Mechano-redox control of integrin de-adhesion
Figures
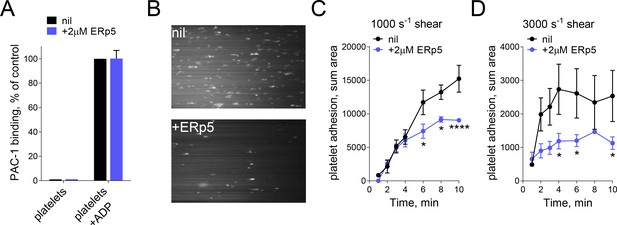
ERp5 triggers αIIbβ3 integrin de-adhesion from fibrinogen.
(A) ERp5 does not induce αIIbβ3 integrin activation on platelets, or influence integrin activation by ADP. The activated αIIbβ3 is measured by PAC-1 antibody binding. The data points and errors (1 SD) are from measurements of four different healthy donor platelets. (B–D) Platelet adhesion to fibrinogen under flow is impaired by ERp5. Part B are representative images of platelet adhesion to immobilized fibrinogen in the absence of presence of 2 µM ERp5. Parts C and D are kinetics of platelet adhesion to fibrinogen in the absence or presence of 2 µM ERp5 at fluid shear rates of 1000 s−1 or 3000 s−1, respectively. The data points and errors (1 SD) are from three measurements each from three different healthy donor platelets. *p<0.05, ****p<0.001; assessed by unpaired, two-tailed Student’s t-test.
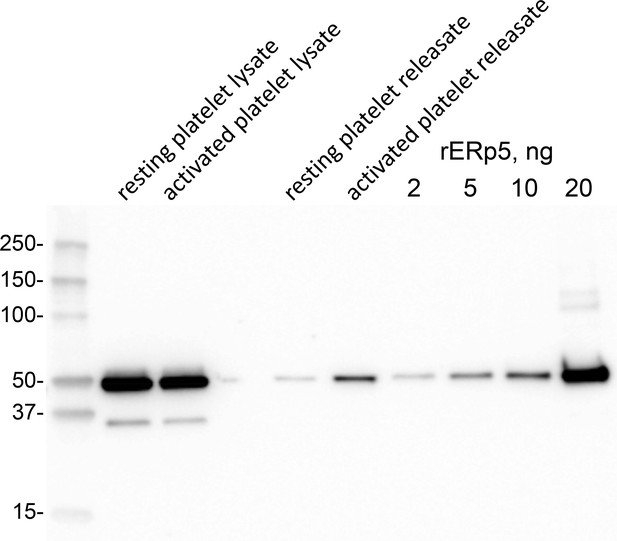
ERp5 level in human platelets and platelet releasate.
Washed human platelets were prepared from a healthy donor. One ml of platelet suspension (363,000 per µL) was untreated or activated with 20 µM ADP for 2 min. Platelet releasate was collected by centrifugation for 15 min at 1000 g. The platelet pellet was lysed with 50 µL 2% NP40, 30 mM Hepes, 150 mM NaCl, 2 mM EDTA, pH 7.4 buffer containing proteinase inhibitor cocktail and clarified by centrifugation at 10,000 g for 20 min at 4°C. One µL of platelet lysate and 10 µL of releasate were resolved on reducing SDS-PAGE and the ERp5 immunoblotted with 1 µg/mL rabbit anti-ERp5 polyclonal antibody. Platelet ERp5 levels were calculated by reference to a standard curve of recombinant ERp5 concentrations. Densitometry of the chemiluminescent bands was performed by ImageQuant TL software (Biorad). Molecular size markers are shown at left.
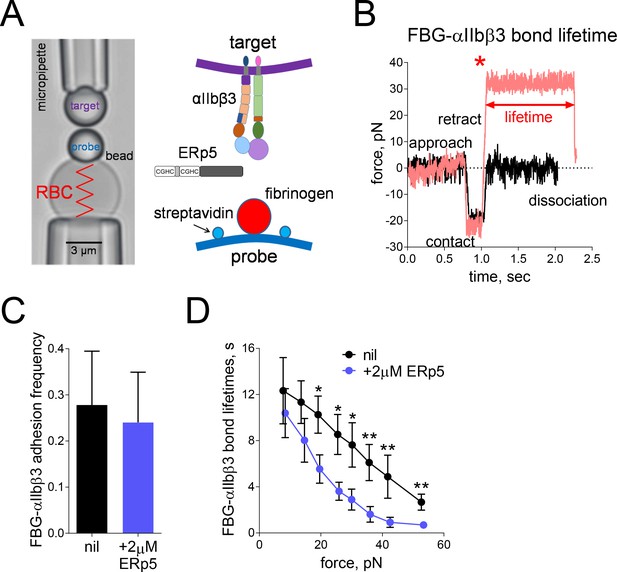
Mechanical force accelerates ERp5-induced fibrinogen de-adhesion.
(A) Biomembrane-Force-Probe (BFP) detection of αIIbβ3–fibrinogen (FBG) binding in a purified system. The probe bead was coated with fibrinogen and streptavidin (SA) for attachment of the bead to biotinylated red blood cell (RBC). Fibrinogen is the focus for interaction with αIIbβ3 on the target bead. The effect of soluble ERp5 on this interaction was measured. (B) Force versus time traces from two representative test cycles. The target bead was driven to approach and contact the probe bead, then retracted. In a 'no bond' event (black), the cycle ended after the probe–target separation. In a 'bond' event (red), the target was clamped (marked by *) at a preset force until dissociation. Lifetime is measured from the point when the clamped force (i.e. 30 pN) was reached to the point when the bond dissociated, signified by a force drop to zero. (C) Soluble ERp5 has no significant effect on αIIbβ3–fibrinogen adhesion frequency. The adhesion frequencies represent mean ± SEM of n = 3 independent experiments in the absence or presence of 2 µM ERp5. For each experiment, five random probe–target pairs with 50 touches were analyzed and averaged. (D) Lifetime of fibrinogen–αIIbβ3 integrin bonds vs. clamp force in the absence or presence of 2 µM ERp5. Results represent mean ±SEM of 10–30 measurements at each force bin. *p<0.05, **p<0.01; assessed by unpaired, two-tailed Student’s t-test.
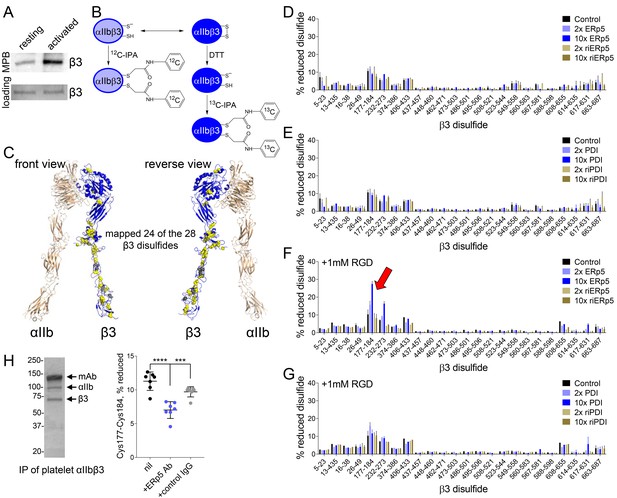
ERp5 cleaves the βI-domain Cys177-Cys184 disulfide bond.
(A) One or more disulfide bonds are cleaved in platelet β3 integrin upon platelet activation. Washed platelets were untreated or activated with ADP and unpaired cysteine thiols in platelet surface proteins labelled with the biotin-linked maleimide, MPB. The β3 integrin was immunoprecipitated from the platelet lysate and blotted for MPB. (B) Differential cysteine alkylation and mass spectrometry method of measuring the redox state of the β3 integrin cysteines. Unpaired cysteine thiols in purified β3 integrin are alkylated with 12C-IPA and the disulfide bonded cysteine thiols with 13C-IPA following reduction with DTT. Sixty-eight peptides (Supplementary file 1) encompassing cysteines representing 24 of the 28 β3 integrin disulfide bonds were analyzed. (C) Positions of the β3 integrin disulfide bonds (yellow and grey spheres) in a modelled open structure of the complete αIIbβ3 integrin ectodomain (blue ribbon). We were able to map 24 of the 28 β3 disulfide bonds. The mapped disulfides are in yellow and the unmapped bonds are in grey. (D–E) Redox state of 24 of the 28 β3 integrin disulfide bonds in the absence or presence of 2- or 10-fold molar excess of ERp5 or redox inactive ERp5 (D), or PDI or redox inactive PDI (E). (F–G) Redox state of 24 of the 28 β3 integrin disulfide bonds in the absence or presence of 2- or 10-fold molar excess of ERp5 or redox inactive ERp5 (D), or PDI or redox inactive PDI (E), and 1 mM RGD peptide. The bars and errors (1 SD) are for 5–15 measurements from three different integrin preparations. The β3 Cys177-Cys184 disulfide bond (indicated by red arrow) is significantly cleaved by 10-fold molar excess of ERp5 (p<0.05) (H) The βI-domain Cys177-Cys184 disulfide bond is cleaved on the platelet surface by platelet ERp5. Washed platelets were incubated with function-blocking anti-ERp5 antibodies or isotype control antibodies and the redox state of the βI-domain disulfide determined in the integrin immunoprecipitated from lysate. The bars and errors (1 SD) are from three different platelet preparations from healthy donors. ***p<0.005; ****p<0.001; assessed by unpaired, two-tailed Student’s t-test.
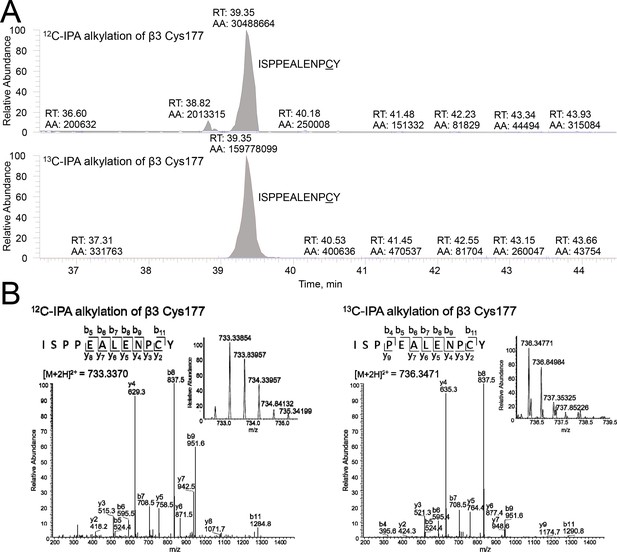
Differential cysteine alkylation and mass spectrometry analysis of the β3 Cys177-Cys184 disulfide bond.
(A) HPLC resolution of the β3 ISPPEALENPCY peptide containing Cys177 labelled with either 12C-IPA or 13C-IPA. (B) Representative tandem mass spectra of the β3 ISPPEALENPCY peptide. At left is an example of 12C-IPA-alkylation of Cys177, and at right is an example of 13C-IPA-alkylation of Cys177. The accurate mass spectrum of the peptide is shown in the inset (left, observed [M + 2 hr]2+ = 733.3370 m/z and expected [M + 2 hr]2+ = 687.3207 m/z; right, observed [M + 2 hr]3+ = 736.3471 m/z and expected [M + 2 hr]2+ = 690.331).
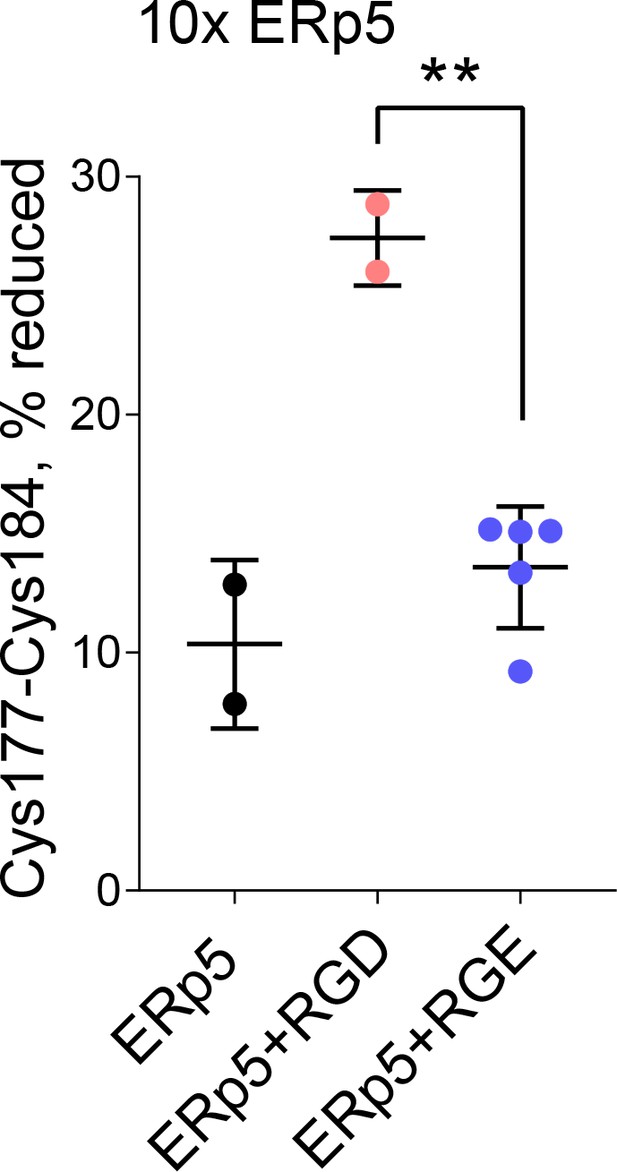
Redox state of the β3 Cys177-Cys184 disulfide bond in the absence or presence of 10-fold molar excess of ERp5 and 1 mM RGD or control RGE peptide.
The bars and errors (1 SD) are for 2–5 biological replicates from a single integrin preparations. **p<0.01; assessed by unpaired, two-tailed Student’s t-test.
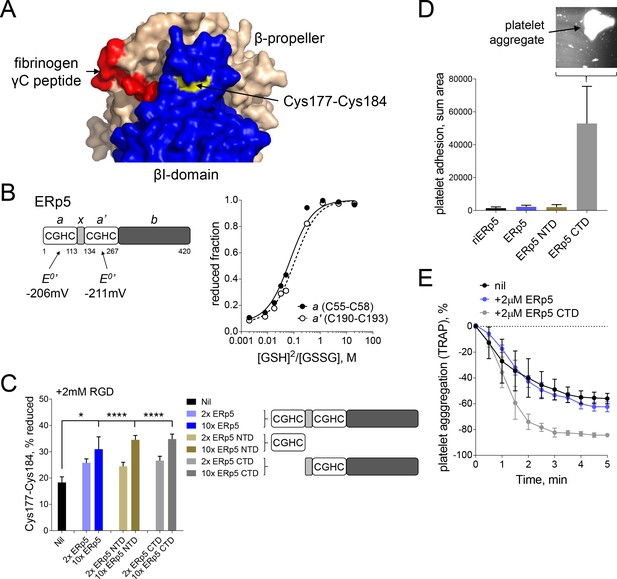
Both catalytic domains of ERp5 are required for substrate specificity.
(A) Surface structure of the extended holo conformation headpiece (PDB code 2vdo) showing the βI-domain (blue), β-propeller domain of the α2b subunit (wheat) and fibrinogen γC peptide (red). Cys184 of the Cys177-Cys184 disulfide (yellow) is surface exposed in a pocket that is removed from the ligand binding pocket. (B) Redox potentials of the a (Cys55-Cys58) and a' (Cys190-Cys193) active-site dithiols/disulfides of ERp5. Plots of the fraction of reduced ERp5 as a function of the ratio of GSH to GSSG. The lines represent the best non-linear least squares fit of the data to Equation 1. The calculated equilibrium constants were used to determine the standard redox potentials from Equation 2. Data points and errors are the mean of 2–4 peptides encompassing the active site cysteine residues. (C) Cleavage of the Cys177-Cys184 disulfide bond by 2- or 10-fold molar excess of full-length ERp5, ERp5 N-terminal domain, or ERp5 C-terminal domain. The bars and errors (1 SD) are for 2–6 measurements. *p<0.05, ****p<0.001; assessed by unpaired, two-tailed Student's t-test. (D) Platelet adhesion to fibrinogen at 4 min in the absence or presence of 2 µM redox inactive ERp5 (riERp5), ERp5 or the N- or C-terminal ERp5 domains at a fluid shear rate of 1000 s−1. The inset is a representative image of platelet adhesion to immobilized fibrinogen in the presence of 2 µM ERp5 CTD. The bars and errors (1 SD) are from three measurements each from six different healthy donor platelets. (E) Aggregation of washed platelets activated with the PAR-1 agonist, TRAP (7 µM), in the absence or presence of 2 µM ERp5 or ERp5 CTD. The data points and errors (1 SEM) are from three different healthy donor platelets.
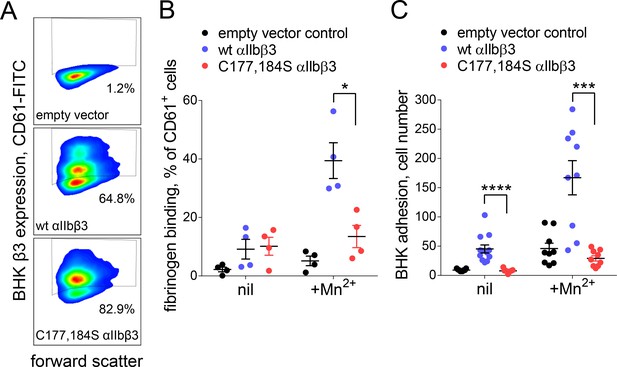
Cleavage of the βI domain disulfide impairs fibrinogen binding.
(A) Expression of wild-type (64.8% of cells) and β3 C177,184S disulfide mutant αIIbβ3 integrin (82.9% of cells) on BHK cells from staining with fluorescene-conjugated anti-CD61 antibody and flow cytometry. Transfection with empty vector served as negative control. (B) Binding of soluble fibrinogen-AF647 (0.1 mg/mL) to BHK cells expressing wild-type or C177,184S mutant αIIbβ3 integrin. Binding is expressed as the % of CD61+ cells that bound fibrinogen. Data points and errors (SD) are from four separate experiments. (C) Adhesion of BHK cells expressing wild-type or C177,184S mutant αIIbβ3 integrin to immobilized fibrinogen. Experiments were performed in triplicate, three fields were analyzed per well and errors are SD. *p<0.05, ***p<0.005, ****p<0.001; assessed by unpaired, two-tailed Student’s t-test.
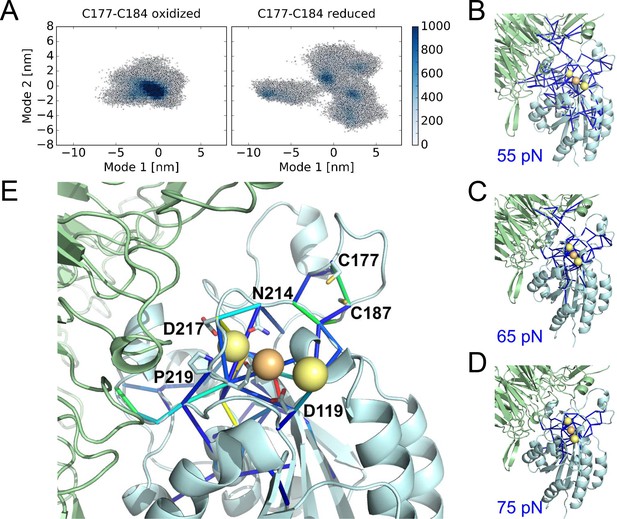
Cleavage of the βI disulfide increases βI-domain flexibility and stresses at the MIDAS site.
(A) Conformational distribution of the oxidized (left) and reduced (right) αIIbβ3 integrin apo headpiece along two major conformational modes as obtained from MD simulations, starting from the extended apo conformation (PDB code 3fcu). The color code shows the density of conformations, ranging from low (white) to high (dark blue) density (in arbitrary units). Disulfide reduction increases the covered area and, thus, the conformational fluctuations. (B–D) Allosteric signaling network upon reduction of the Cys177-Cys184 disulfide bond. Differences in Fpair-wise force between the oxidized and reduced integrin headpiece are shown as blue sticks for the indicated force cut-off values. Calcium and magnesium ions are presented as yellow and orange spheres, respectively, the β-propeller domain as the green cartoon and the βI-domain as the cyan cartoon. (E) Details of the Fpair-wise force difference network around the metal-binding sites in the βI domain. Stress intensity is indicated by the color spectrum as used for b-factors (spectrum ranging from 75 pN – 399 pN).
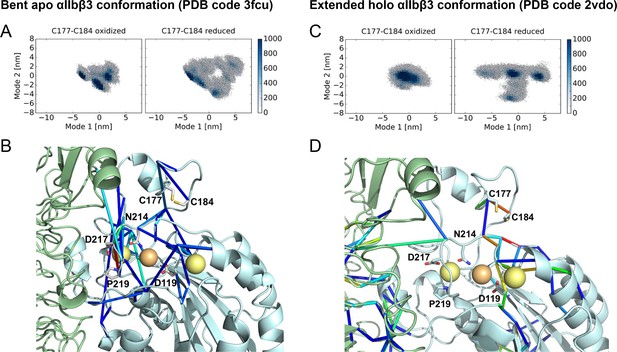
Cleavage of the βI disulfide results in increased βI domain flexibility and high stresses at the MIDAS site in the bent apo and extended holo αIIbβ3 structures.
Conformational distribution of the oxidized (left) and reduced (right) αIIbβ3 integrin, obtained from MD simulations, starting from the bent apo (A) and extended holo (C) conformation. The MD trajectories are projected along two major conformational modes, obtained from principal component analysis. The color code shows the density of conformations, ranging from low (white) to high (dark blue) density (in arbitrary units). Disulfide reduction increases the covered area and, thus, the conformational fluctuations. Allosteric signaling network upon reduction of the Cys177-Cys184 disulfide bond in the bent apo (B) and extended holo (D) conformations, recovered from force distribution analysis. Only pairs of residues for which the pair-wise force difference was larger than a 55 pN cutoff value are shown. Differences in pairwise forces are shown as blue sticks, calcium and magnesium ions as yellow and orange spheres, the β propeller domain as a green cartoon and the βI domain as a cyan cartoon. The peptide ligand is not shown. Stress intensity is indicated by the color spectrum as used for b-factors (spectrum ranging from 55 pN - 268 pN force difference between the oxidized and reduced integrin headpiece in (B) and from 55 pN - 165 pN in (C).
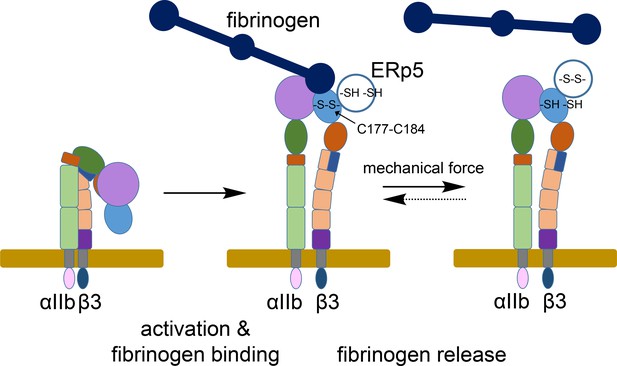
ERp5 regulation of fibrinogen release from activated αIIbβ3 integrin.
Activation of αIIbβ3 integrin results in transition from a bent (low affinity) to an extended (high affinity) structure. While much is known about the structure and function of resting and activated integrins, little is known about how integrins disengage from their ligands. Our findings indicate that ERp5 regulates fibrinogen release from extended αIIbβ3 integrin by reducing the βI domain Cys177-Cys184 disulfide bond (light blue oval). Cleavage of the disulfide is controlled by both ligand binding and mechanical force. It is feasible that the disulfide bond could reform and the integrin reengage fibrinogen (dotted arrow).
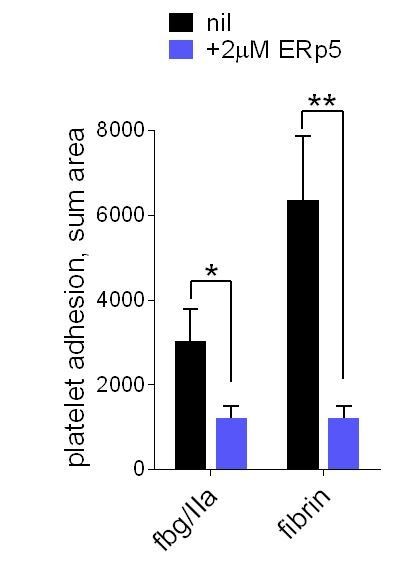
ERp5 triggers αIIbβ3 integrin de-adhesion from fibrin.
Platelet adhesion to immobilized fibrin in the absence of presence of 2 µM ERp5 was measured. Fibrin was immobilized in two ways: immobilized fibrinogen was incubated with thrombin and residual thrombin inactivated with hirudin (fbg/IIa), or pre-formed fibrin was directly immobilized (fibrin). Adhesion was measured in the first 4 minutes at a fluid shear rate of 1000 s-1. The bars and errors (1 SD) are from three measurements each from three different healthy donor platelets. *, p < 0.05, **, p < 0.01; assessed by unpaired, two-tailed Student’s t-test. Additional experimental details are as described in the manuscript for platelet adhesion to fibrinogen.
Additional files
-
Supplementary file 1
List of β3 integrin cysteine containing peptides detected by mass spectrometry.
- https://doi.org/10.7554/eLife.34843.014
-
Supplementary file 2
Structural features of the β3 integrin Cys177-Cys184 disulfide bond.
- https://doi.org/10.7554/eLife.34843.015
-
Supplementary file 3
The βI Cys177-Cys184 disulfide bond is conserved in 7 of 8 β integrins.
Phylogenetic tree for the integrin β-subunit Cys177-Cys184 disulfide bond. The cysteines forming the disulfide are highlighted in yellow.
- https://doi.org/10.7554/eLife.34843.016
-
Transparent reporting form
- https://doi.org/10.7554/eLife.34843.017





