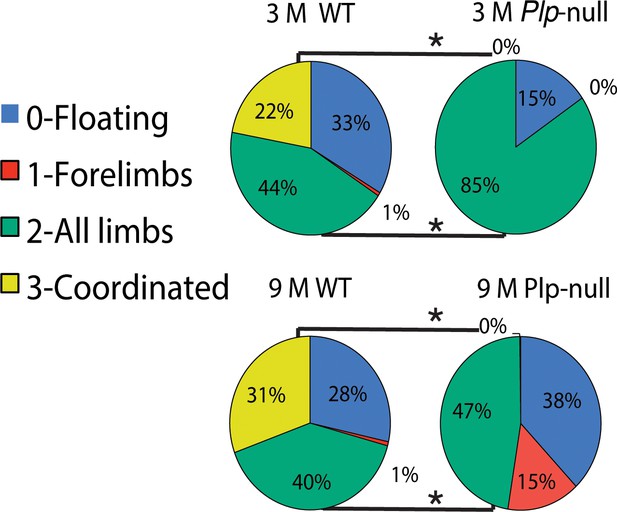
Swim scale evaluation of coordinated swimming.
Results were calculated using repeated-measures ANOVA. Floating was similar between WT and Plp1-null mice (score = 0, 3M: 33 ± 5.1 vs 14 ± 9.9, 9M: 8.2 ± 3.8 vs 6.7 ± 4.5 s; n = 10/genotype). Plp1-null mice swam with all limbs active (score = 2; 3M: 44 ± 6.7 vs 84 ± 6.7, 9M: 55 ± 6.5 vs 87 ± 4.6 s *p<0.01). A coordinated swimming pattern was never observed in Plp1-null mice (score = 3, 3M: 0 ± 0 vs 22 ± 7.2, 9M: 6.2 ± 3.0 vs 37 ± 6.1; *p<0.01).
Movie showing the swimming pattern for a WT mouse.
https://doi.org/10.7554/eLife.34783.019Movie showing the swimming pattern for a Plp1-null mouse.
https://doi.org/10.7554/eLife.34783.020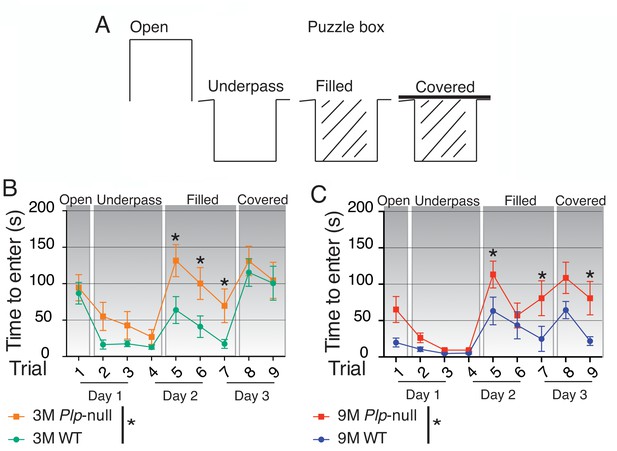
Cognitive function is impaired in Plp1-null mice.
(A) Diagram of entry conditions in the Puzzle Box task. (B) Time to enter the goal box differed significantly between 3M WT and Plp1-null mice (F(1,161) =12.2, ANOVA, n = 10/genotype, p<0.01). 3M WT and Plp1-null differed in trials 5, 6, and 7 (trial 5: p<0.01, trial 6: p<0.01, trial 7: p=0.03, Sidak’s multicomparison, n = 10/genotype). (C) Time to enter differed significantly overall between WT and Plp1-null at 9M (F(1,170) =22.7, ANOVA, n = 10/genotype, p<0.01). 9M WT and Plp1-null differed in trials 5, 7, and 9 (trial 5: p<0.01, trial 7: p<0.01, trial 9: p=0.03, Sidak’s multicomparison, n = 10/genotype).
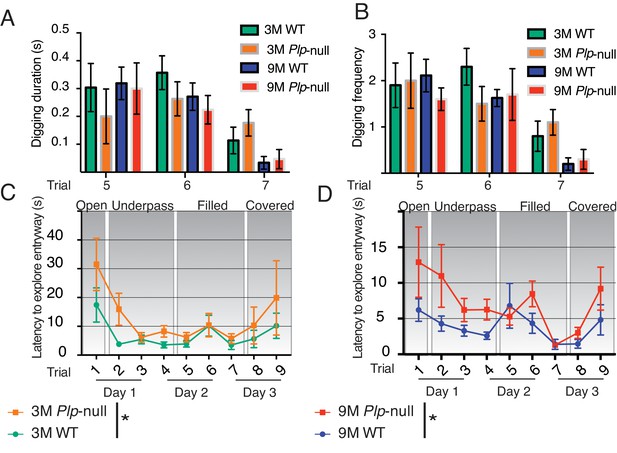
Plp1-null mice exhibit no alteration in digging behavior in the Puzzle Box task.
(A) Digging behavior was manually scored by a blind observer using the EthoVisionXT tracking software. There was no significant difference between WT and Plp1-null mice in the duration spent digging in the entryway (repeated-measures ANOVA, n = 10/genotype, p=0.42). (B) No difference was observed in digging frequency in WT versus Plp1-null mice entryway (repeated-measures ANOVA, n = 10/genotype, p=0.48). (C) The latency to explore the entryway was significantly different between 3M WT and Plp1-null mice overall (F(1,161) = 5.9, repeated measures ANOVA, n = 10/genotype, p=0.02). (D) The latency to explore the entryway was significantly different between 9M WT and Plp1-null mice overall (F(1,159) = 9.5, repeated measures ANOVA, n = 10/genotype, p<0.01).
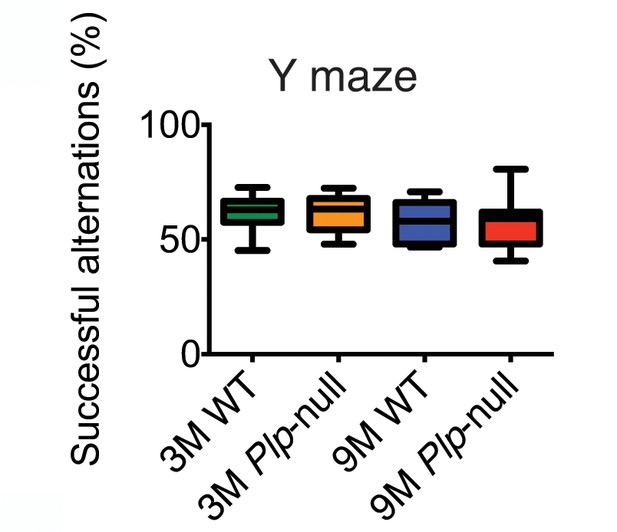
The percent successful alternation patterns in Y maze was similar in WT and Plp1-null mice (two-way ANOVA, p=0.99, n = 10/ genotype).
https://doi.org/10.7554/eLife.34783.024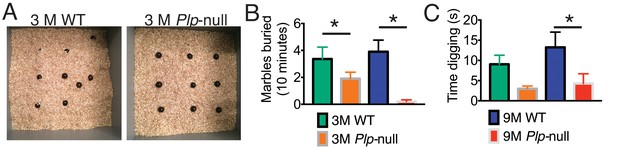
Marble burying task.
(A) Image of marble distribution at the end of marble burying task in 9 month WT and Plp1-null mice. (B) The number of marbles buried differed significantly between WT and Plp1-null mice at 3 months (1.6 ± 0.4 vs 4.8 ± 1.1; *p<0.01) and 9 months (3.9 ± 0.9 vs 0.2 ± 0.1; *p<0.01). (C) 9 month Plp1-null mice spent less time digging then WT (4.3 ± 2.4 vs 13 ± 3.8 s; *p<0.01)n = 10/genotype. Values are means ± s.e.m.
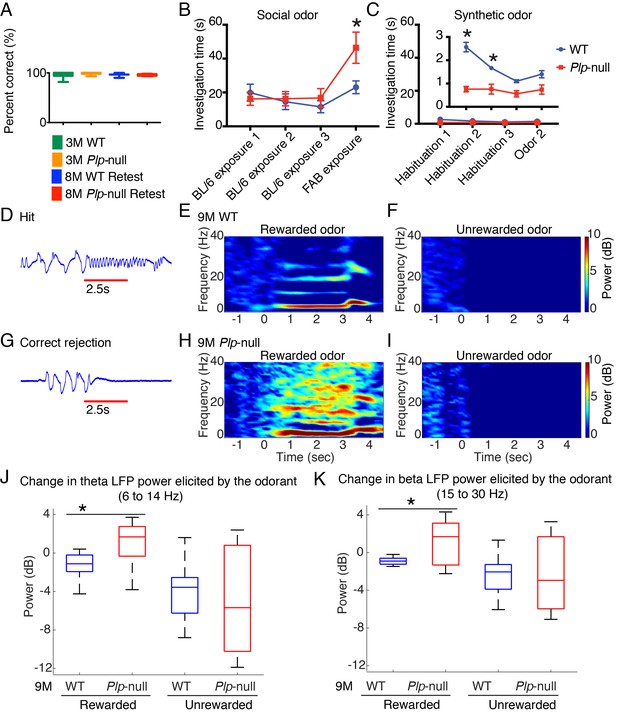
Olfactory function.
(A) Plp1-null mice (3M and 8M) performed similarly to WT in go-no-go task (p=0.69, one-way ANOVA, n = 10/genotype). (B) 9M Plp1-null mice exhibit increased exploration of novel social odor (46 ± 9 vs 23 ± 4 s; 2-way ANOVA, n = 10/genotype, *p<0.01). (C) Synthetic odor investigation was reduced in Plp1-null mice (F(Browne et al., 2014; Almeida and Lyons, 2017) = 30.9; two-way ANOVA, n = 10/genotype, *p<0.01). (D) Representative extracellular voltage trace recorded from piriform cortex when the animal is responding correctly to rewarded odor (hit). Red line = odor delivery. (E-F) Heat maps displaying the stimulus-induced change in power of oscillations during the rewarded (E) and unrewarded trials (F) in 9M WT mice. (G) Voltage trace recorded from piriform cortex during the unrewarded trial (correct rejection). (H-I) Heat maps showing the change in the power of oscillations during rewarded trial (H) and unrewarded trial (I) in 9M Plp1-null mice. (J) During rewarded trials, 9M Plp1-null mice demonstrated odorant-induced increase of power in theta (6 to 14 Hz) (p<0.01, Mann-Whitney U test, n = 64 LFP recordings/genotype). (K) 9M Plp1-null mice demonstrated a greater increase in the change in beta power during rewarded trials (15–30 Hz) (p<0.01, Mann-Whitney U test, n = 64 LFP recordings/genotype).
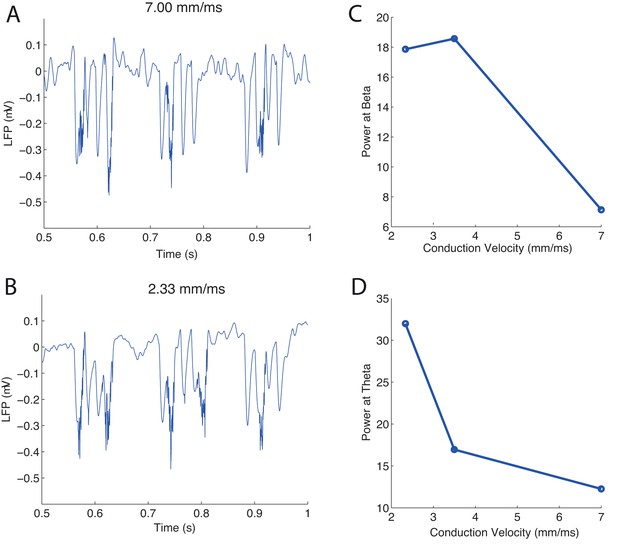
Piriform cortex modeling shows an increase in LFP power when LOT conduction velocity is decreased.
(A, B) Simulated LFPs of the piriform cortex for conduction velocities of 7 mm/ms (A) and 2.33 mm/ms (B). The peak of the power spectral frequencies of the simulated LFP oscillations varies with the CV of the LOT. (C) Power of beta oscillations (6 to 14 Hz). (D) Power of theta oscillations (15 to 30 Hz).
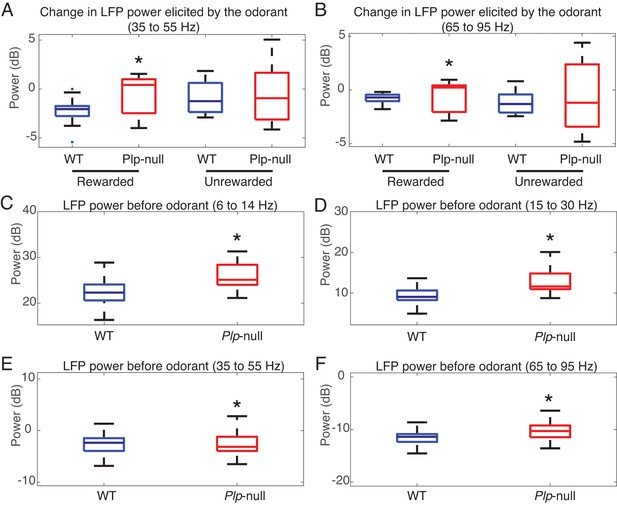
LFP power in WT and Plp1-null mice.
(A and B) Odorant-induced change in LFP power for low gamma and high gamma. (A) Low gamma (35 to 55 Hz) change in power differs for the rewarded odorant between WT and Plp1-null mice (p=1.00e-06, Mann-Whitney U test, n = 64 LFP recordings/genotype). (B) High gamma (65 to 95 Hz) change in power differs for the rewarded odorant between WT and Plp1-null mice (p=5e-05, Mann-Whitney U test, n = 64 LFP recordings/genotype). (C-F) LFP power before odorant application. C. LFP power for the theta band (6 to 14 Hz) differs between WT and Plp1-null mice (p=3e-06, Mann-Whitney U test, n = 64 LFP recordings/genotype). (D) LFP power for the beta band (15 to 30 Hz) differs between WT and Plp1-null mice (p=0.001, Mann-Whitney U test, n = 64 LFP recordings/genotype). (D) LFP power for the low gamma band (35 to 55 Hz) differs between WT and Plp1-null mice (p=0.003, Mann-Whitney U test, n = 64 LFP recordings/genotype). (E) LFP power for the high gamma band (65 to 95 Hz) differs between WT and Plp1-null mice (p=0.01, Mann-Whitney U test, n = 64 LFP recordings/genotype).
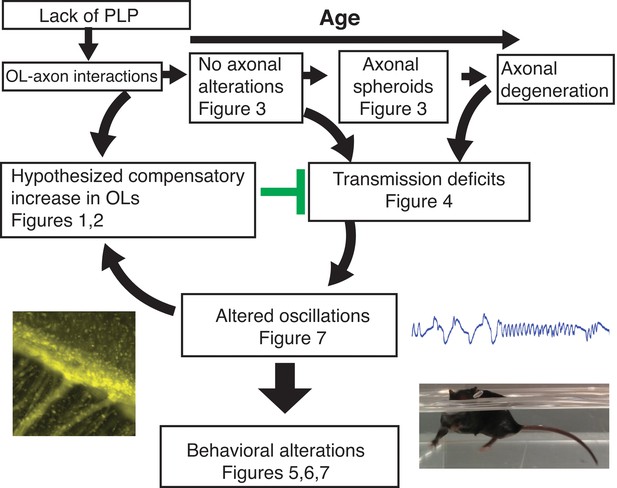
The lack of PLP1 dependent signaling led to early (P18) changes in the number of OPCs in the SVZ and CC, followed by formation of axonal spheroids, increased number of OPCs in OB, and glial inflammation at 2M.
The cellular changes at 2M were accompanied by loss of the N1 fast conduction velocity peak in the CAP of the CC, which was followed by motor coordination and cognitive deficits at 3M. By 4M, mature oligodendrocytes were increased in the OB and CC. Note: Changes in olfactory function were assayed at 9M and may or may not happen at earlier ages. The images included are Plp1-eGFP in the CC (Figure 1D) (left), the raw LFP in piriform cortex (Figure 7D)(top right) and the Plp1-null performing uncoordinated swimming by extending the front paws (Figure 5—videos 1 and 2)(bottom right).
Tables
Abbreviations for brain areas.
https://doi.org/10.7554/eLife.34783.006| AC | Anterior commissure |
|---|---|
| BS | Brainstem |
| CB | Cerebellum |
| CC | Corpus callosum |
| CX | Cortex |
| EPL | External plexiform layer |
| FI | Fimbria |
| HP | Hippocampus |
| LOT | Lateral olfactory tract |
| M1 | Motor cortex |
| OB | Olfactory bulb |
| PC | Piriform cortex |
| ST | Striatum |
| TH | Thalamus |
Accumulation of EDU+ oligodendrocytes in Plp1-null mice.
WT and Plp1-null mice were compared with a repeated measures ANOVA, n = 3 per genotype. Significant differences (p<0.05) are shown in red font.
| Region | Mean(SEM), p value | ||||
|---|---|---|---|---|---|
| 3 weeks | Plp1-eGFP + EDU + | WT | Null | p value | |
| (cells/mm3) | OB | 0.07 (0.07) | 0.60 (0.14) | 0.02 | |
| CC | 0.81 (0.15) | 2.5 (0.20) | <0.01 | ||
| M1 | 0.19 (0.07) | 0.52 (0.24) | 0.11 | ||
| HP | 0.11 (0.06) | 0.07 (0.04) | 0.85 | ||
| 3 weeks | % Plp1-eGFP (EDU+) | ||||
| OB | 0.11 (0.11) | 0.75 (0.13) | 0.11 | ||
| CC | 0.92 (0.21) | 2.00 (0.20) | <0.01 | ||
| M1 | 0.31 (0.12) | 0.81 (0.36) | 0.27 | ||
| HP | 0.23 (0.14) | 0.20 (0.10) | 0.99 | ||
| 3 weeks | SOX10+ EDU+ | ||||
| (cells/mm3) | OB | 0.35 (0.07) | 1.1 (0.02) | 0.04 | |
| CC | 0.81 (0.15) | 2.5 (0.20) | <0.01 | ||
| M1 | 0.46 (0.15) | 1.2 (0.07) | 0.07 | ||
| HP | 0.81 (0.13) | 1.4 (0.43) | 0.13 | ||
| 3 weeks | % SOX10 (EDU+) | ||||
| OB | 0.95 (0.25) | 2.1 (0.11) | 0.19 | ||
| CC | 0.90 (0.25) | 1.9 (0.15) | 0.36 | ||
| M1 | 1.8 (0.36) | 3.0 (0.8) | 0.17 | ||
| HP | 1.6 (0.57) | 3.0 (0.24) | 0.09 | ||
| 3 weeks | % EDU (SOX10+) | ||||
| OB | 1.6 (0.45) | 6.6 (1.0) | 0.86 | ||
| CC | 70 (2.8) | 94 (4.5) | <0.01 | ||
| M1 | 91 (4.8) | 89 (4.7) | 0.99 | ||
| HP | 17 (5.2) | 43 (4.7) | <0.01 | ||
Distance traveled and velocity did not differ between WT and Plp1-null mice in a subset of behavioral tasks.
WT and Plp1-null mice were compared with a repeated measures ANOVA, n = 10 per genotype. There were no significant differences (p>0.05).
| 3 months | 9 months | |||||
|---|---|---|---|---|---|---|
| Behavioral performance | WT (n = 10) Mean(SEM) | Null (n = 10) Mean(SEM) | p value | WT (n = 10) Mean(SEM) | Null (n = 10) Mean(SEM) | p value |
| Open field (duration in center (s)) | 217.0 (15.52) | 203.71 (0.336) | 0.88 | 238.5 (25.14) | 158.6 (22.99) | 0.02 |
| Zero maze (duration open arms (s)) | 92.65 (17.03) | 175.7 (21.24) | 0.03 | 126.5 (27.41) | 117.2 (26.09) | 0.95 |
| Y maze (% successful alternations) | 61.48 (2.484) | 61.56 (2.220) | 0.99 | 57.33 (2.888) | 57.32 (3.726) | 0.99 |
| Y maze (arm entries) | 32.18 (2.529) | 35.91 (2.387) | 0.60 | 32.20 (3.608) | 29.80 (3.252) | 0.82 |
| Marble burying (marbles buried) | 4.800 (1.052) | 1.600 (0.400) | <0.01 | 3.900 (0.862) | 0.200 (0.133) | <0.01 |
| Marble burying (time digging (s)) | 8.428 (2.245) | 2.994 (0.666) | 0.25 | 13.24 (3.767) | 4.294 (2.390) | 0.04 |
| Locomotion | ||||||
| Rotarod (mean latency to fall (s)) | 196.2 (25.08) | 231.2 (28.41) | 0.88 | 145.1 (17.38) | 129.7 (7.948) | 0.94 |
| Distance traveled (cm) | ||||||
| Open field | 5255 (466.3) | 4589 (229.8) | 0.39 | 4510 (444.4) | 5205 (322.4) | 0.36 |
| Zero maze | 9532 (3170) | 10831 (2210) | 0.92 | 6148 (1708) | 7030 (1694) | 0.96 |
| Y maze | 27693 (1346) | 15484 (11985) | 0.68 | 3931 (734.5) | 3812 (324.3) | 0.99 |
| Marble burying | 3729 (379.1) | 2931 (361.1) | 0.16 | 5591 (204.0) | 5566 (289.0) | 0.99 |
| Habituation | 845.2 (78.3) | 1041 (56.31) | 0.06 | |||
| Velocity (cm/s) | ||||||
| Open field | 9.551 (0.673) | 9.287 (0.336) | 0.94 | 7.518 (0.741) | 8.667 (0.537) | 0.32 |
| Zero maze | 16.34 (6.224) | 10.25 (2.864) | 0.52 | 11.79 (2.815) | 18.38 (3.695) | 0.46 |
| Y maze | 65.95 (43.50) | 33.15 (24.92) | 0.61 | 8.987 (1.585) | 8.198 (0.885) | 0.99 |
| Marble burying | 6.766 (0.407) | 6.188 (0.595) | 0.61 | 9.323 (0.339) | 9.298 (0.482) | 0.99 |
| Habituation | 7.859 (0.619) | 6.986 (1.028) | 0.47 | |||
| Distance traveled (cm) | ||||||
| Open field | 5255 (466.3) | 4589 (229.8) | 0.39 | 4510 (444.4) | 5205 (322.4) | 0.36 |
| Zero maze | 9532 (3170) | 10831 (2210) | 0.92 | 6148 (1708) | 7030 (1694) | 0.96 |
| Y maze | 27693 (1346) | 15484 (11985) | 0.68 | 3931 (734.5) | 3812 (324.3) | 0.99 |
| Marble burying | 3729 (379.1) | 2931 (361.1) | 0.16 | 5591 (204.0) | 5566 (289.0) | 0.99 |
| Habituation | 845.2 (78.3) | 1041 (56.31) | 0.06 | |||
| Velocity (cm/s) | ||||||
| Open field | 9.551 (0.673) | 9.287 (0.336) | 0.94 | 7.518 (0.741) | 8.667 (0.537) | 0.32 |
| Zero maze | 16.34 (6.224) | 10.25 (2.864) | 0.52 | 11.79 (2.815) | 18.38 (3.695) | 0.46 |
| Y maze | 65.95 (43.50) | 33.15 (24.92) | 0.61 | 8.987 (1.585) | 8.198 (0.885) | 0.99 |
| Marble burying | 6.766 (0.407) | 6.188 (0.595) | 0.61 | 9.323 (0.339) | 9.298 (0.482) | 0.99 |
| Habituation | 7.859 (0.619) | 6.986 (1.028) | 0.47 | |||
Behavioral performance in the Y maze was similar in WT and Plp1-null mice (two-way ANOVA, p=0.99, n = 10/ genotype, p>0.05).
https://doi.org/10.7554/eLife.34783.026| 3 month | 9 month | |||||
|---|---|---|---|---|---|---|
| Behavioral performance | WT (n = 10) Mean(SEM) | Null (n = 10) Mean(SEM) | p value | WT (n = 10) Mean(SEM) | Null (n = 10) Mean(SEM) | p value |
| Y maze (% successful alternations) | 61.48 (2.484) | 61.56 (2.220) | 0.99 | 57.33 (2.888) | 57.32 (3.726) | 0.99 |
| Y maze (arm entries) | 32.18 (2.529) | 35.91 (2.387) | 0.60 | 32.20 (3.608) | 29.80 (3.252) | 0.82 |
Behavioral characterization of anxiety-like behavior in Plp1-null mice.
WT and Plp1-null mice were compared with a repeated measures ANOVA, n = 10 per genotype. Significant differences (p<0.05) are shown in red font.
| 3 months | 9 months | |||||
|---|---|---|---|---|---|---|
| Behavioral performance | WT (n = 10) Mean(SEM) | Null (n = 10) Mean(SEM) | p value | WT (n = 10) Mean(SEM) | Null (n = 10) Mean(SEM) | p value |
| Open field (duration in center (s)) | 217.0 (15.52) | 203.71 (0.336) | 0.88 | 238.5 (25.14) | 158.6 (22.99) | 0.02 |
| Zero maze (duration open arms (s)) | 92.65 (17.03) | 175.7 (21.24) | 0.03 | 126.5 (27.41) | 117.2 (26.09) | 0.95 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.34783.032





