The plant-specific transcription factors CBP60g and SARD1 are targeted by a Verticillium secretory protein VdSCP41 to modulate immunity
Abstract
The vascular pathogen Verticillium dahliae infects the roots of plants to cause Verticillium wilt. The molecular mechanisms underlying V. dahliae virulence and host resistance remain elusive. Here, we demonstrate that a secretory protein, VdSCP41, functions as an intracellular effector that promotes V. dahliae virulence. The Arabidopsis master immune regulators CBP60g and SARD1 and cotton GhCBP60b are targeted by VdSCP41. VdSCP41 binds the C-terminal portion of CBP60g to inhibit its transcription factor activity. Further analyses reveal a transcription activation domain within CBP60g that is required for VdSCP41 targeting. Mutations in both CBP60g and SARD1 compromise Arabidopsis resistance against V. dahliae and partially impair VdSCP41-mediated virulence. Moreover, virus-induced silencing of GhCBP60b compromises cotton resistance to V. dahliae. This work uncovers a virulence strategy in which the V. dahliae secretory protein VdSCP41 directly targets plant transcription factors to inhibit immunity, and reveals CBP60g, SARD1 and GhCBP60b as crucial components governing V. dahliae resistance.
https://doi.org/10.7554/eLife.34902.001eLife digest
Like animals, plants have an immune system to protect themselves from disease. When a plant detects a disease-causing microbe, proteins that serve as master regulators of its immune system activate defense-related genes. Yet some microbes can overcome these defenses and successfully infect plants. Verticillium dahliae is a fungus, found in soil, that infects the roots of many plants – including cotton, tomatoes and potatoes. Infection by this fungus causes the leaves to curl and discolor, and the plant to wilt.
The V. dahliae fungus releases, or secretes, nearly 800 proteins during an infection. Yet it remains unknown if and how any of these proteins help the fungus to infect plants. A better understanding of how V. dahliae impairs plant immunity to infect its hosts could give insights into ways to improve plant resistance against this fungus.
Now, Qin et al. show that a secreted protein called VdSCP41 promotes V. dahliae infection in both cotton and Arabidopsis plants. Further experiments showed that after leaving the fungus, VdSCP41 enters into the plant’s own cells. Protein-protein interaction and biochemical studies then indicated VdSCP41 associates with a master immune regulator in Arabidopsis called CBP60g. This interaction interferes with CBP60g’s ability to activate the defense-related genes.
Now that this role for VdSCP41 has been confirmed, the next step would be to see if targeting it would make plants more resistant to this fungus. One approach would be to genetically engineer plants so that they can specifically shut down, or ‘silence’, the fungal gene that encodes for this protein. Further experiments are required to see whether using this technique – known as host-induced gene silencing (or HIGS for short) – against VdSCP41would enhance plant resistance to V. dahliae. If it does prove effective, this approach may eventually reduce the need for chemical pesticides to protect crop plants.
https://doi.org/10.7554/eLife.34902.002Introduction
The vascular pathogen Verticillium dahliae infects a broad range of plants and causes devastating diseases. Verticillium dahliae can survive in the form of microsclerotia in soil for over ten years (Schnathorst, 1981). During V. dahliae colonization, microsclerotia germinate and develop hyphae that, upon perception of plant roots, adhere tightly to the root surface (Zhao et al., 2014). A few hyphae form hyphopodia at the infection site and further differentiate into penetration pegs that penetrate into plant cells and colonize the vascular tissue (Fradin and Thomma, 2006; Schnathorst, 1981; Vallad and Subbarao, 2008; Zhao et al., 2014Zhao et al., 2016). Although a few key steps mediating these infection processes have been elucidated, the molecular mechanisms underlying V. dahliae virulence remain largely unknown.
Progress has been made in isolating virulence factors that are crucial for V. dahliae virulence. Several genes that regulate V. dahliae development have been characterized as contributing to virulence. VDH1 and VdGARP1, which regulate microsclerotial development, are required for V. dahliae virulence in cotton plants (Gao et al., 2010; Klimes and Dobinson, 2006). VdRac1 and VdPKAC1 regulate both the development and the pathogenicity of the fungus in host plants (Tian et al., 2015; Tzima et al., 2010, 2012). VdEG1, VdEG3, VdCYP1, and VdSNF1 also function as factors that contribute to V. dahliae virulence as it infects cotton (Gui et al., 2017; Tzima et al., 2011; Zhang et al., 2016).
In addition to development-associated virulence factors, fungal pathogens deliver effectors that act as virulence factors, thereby inhibiting host defense and promoting pathogenesis. Bacterial and oomycete pathogens also deliver such effectors. The race 1 strain-specific effector Ave1 contributes to V. dahliae virulence on tomato plants not carrying Ve1 (de Jonge et al., 2012). Necrosis and ethylene-inducing peptide 1 (Nep1)-like proteins (NLPs) (NLP1 and NLP2) secreted by V. dahliae strain JR2 are required for its pathogenicity in tomato and Arabidopsis plants (Santhanam et al., 2013). VdSge1 encodes a transcriptional regulator that controls the expression of six putative effector genes. Deletion of VdSge1 in V. dahliae significantly impairs its pathogenicity in tomato, suggesting an important role for secreted effectors in suppressing host immunity in V. dahliae (Santhanam and Thomma, 2013).
Nevertheless, only two secreted effectors of V. dahliae have been indicated to function inside the plant cell to modulate host immunity to date. VdIsc1, a V. dahliae effector lacking a known signal peptide, is thought to be delivered into host cells to hydrolyze a salicylic acid (SA) precursor and thereby inhibit salicylate metabolism (Liu et al., 2014). The small cysteine-containing protein (SCP) VdSCP7 translocates into the nucleus of plant cells to either suppress or induce defense in plants through unknown mechanisms (Zhang et al., 2017).
Plants are equipped with immune components to counteract V. dahliae virulence. Tomato Ve1 has been identified as an effective resistance locus that recognizes Ave1 secreted by Race 1 strains (Fradin et al., 2009; Schaible et al., 1951). Genetic analyses indicated that EDS1, NDR1 and SERK3/BAK1 are required for Ve1-mediated resistance in both tomato and Arabidopsis (Fradin et al., 2009, 2011). Studies in cotton also showed that GhBAK1 and GhNDR1 are crucial components in regulating defense against V. dahliae (Gao et al., 2013b; Gao et al., 2011), demonstrating that both NDR1 and SERK3/BAK1 are required in a conserved mechanism for defense against V. dahliae in these plants. GbWRKY1, GhSSN, GbERF, GhMLP28, GhMKK2 and GbNRX1 have also been shown to be required for cotton resistance against V. dahliae (Gao et al., 2011; Li et al., 2014a, 2016; Qin et al., 2004; Sun et al., 2014; Yang et al., 2015). A comparative proteomic analysis indicated the involvement of both brassinosteroids and jasmonic acid signaling pathways in the regulation of cotton resistance to V. dahliae (Gao et al., 2013a).
Although several regulators have been identified, the mechanisms through which plants defend against V. dahliae remain obscure, and further investigation is required to isolate more host immune components governing V. dahliae resistance. Targeting key immune components is a common strategy employed by pathogenic effectors to promote pathogenicity (Boller and He, 2009; Cui et al., 2009; Dou and Zhou, 2012); thus, the screening of host proteins targeted by pathogenic effectors provides an efficient way to identify crucial host defense components.
The calmodulin-binding proteins (CBPs) function to bind calmodulin and thus to transduce calcium signals (Bouché et al., 2005). The plant-specific CALMODULIN BINDING PROTEIN 60 (CBP60) protein family contains eight family members, including CBP60a–g and SYSTEMIC ACQUIRED RESISTANCE DEFICIENT 1 (SARD1) (Bouché et al., 2005). CBP60g and SARD1 are two closely related members that function partially redundantly in both SA signaling and bacterial resistance (Zhang et al., 2010; Wang et al., 2011). CBP60g contains a calmodulin-binding domain (CBD) that is essential for its function in defense, whereas SARD1 does not bind calmodulin (Wang et al., 2009Wang et al., 2011; Zhang et al., 2010). Both CBP60g and SARD1 function as transcription factors that directly bind to the promoters of genes that control SA synthesis, such as Isochorismate Synthase 1 (ICS1), ENHANCED DISEASE SUSCEPTIBILITY 5 (EDS5), and NON-EXPRESSOR OF PATHOGENESIS RELATED GENES1 (NPR1) (Dong, 2004; Nawrath et al., 2002; Sun et al., 2015; Wildermuth et al., 2001). Moreover, ChIP-seq analyses have revealed that CBP60g and SARD1 directly bind to the promoters of a number of genes, thereby regulating pathogen-associated molecular patterns (PAMPs)-triggered immunity (PTI), effector-triggered immunity (ETI) and systemic acquired resistance (SAR) (Sun et al., 2015), indicating their broad role in the regulation of plant immunity. In this study, we identified VdSCP41 as a virulence effector that suppresses plant immunity induced by PAMPs. VdSCP41 interacts with Arabidopsis CBP60g and SARD1 and modulates their transcription factor activity. The contribution of VdSCP41 to V. dahliae virulence is significantly reduced during the infection of the cbp60g-1/sard1-1 double mutant. GhCBP60b, the closest protein homolog of CBP60g in cotton, is also targeted by VdSCP41 and contributes to cotton resistance against V. dahliae. Taken together, our findings revealed that CBP60g, SARD1 and GhCBP60b are novel components that govern V. dahliae resistance and that these proteins are modulated by a secretory effector VdSCP41.
Results
VdSCP41 contributes to V. dahliae virulence in Arabidopsis and cotton plants
Secretome analyses revealed more than 700 potential secreted proteins in V. dahliae (Klosterman et al., 2011), but to date, relatively few of these secreted proteins have been characterizes as carrying virulence function. We performed a reverse genetic screen to identify secreted proteins that are crucial for V. dahliae virulence. Using the newly developed USER-ATMT-DS binary vector (Wang et al., 2016), we constructed 56 gene deletion mutants in V. dahliae strain 592 (V592), each of which targets an individual potentially secreted protein. The resulting mutants were subjected to virulence assessment in host plants, including upland cotton (Gossypium hirsutum) and the model plant Arabidopsis. A mutant carrying a targeted deletion of VdSCP41 (VdΔscp41) was isolated (Figure 1A) and shown to display significantly reduced virulence compared with WT strain V592.
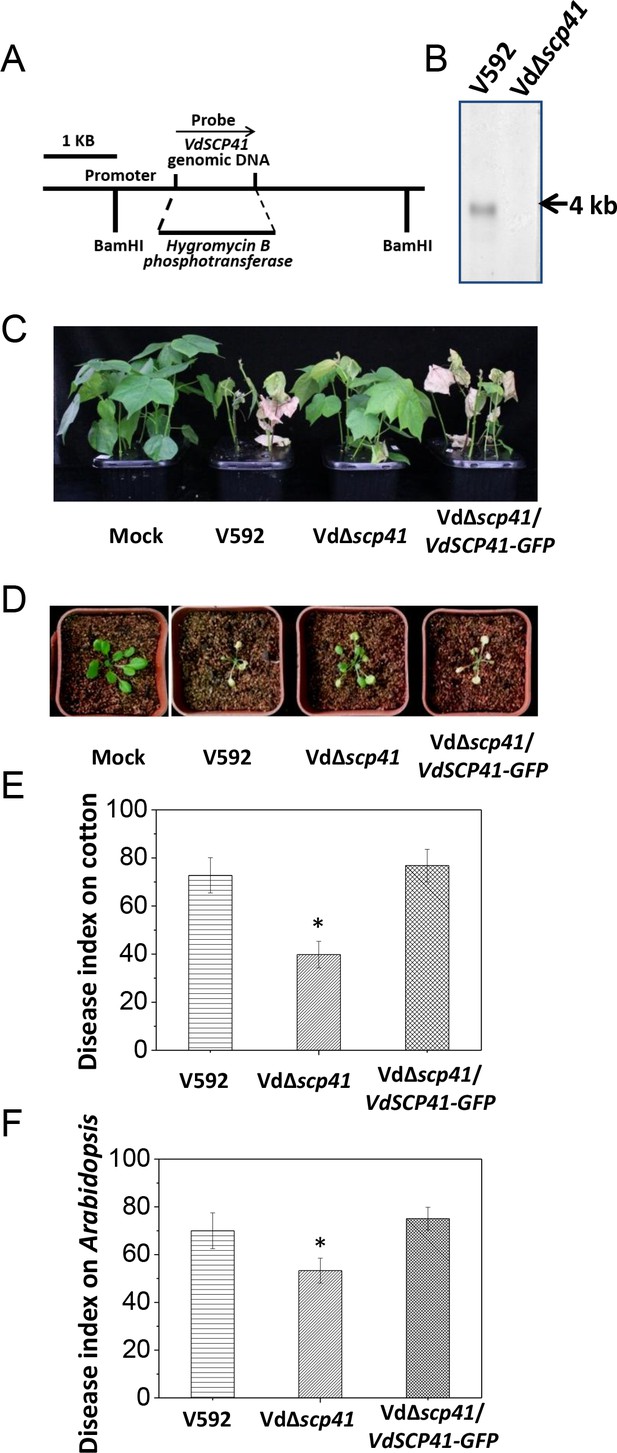
VdSCP41 contributes to V. dahliae virulence in host plants.
(A) Schematic description of the generation of the VdΔscp41 mutant. (B) Southern blot analysis of the VdSCP41 gene deletion in the VdΔscp41 mutant. Genomic DNA samples isolated from V592 and VdΔscp41 strains were digested by BamHI and subjected to Southern blot analysis. (C–D) Disease symptoms of upland cotton (C) and Arabidopsis (D) plants infected with the wildtype (V592), VdΔscp41, and VdΔscp41/VdSCP41-GFP strains. (E–F) Disease index analyses of upland cotton (E) and Arabidopsis (F) infected with the indicated strains. The plants were photographed and subjected to disease index analyses 3–4 weeks post inoculation. The disease indexes were evaluated with three replicates generated from 24 plants for each inoculum. Error bars indicate the standard deviation of three biological replicates. Student’s t-test was carried out to determine the significance of difference. *Indicates significant difference at a P-value of < 0.05. The experiments were repeated three times with similar results.
-
Figure 1—source data 1
Source data for Figure 1.
- https://doi.org/10.7554/eLife.34902.005
VdSCP41 encodes a hypothetical protein in the secretome of the V. dahliae Ls.17 strain (VdLs.17) (Klosterman et al., 2011). The expression of this gene is significantly upregulated at 2 days post-inoculation (dpi) in Arabidopsis (Figure 1—figure supplement 1), suggesting a putative function of VdSCP41 in V. dahliae infection. Targeted gene deletion of VdSCP41 in the VdΔscp41 mutant was verified by southern blotting (Figure 1B). The VdΔscp41 mutant displayed much weaker disease symptoms than WT V592 in both upland cotton (Figure 1C) and Arabidopsis (Figure 1D). Disease index analyses indicated significantly reduced virulence of the VdΔscp41 mutant compared with that of V592 in both upland cotton and Arabidopsis (Figure 1E–F, Figure 1—source data 1). The reduced virulence of the VdΔscp41 mutant was restored upon complementation with GFP-tagged VdSCP41 (VdΔscp41/VdSCP41-GFP) (Figure 1C–F). Thus, VdSCP41 functions as a virulence effector that contributes to V. dahliae virulence on host plants.
VdSCP41 acts as an intracellular effector
In V. dahliae, some signal-peptide-containing SCPs are delivered to the septin-organized hyphal neck, which develops from the base of the hyphopodia and functions as a fungus–host penetration interface for the dynamic delivery of secretory proteins (Zhou et al., 2017). VdSCP41 contains an N-terminal signal peptide predicted by SignalP (http://www.cbs.dtu.dk/services/SignalP/) (Figure 2—figure supplement 1A). Therefore, we examined the localization of VdSCP41 in V. dahliae. GFP-tagged VdSCP41 (VdSCP41-GFP) was found to localize to the base of the hyphopodium and showed ring signals surrounding the hyphal neck when the Vd∆scp41 mutant strain complemented with VdSCP41-GFP (Vd∆scp41/VdSCP41-GFP) was cultured on a cellophane membrane for hyphopodium induction (Figure 2A). By contrast, VdSCP41 lacking signal peptide (ΔspVdSCP41-GFP) showed diffused signal at the base of the hyphopodium without clear ring signals surrounding the hyphal neck. The results demonstrate that VdSCP41-GFP was delivered to the penetration interface for secretion.
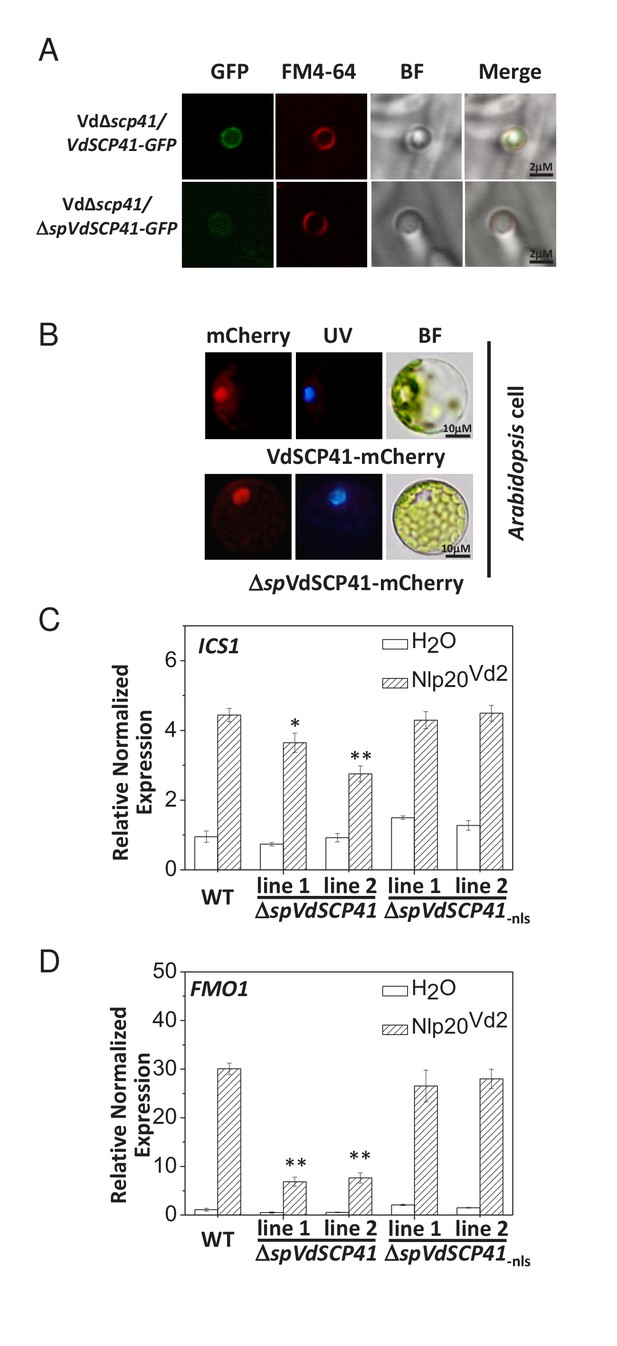
VdSCP41 functions to inhibit immunity in plants.
(A) VdSCP41 localizes to the base of the hyphopodia and forms ring signals surrounding the hyphal neck. The Vd∆scp41/VdSCP41-GFP strain and Vd∆scp41/ΔspVdSCP41-GFP was cultured on a cellophane membrane for 5 days to induce the formation of hyphopodia. Localization of VdSCP41-GFP was visualized using a Leica SP8 microscope. (B) Transiently expressed VdSCP41 preferentially localizes to the nucleus of Arabidopsis cells. Arabidopsis protoplasts were transfected with either a VdSCP41-mCherry or a ΔspVdSCP41-mCherry plasmid as indicated. mCherry fluorescence was visualized 16 hr post transfection. DAPI staining of the nucleus was visualized under UV light. (C–D) Expression of ΔspVdSCP41 in Arabidopsis inhibits nlp20Vd2-induced ICS1 (C) and FMO1 (D) expression. Wildtype (WT) and transgenic plants expressing ΔspVdSCP41 or ΔspVdSCP41-nls were treated with H2O or nlp20Vd2 as indicated. RNA was extracted for real-time PCR analyses. The experiments were repeated three times with similar results. Error bars indicate standard deviations. Student’s t-test was carried out to determine the significance of difference. * Indicates significant difference at a P-value of < 0.05, whereas ** indicates significant difference at a P-value of < 0.01.
-
Figure 2—source data 1
Source data for Figure 2.
- https://doi.org/10.7554/eLife.34902.009
Nuclear localization signal (NLS) sequence prediction (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi) identified a potential NLS in VdSCP41 (Figure 2—figure supplement 1A). We therefore took advantage of this NLS to examine the putative translocation of V. dahliae-delivered VdSCP41 into the nucleus of plant cells. The Vd∆scp41/VdSCP41-GFP and GFP-expressing V592 (V592-GFP) strains were separately inoculated onto onion epidermal cells. Although GFP fluorescence from the V592-GFP strain was observed in conidial spores, VdSCP41-GFP secreted by V. dahliae was capable of translocating into plant cells, and of localizing to the nucleus in addition to the pericellular space of onion epidermal cells (Figure 2—figure supplement 1B). The potential NLS sequence of SCP41 was mutated (SCP41-nls-GFP) and then complemented into Vd∆scp41 to construct the VdΔscp41/VdSCP41-nls-GFP strain. In contrast to VdSCP41-GFP, VdSCP41-nls-GFP failed to translocate into the nucleus of onion epidermal cells (Figure 2—figure supplement 1B). These results suggested that VdSCP41 delivered by V. dahliae translocates into the nucleus of the plant cell, which requires the NLS predicted within its sequence.
We next transiently expressed mCherry-tagged VdSCP41 in plants to further verify its nuclear localization in plant cells. As the signal peptide located at the N terminus may guide secreted proteins into the plant extracellular space in some cases, we fused mCherry to VdSCP41 both with and without (ΔspVdSCP41) the signal peptide in order to analyze subcellular localization. VdSCP41-mCherry and ΔspVdSCP41-mCherry were individually transiently expressed in either Arabidopsis protoplasts or in Nicotiana benthamiana (N. b.) leaves. mCherry fluorescence imaging revealed that both VdSCP41 and ΔspVdSCP41 localized to the nucleus in Arabidopsis cells (Figure 2B). The protein expression level of VdSCP41-mCherry and ΔspVdSCP41-mCherry was detected by immunoblot (Figure 2—figure supplement 1C). Similar nuclear localization was observed for both VdSCP41-mCherry and ΔspVdSCP41-mCherry in N. b. cells (Figure 2—figure supplement 1D). These results are consistent with the nuclear localization of the V. dahliae-delivered VdSCP41 in onion epidermal cells.
VdSCP41 expression in Arabidopsis inhibits plant immunity
It is believed that the initial function of a fungal effector protein is to suppress PTI. To investigate whether VdSCP41 is capable of inhibiting plant immunity when it is directly expressed in plants, Arabidopsis transgenic lines expressing ΔspVdSCP41 were constructed and assessed for PAMP-induced defense responses. Flg22 is the best-characterized PAMP derived from a bacterial pathogen, and it induces the expression of PTI-responsive genes in wildtype (WT) Arabidopsis plants. We observed reduced induction of flg22-induced ICS1 in two independent VdSCP41-expressing lines (Figure 2—figure supplement 1E), suggesting inhibition of PTI conferred by VdSCP41.
NLP proteins derived from bacterial, oomycete and fungal organisms have recently been characterized as PAMPs. A conserved 20-amino-acid peptide (nlp20) within NLP represents the active immunogenic motif that induces PTI in plants (Albert et al., 2015; Böhm et al., 2014; Ottmann et al., 2009). We previously reported the immune-inducing activity of VdNLP1 and VdNLP2 derived from V592 in N. b., Arabidopsis, and cotton plants (Zhou et al., 2012). A corresponding peptide located in VdNLP2 derived from V592 (nlp20Vd2) was synthesized and shown to cause upregulation of cotton PR genes (Du et al., 2017), indicating the immunogenic activity of this peptide in cotton. Treatment of Arabidopsis with nlp20Vd2 significantly induced ICS1 and FMO1 expression (Figure 2C–D, Figure 2—source data 1), indicating that nlp20Vd2 also exhibits the immunogenic activity characteristic of a PAMP in Arabidopsis. In transgenic lines expressing VdSCP41, but not transgenic lines expressing NLS mutated VdSCP41 (ΔspVdSCP41-nls), suppression of nlp20Vd2-induced ICS1 (Figure 2C) and FMO1 (Figure 2D) was observed, suggesting that VdSCP41 expression in plants inhibits nlp20Vd2-triggered immunity. ICS1 encodes the key enzyme controlling SA production and is required for pathogen-induced SA accumulation (Wildermuth et al., 2001). We next quantified SA production in response to a nonpathogenic bacterial pathogen, Pst DC3000 hrcC–, in both WT and ΔspVdSCP41-expressing plants. Consistent with the suppression of PAMP-induced ICS1 expression, transgenic lines expressing ΔspVdSCP41 accumulated less free SA in response to Pst hrcC– than did WT plants (Figure 2—figure supplement 2A).
Inoculation of Arabidopsis with the WT V. dahliae strain V592 induced the expression of ICS1 and FMO1 (Figure 2—figure supplement 2B–C). This induced expression was further enhanced when the plants were inoculated with the VdΔscp41 mutant instead of V592 (Figure 2—figure supplement 2B–C), indicating that VdSCP41 suppresses the induced expression of ICS1 and FMO1 during V. dahliae infection. Taken together, these results reveal an inhibitory role of VdSCP41 in modulating plant immunity.
Arabidopsis master immune regulators CBP60g and SARD1 are targeted by VdSCP41
Modulating the activity of plant immune components is a strategy commonly used by effectors to suppress host immunity. To explore the virulence mechanisms employed by VdSCP41 in inhibiting plant immunity, we next searched for plant components that are targeted by VdSCP41. ΔspVdSCP41 was fused with a 3 × FLAG tag and transiently expressed in Arabidopsis protoplasts, before the VdSCP41-containing protein complexes were purified. Protein lysates were immuno-precipitated using anti-FLAG-conjugated beads and subjected to tandem mass spectrometry. The plant CBP CBP60g was identified as a candidate interactor of VdSCP41 (Supplementary file 1).
CBP60g was then fused with a 3 × HA tag and used for reverse co-immunoprecipitation (Co-IP) analysis to verify its interaction with VdSCP41. FLAG-tagged VdSCP41 was transfected, either alone or together with HA-tagged CBP60g, into Arabidopsis protoplasts for transient expression. Anti-HA IP followed by an anti-FLAG immunoblot revealed that VdSCP41 was co-purified with CBP60g from plant cells (Figure 3—figure supplement 1A). VdSCP41 was further divided into an N-terminal portion (VdSCP41N, amino acids 1–213) and a C-terminal portion (VdSCP41C, amino acids 163-end), which exhibited an overlap of 50 amino acids, which were used to test interactions with CBP60g. Co-IP analysis revealed that VdSCP41C is sufficient for interaction with CBP60g (Figure 3A).
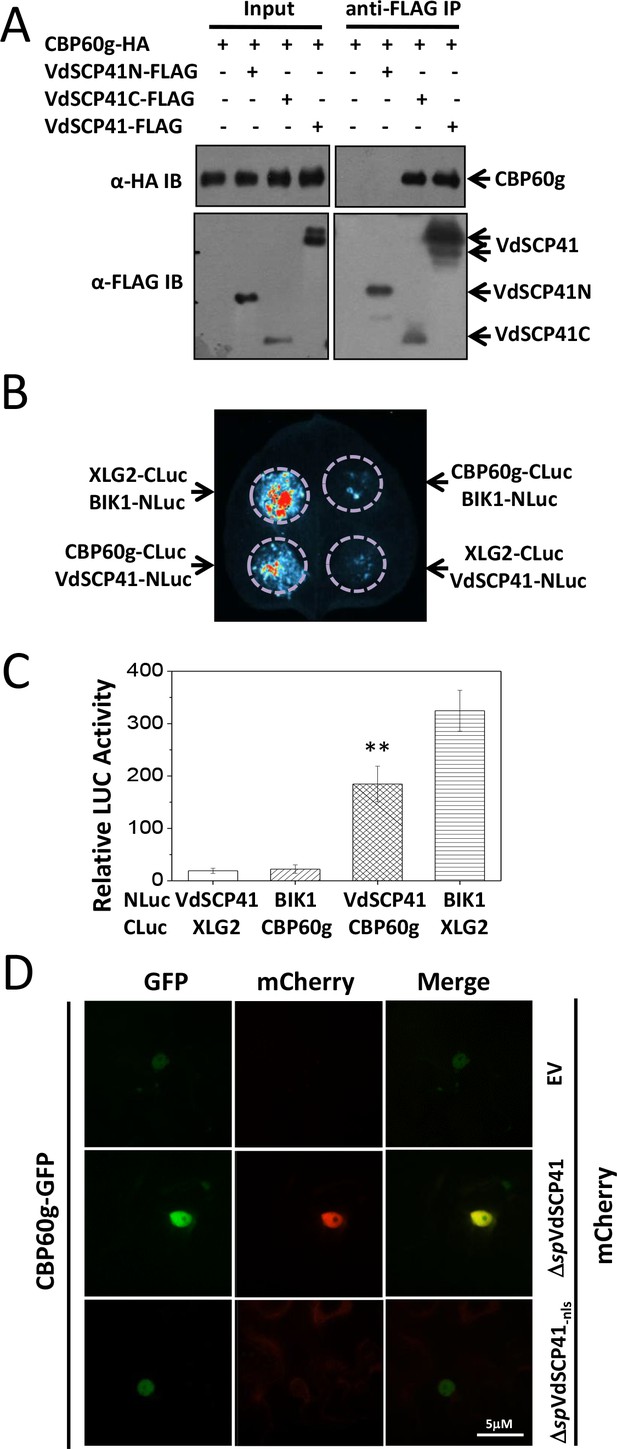
VdSCP41 interacts with CBP60g in plants.
(A) VdSCP41C interacts with CBP60g. Arabidopsis protoplasts were transfected with the indicated constructs. Protein was extracted 16 hr post transfection and immunoprecipitated with anti-FLAG. The presence of VdSCP41-FLAG or CBP60g-HA in the purified complex was detected by anti-FLAG or anti-HA immunoblot as indicated. (B) VdSCP41 interacts with CBP60g in N. b.. Luciferase imaging of VdSCP41 and CBP60g interaction in N. b. leaves. N. b. leaves infiltrated with an Agrobacterium strain carrying constructs as indicated were subjected to luciferase complementation imaging assay. (C) Quantitative luminescence of VdSCP41 and CBP60g interaction. N. b. leaves infiltrated with the indicated constructs were sliced into strips, and their relative luminescence was determined using a microplate luminometer. Error bars indicate standard deviations of three technical repeats. ** Indicates significant difference at P-value < 0.01. The experiments were repeated three times with similar results. (D) VdSCP41 co-localizes with CBP60g and increases its nuclear accumulation. Representative confocal images of CBP60g-GFP subcellular accumulation were visualized using a spin-disk microscope. An Agrobacterium strain carrying CBP60g-GFP was Agro-infiltrated into N. b. leaves alone, or together with an Agrobacterium strain carrying ΔspVdSCP41-mCherry, ΔspVdSCP41-nls-mCherry. An overlay of GFP and mCherry fluorescence imaging was visualized 48 hr post Agro-infiltration in N. b. leaves. The experiments were repeated three times with similar results.
-
Figure 3—source data 1
Source data for Figure 3.
- https://doi.org/10.7554/eLife.34902.014
Quantitative luciferase complementation imaging assays were performed to further verify the interaction between VdSCP41 and CBP60g in N. b.. Co-expression of the N-terminal region of luciferase (NLuc)-tagged BIK1 and the C-terminal region of luciferase (CLuc)-tagged XLG2 driven by the35S promoter was performed as a positive interaction control (Liang et al., 2016). The co-expression of NLuc-VdSCP41 and CLuc-CBP60g driven by the 35S promoter in N. b. resulted in much higher luciferase activity than did the co-expression of NLuc-VdSCP41 and CLuc-XLG2 or of CLuc-CBP60g and NLuc-BIK1 (Figure 3B–C, Figure 3—source data 1), confirming the interaction between VdSCP41 and CBP60g in the plants. The expression levels of the NLuc- and CLuc-fusion proteins were further detected by immunoblotting (Figure 3—figure supplement 1B).
The nuclear localization of VdSCP41 prompted us to examine whether CBP60g co-localizes with VdSCP41 in the nucleus. GFP-tagged CBP60g mainly localized in the nucleus when it was expressed alone in N. b. cells (Figure 3—figure supplement 1C). GFP-tagged CBP60g was co-expressed with mCherry-tagged VdSCP41 without signal peptide (ΔspVdSCP41-mCherry) in N. b. leaves through Agrobacterium-mediated transient expression. An overlay of the results of GFP and mCherry fluorescence imaging indicated co-localization of ΔspVdSCP41 and CBP60g in the nucleus of N. b. cells (Figure 3D). The protein expression level of GFP-CBP60g, ΔspVdSCP41-mCherry and ΔspVdSCP41-nls-mCherry was detected by immunoblot with the indicated antibodies (Figure 3—figure supplement 1D). In addition to co-localization, co-expression of ΔspVdSCP41-mCherry significantly increased the nuclear accumulation of CBP60g-GFP (Figure 3D), which was not observed when ΔspVdSCP41-nls–mCherry with a mutated NLS was co-expressed with CBP60g-GFP (Figure 3D).
CBP60g was induced by pathogen and PAMP treatments and was required for full resistance against bacterial pathogens (Wang et al., 2011; Zhang et al., 2010). A closely related protein in the CBP family, SARD1, functions partially redundantly with CBP60g in bacterial resistance (Sun et al., 2015; Wang et al., 2011). We also detected an interaction between VdSCP41 and SARD1 by Co-IP in Arabidopsis protoplasts (Figure 3—figure supplement 2A). VdSCP41 co-localized with SARD1 in N. b. leaves and increased its nuclear accumulation (Figure 3—figure supplement 2B), indicating similar targeting of SARD1 by VdSCP41 in N. b.. It is unlikely to be an interaction between CBP60g and SARD1 because the luciferase complementation assay did not show interaction between CLuc-tagged CBP60g and NLuc-tagged SARD1 (Figure 3—figure supplement 3). Taken together, the results described above demonstrated that both CBP60g and SARD1 are targeted by VdSCP41.
VdSCP41 interferes with the transcription factor activity of CBP60g
CBP60g encodes a plant-specific transcription factor that regulates the expression of a number of defense-related genes. The fact that CBP60g functions as a master transcription regulator (Sun et al., 2015; Wang et al., 2011) prompted us to examine whether the targeting of CBP60g by VdSCP41 affects the induction of its target genes. Dual reporter analyses revealed that CBP60g expression in Arabidopsis protoplasts significantly enhanced the expression of ICS1::LUC or FMO1::LUC (firefly luciferase) (Figure 4A–B, Figure 4—source data 1). Co-expression of VdSCP41 inhibited CBP60g-induced ICS1::LUC (Figure 4A) and FMO1::LUC (Figure 4B) expression, whereas the co-expression of VdSCP41N, which is unable to bind CBP60g, was impaired in this inhibition (Figure 4A–B). CBP60g was next divided into an N-terminal portion (CBP60gN, amino acids 1–361) containing its DNA-binding domain and a C-terminal portion (CBP60gC, amino acids 211-end) lacking the functional DNA-binding domain (Zhang et al., 2010), which exhibited an overlap of 150 amino acids. CBP60gN and CBP60gC were then used to test interactions with VdSCP41. Co-IP analysis showed that VdSCP41 binds to CBP60gC, but not CBP60gN (Figure 4C). The results indicated that VdSCP41 binds the C-terminal portion of CBP60g to interfere with its activity.
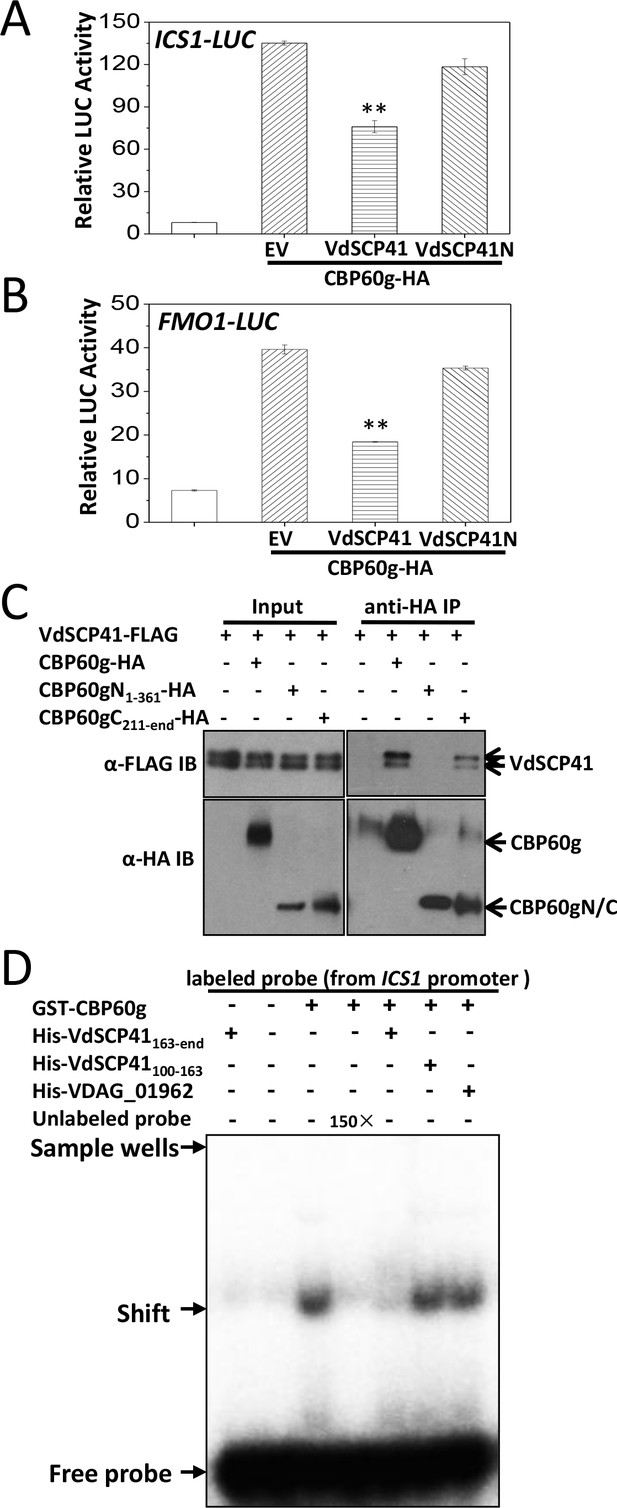
VdSCP41 binds the C-terminal portion of CBP60g to interfere with its activity.
(A–B) Expression of VdSCP41 inhibits the CBP60g-activated ICS1::LUC (A) and FMO1::LUC (B) reporter. ICS1::LUC or FMO1::LUC were transfected alone, or together with CBP60g and VdSCP41 or its variants, as indicated. 35S::RLUC was co-transfected as internal control. Error bars indicate standard deviations of three technical repeats. ** Indicates significant difference between VdSCP41 and EV (empty vector) at a P-value of < 0.01. (C) CBP60gC interacts with VdSCP41. Arabidopsis protoplasts were transfected with VdSCP41-FLAG alone or together with CBP60gN-HA, CBP60gC-HA, or CBP60g-HA. Protein was extracted 16 hr post transfection and immunoprecipitated with anti-HA antibody. The presence of VdSCP41-FLAG in the purified complex was detected by anti-FLAG immunoblot. The experiments were repeated three times with similar results. (D) VdSCP41 inhibits the DNA-binding activity of CBP60g. GST-CBP60g was incubated with [γ−32P]ATP-labeled 60-bp double-stranded DNA probe within the ICS1 promoter, and subjected to electrophoretic mobility shift assay (EMSA). Unlabeled probe was used as competitors (150×) for binding. His-tagged VdSCP41C, VdSCP41100-163 or VDAG_01962 was preincubated with GST-CBP60g for 30 min at room temperature where needed (as indicated) before EMSA. The experiments were repeated three times with similar results.
-
Figure 4—source data 1
Source data for Figure 4.
- https://doi.org/10.7554/eLife.34902.017
To test whether VdSCP41 targeting directly affects the DNA-binding activity of CBP60g, recombinant GST-tagged CBP60g and His-tagged VdSCP41C were purified and used for electrophoretic mobility shift assays (EMSAs). GST-CBP60g showed a specific binding to a 60-bp DNA fragment (ICS1 promoter probe) within the ICS1 promoter, which is reduced by the addition of unlabelled probe, as previously reported (Zhang et al., 2010) (Figure 4D). The preincubation of VdSCP41C with CBP60g significantly reduced the DNA-binding activity of CBP60g, whereas a soluble fragment of His-tagged VdSCP41 which does not contain the C-terminal portion (VdSCP41100-163) did not (Figure 4D). Another His-tagged V. dahliae protein (VDAG_01962) also did not affect the DNA-binding activity of CBP60g (Figure 4D). Coexpression of VdSCP41 did not lead to cleavage or mobility shift of CBP60g (Figure 4—figure supplement 1), suggesting that VdSCP41 is unlikely to act as a protease to target CBP60g. The results proved that binding of VdSCP41C to CBP60g directly inhibits the DNA-binding activity of CBP60g.
CBP60gC harbors a transcription activation domain that is required for interaction with VdSCP41
The binding of VdSCP41 to the C-terminal portion of CBP60g prompted us to test the role of CBP60gC in CBP60g-mediated gene activation. Compared to the full induction of ICS1::LUC or FMO1::LUC by CBP60g (Figure 5A, Figure 5—source data 1), the deletion of the C-terminal portion (ΔC-CBP60g) dramatically compromised its activity to induce both ICS1::LUC and FMO1::LUC (Figure 5A), indicating that CBP60gC is required for CBP60g-mediated gene activation. The results suggest that putative transcription activator activity may be harbored within the CBP60gC. The basic helix-loop-helix (bHLH) transcription factor MYC2 directly binds to the G-box-like (CANNTG) element (Dombrecht et al., 2007; Godoy et al., 2011; Lian et al., 2017) via its bHLH domain to regulate the expression of its target genes, such as the TERPENE SYNTHASE gene 10 (TPS10) (Li et al., 2014b). We therefore took advantage of bHLH-mediated binding to the TPS10 promoter to examine the putative transcription activator activity of CBP60gC. A fragment within the C-terminal portion of CBP60g (amino acids 211–440) activated TPS10::LUC reporter when it was fused with the bHLH domain of MYC2 (bHLH-CBP60g211-440) (Figure 5B, Figure 5—source data 1) rather than bHLHMYC2 alone, indicating that the CBP60g211-440 harbors transcription activator activity. Moreover, Co-IP analysis showed that CBP60g211-end but not CBP60g441-end co-purified with VdSCP41, indicating the requirement for CBP60g211-440 for binding to VdSCP41 (Figure 5C). Thus, CBP60gC harbors a transcription activation domain, CBP60g211-440, that is required for VdSCP41 targeting.
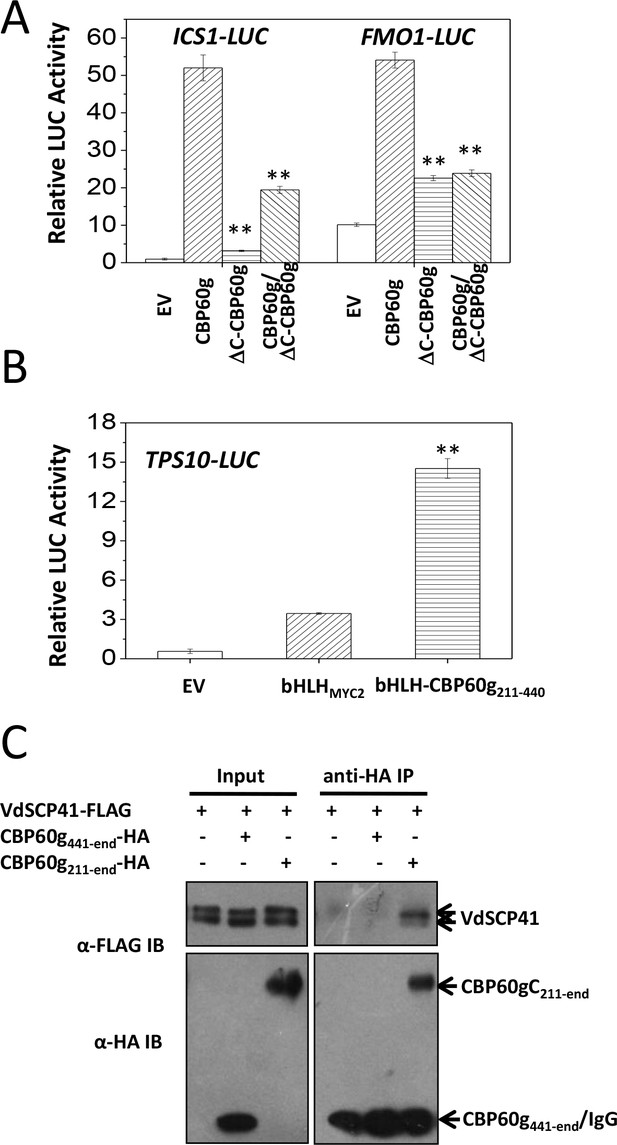
CBP60gC harbors a transcription activation domain that is required for interaction with VdSCP41.
(A) CBP60gC is required for CBP60g-mediated ICS1 and FMO1 activation. ICS1::LUC or FMO1::LUC was transfected alone, or together with CBP60g or its variants, as indicated. ** Indicates significant difference between ΔC-CBP60g or ΔC-CBP60g/CBP60g and CBP60g at a P-value of < 0.01. (B) CBP60g211-440 harbors transcription activator activity. TPS10::LUC was transfected alone, or together with bHLHMYC2 or bHLH-CBP60g211-440, as indicated. 35S::RLUC was co-transfected as internal control. ** Indicates significant difference between bHLH-CBP60g211-440 and bHLHMYC2 at a P-value of < 0.01. LUC reporter activity was determined 16 hr post transfection. The experiments were repeated three times with similar results. (C) CBP60g211-440 is required for interacting with VdSCP41. Arabidopsis protoplasts were transfected with the indicated constructs. Protein was extracted 16 hr post transfection and immunoprecipitated with anti-HA. The presence of VdSCP41-FLAG or CBP60g variants in the purified complex was detected by anti-FLAG or anti-HA immunoblot as indicated.
-
Figure 5—source data 1
Source data for Figure 5.
- https://doi.org/10.7554/eLife.34902.019
The ΔC-CBP60g was further co-transfected with CBP60g, together with ICS1::LUC or FMO1::LUC. Dual reporter analyses indicated that co-expression of ΔC-CBP60g significantly suppressed CBP60g-induced ICS1::LUC and FMO1::LUC (Figure 5A), suggesting a dominant-negative effect of ΔC-CBP60g on the activity of CBP60g.
CBP60g and SARD1 contribute to Arabidopsis resistance against V. dahliae
SARD1 functions both redundantly and differentially with CBP60g (Sun et al., 2015; Wang et al., 2011). The finding that VdSCP41 targeted both CBP60g and SARD1 prompted us to assess the roles of CBP60g and SARD1 in resistance to V. dahliae. The WT and cbp60g-1/sard1-1 double mutant plants were inoculated with V592 for the assessment of disease symptoms. As shown in Figure 6, the cbp60g-1/sard1-1 double mutant displayed compromised resistance compared with the WT plants, demonstrating a contribution of CBP60g and SARD1 to V. dahliae resistance.
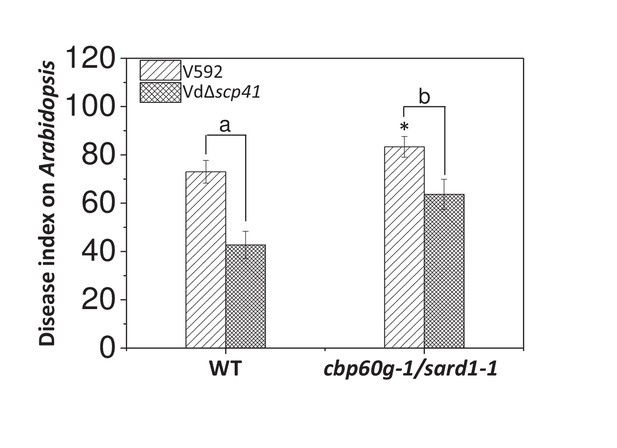
CBP60g and SARD1 are required for VdSCP41-mediated virulence.
Wildtype (WT) and cbp60g-1/sard1-1 double mutant plants were inoculated with V592 and VdΔscp41 mutant strains. The plants were subjected to disease index analyses 3–4 weeks post inoculation. The disease indexes were evaluated with three replicates generated from 24 plants for each inoculum. Error bars indicate standard deviations of three biological replicates. *Indicates significant difference of V592 virulence at a P-value of < 0.05 between WT and cbp60g-1/sard1-1 double mutant plants. Student’s t-test was carried out to determine the significance of difference between indexes collected from three biological replicates. Lower case letters indicate a significant difference at a P-value of < 0.05. The experiments were repeated three times with similar results.
-
Figure 6—source data 1
Source data for Figure 6.
- https://doi.org/10.7554/eLife.34902.021
We next investigated the requirement for CBP60g and SARD1 for VdSCP41-mediated virulence. The WT and cbp60g-1/sard1-1 double mutant plants were inoculated with the VdΔscp41 mutant. When compared with V592, the VdΔscp41 mutant displayed reduced virulence on the WT plants. The reduced virulence arising from VdSCP41 deletion was partially impaired in the cbp60g-1/sard1-1 double mutant plants compared with that in the WT plants (Figure 6, Figure 6—source data 1). The results indicated that both CBP60g and SARD1 are required for full virulence conferred by VdSCP41. However, we still observed reduced virulence of the VdΔscp41 mutant compared with that of V592 on the cbp60g-1/sard1-1 double mutant plants, suggesting the existence of additional targets for VdSCP41 during V. dahliae infection.
VdSCP41 interacts with and co-localizes with GhCBP60b
As in Arabidopsis, we observed similar compromised virulence of the VdΔscp41 mutant in upland cotton (Figure 1E) compared with V592. The results prompted us to test the putative VdSCP41 targeting of GhCBP60b, the closest protein homolog of CBP60g and SARD1 in cotton. Co-IP analysis revealed an interaction between VdSCP41 and GhCBP60b (Figure 7A). To examine the subcellular localization of GhCBP60b, GhCBP60b-GFP was constructed for transient expression in N. b. leaves. GhCBP60b-GFP localized to the nucleus of N. b. cells, and co-expression of GhCBP60b-GFP with VdSCP41-mCherry revealed co-localization of VdSCP41 and GhCBP60b in the nucleus of N. b. cells (Figure 7B), suggesting conserved targeting of Arabidopsis CBP60g and SARD1 and of cotton GhCBP60b by VdSCP41.
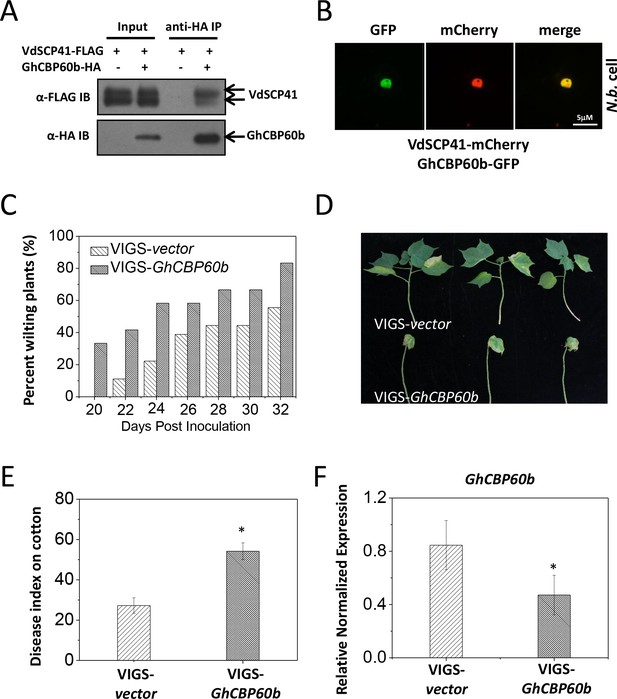
GhCBP60b is targeted by VdSCP41 and required for cotton resistance against V. dahliae.
(A) VdSCP41 is co-purified with GhCBP60b in Arabidopsis protoplasts. Arabidopsis protoplasts were transfected with VdSCP41-FLAG alone or together with GhCBP60b-HA. Protein was extracted 16 hr post transfection and immunoprecipitated with anti-HA. The presence of VdSCP41-FLAG in the purified complex was detected by anti-FLAG immunoblot. (B) VdSCP41 co-localizes with GhCBP60b. GFP-tagged GhCBP60b and mCherry-tagged VdSCP41 were co-expressed in N. b. leaves. An overlay of GFP and mCherry fluorescence imaging was visualized 48 hr post Agro-infiltration. The experiments were repeated three times with similar results. (C) Cotton VIGS-GhCBP60b plants develop symptoms more rapidly than VIGS-vector plants. The percentage of plants showing the Verticillium wilt phenotype at the indicated time after infection is shown. The disease ratio was scored using 15 plants per treatment and the assays were repeated three times with similar results. (D–E) GhCBP60b is required for full resistance against V. dahliae. Cotton seedlings were infiltrated with Agrobacterium carrying pTRV1 together with pTRV2 or pTRV2-GhCBP60b as indicated. Verticillium dahliae strain V592 was inoculated 10 days post Agrobacterium infiltration. Plants showing disease symptoms were photographed 30 days post V. dahliae inoculation (D). Disease index analyses of VIGS-vector or VIGS-GhCBP60b plants infected with V592 (E). The plants were subjected to disease index analyses 3–4 weeks post inoculation. The disease indexes were evaluated in three replicates generated from 24 cotton plants for each inoculum. The experiments were repeated three times with similar results. Error bars indicate standard deviations of three technical repeats within one biological experiment. The Student’s t-test was carried out to determine the significance of difference. *Indicates significant difference at a P-value of < 0.05. (F) Expression of GhCBP60b is reduced in plants that are infiltrated with Agrobacterium carrying pTRV1 together with pTRV2-GhCBP60b (VIGS-GhCBP60b). Total RNA from infiltrated plants was extracted for RT-PCR analyses of GhCBP60b expression 20 days post V. dahliae inoculation. Error bars indicate standard deviations. *Indicates significant difference at a P-value of < 0.05.
-
Figure 7—source data 1
Source data for Figure 7.
- https://doi.org/10.7554/eLife.34902.023
GhCBP60b is required for cotton resistance against V. dahliae
To further examine whether GhCBP60b functions in resistance to V. dahliae in cotton, we generated a virus-induced gene silencing (VIGS) vector (Liu et al., 2002) targeting GhCBP60b (pTRV2-GhCBP60b). Cotton plants were infiltrated with pTRV1 together with pTRV2 or pTRV2-GhCBP60b and further inoculated with V. dahliae. When compared with pTRV2, pTRV2-GhCBP60b-infiltrated cotton plants exhibited higher ratios of wilting (Figure 7C, Figure 7—source data 1) and more severe disease symptoms (Figure 7D–E, Figure 7—source data 1), indicating a role for GhCBP60b in cotton resistance to V. dahliae. The reduced expression of GhCBP60b in pTRV2-GhCBP60b-infiltrated plants was verified using RT-PCR (Figure 7F, Figure 7—source data 1). The results support the targeting of GhCBP60b by VdSCP41 for virulence during V. dahliae infection in cotton.
Discussion
In this study, we identified VdSCP41 as an intracellular effector that is crucial for V. dahliae virulence. VdSCP41 targets Arabidopsis CBP60g and SARD1 to interfere with the induction of their target genes. CBP60g and SARD1 are required for Arabidopsis resistance to V. dahliae and for VdSCP41-mediated virulence. Silencing of GhCBP60b compromised cotton resistance to V. dahliae. The results revealed a conserved and important role for Arabidopsis CBP60g and SARD1 and for cotton GhCBP60b, which are modulated by an intracellular effector, in plant resistance against V. dahliae.
VdSCP41 is secreted by V. dahliae and translocates into plant cells
To suppress plant defense actively or to modulate host physiology to benefit pathogenic fitness, oomycete and fungal pathogens deliver hundreds of effectors through specialized intracellular fungal structures, such as haustoria and infection hyphaea, into the host apoplastic space or directly into plant cells (Lo Presti et al., 2015). Although haustoria have not been observed during infection, V. dahliae develops a penetration peg from a hyphopodium when infecting plant roots (Zhao et al., 2016). During V. dahliae infection, the penetration peg that developed from the hyphopodium further develops a hyphal neck and forms a septin ring that partitions the hyphopodium and invasive hyphae. This septin-organized apparatus functions as a fungus–host interface for the dynamic delivery of secretory proteins, such as SCPs (Zhao et al., 2016Zhou et al., 2017).
To date, relatively few V. dahliae-secreted effectors have been characterized as functioning inside host cells to modulate host immunity. Here, we provided evidence of the secretion and translocation of VdSCP41, which is secreted by V. dahliae, into plant cells. VdSCP41 contains a signal peptide and localizes to the base of the hyphopodium to form a septin-like ring during infection (Figure 2A), indicating that it is secreted via the septin-organized apparatus at the fungus–host interface. The localization of the VdSCP41 delivered by V. dahliae at the nucleus of onion epidermal cells (Figure 2—figure supplement 1B) suggested the translocation of VdSCP41 into plant cells. A few conserved motifs have been indicated to serve as signals for the uptake of fungal or oomycete effectors into plant cells. For example, a few effectors secreted by cereal powdery mildew and rust pathogens possess a Y/F/WxC motif that serves as a signal for translocation into the plant cell (Godfrey et al., 2010; Spanu et al., 2010). A set of oomycete effectors contain a RXLR-dEER motif that appears to assist in targeting effectors into plant cells (Dou et al., 2008; Whisson et al., 2007). Another set of oomycete effectors possess a FLAK motif for translocation (Schornack et al., 2010). However, VdSCP41 lacks any of the above conserved motifs, and the mechanisms that assist the uptake of VdSCP41 into plant cells remain unclear.
VdSCP41 targets CBP60g and SARD1 and interferes with plant defense
CBP60g and SARD1 are master regulators in immunity. ChIP-seq analyses have allowed the identification of a large number of target genes for CBP60g and SARD1 and support a model in which CBP60g and SARD1 accumulate in the plant nucleus and act as master regulators, which activate both positive and negative immune regulators (Sun et al., 2015; Wang et al., 2009; Zhang et al., 2010). Genetic evidence revealed that CBP60g and SARD1 are positive immune regulators required for immune responses and bacterial resistance (Wang et al., 2009; Zhang et al., 2010). We demonstrated that VdSCP41 targets CBP60g and SARD1 and interferes with their activity, thus supporting the virulence function of VdSCP41 for immune suppression. Consistent with the inhibition of CBP60g and SARD1 activity, we observed less pathogen-induced SA accumulation in transgenic lines that expressed VdSCP41 than in WT plants (Figure 2—figure supplement 2A). As CBP60g and SARD1 also bind directly to the promoters of both ALD1 and SARD4 (Sun et al., 2018) (two major genes encoding pipecolic acid (Pip) biosynthesis enzymes) to regulate Pip biosynthesis, a potential contribution of Pip to V. dahliae resistance will be of interest for further investigation.
We showed that VdSCP41 binds the C-terminal portion of CBP60g to interfere with its transcription factor activity (Figure 4A–B). Coexpression of VdSCP41 did not lead to cleavage or mobility shift of CBP60g (Figure 4—figure supplement 1), suggesting that VdSCP41 is unlikely to act as a protease to target CBP60g. In addition, CBP60g211-440, a domain within the C-terminal portion of CBP60g, harbors transcription activator activity (Figure 5B) and is required for interaction with VdSCP41 (Figure 5C). It is likely that CBP60gC functions to promote transcriptional activation by recruiting additional activators, and that binding of VdSCP41 interrupts either the activity of this domain or the recruitment of associated activators via this domain. The dominant-negative function of ΔC-CBP60g (Figure 5A) supports a dominant-negative effect on CBP60g activity arising from increased nuclear accumulation of VdSCP41-impaired CBP60g. Thus, VdSCP41-mediated over-accumulation of CBP60g provides an additional strategy to further interfere with CBP60g activity as a transcription factor. The results suggest a novel virulence strategy in which a pathogenic effector directly targets host transcription factors to interfere with their activity and to modulate plant immunity. However, it is equally possible that the increased nuclear accumulation of CBP60g results from feed-back regulation as a result of impaired CBP60g function.
VdSCP41 functions to suppress immunity
CBP60g and SARD1 are key components of the SA signaling pathway, which serves as an attractive target for bacterial and fungal effectors (DebRoy et al., 2004; Djamei et al., 2011; Nomura et al., 2011). Targeting of CBP60g and SARD1 by VdSCP41 may therefore interfere with plant resistance to V. dahliae by manipulating SA signaling. On the other hand, ChIP-seq analyses identified a number of SA-independent regulators that are directly targeted by CBP60g and SARD1 (Sun et al., 2015), revealing broader and SA-independent functions of CBP60g and SARD1 in immunity. It is likely that signaling pathways other than SA are also targeted by VdSCP41 through CBP60g and SARD1 to interfere with plant immunity. Consistent with this assumption, we showed that nlp20Vd2 functions to trigger the induction of both SA-dependent and SA-independent defense genes during V. dahliae infection (Figure 2C–D). Furthermore, VdSCP41 expression suppressed the induction of both SA-dependent ICS1 and SA-independent FMO1 by nlp20Vd2 (Figure 2C–D). Modulation of CBP60g and SARD1 may result in interference with their function as master transcription factors in PTI and in other defense pathways.
Taken together, our results support a model in which PAMPs from V. dahliae, such as NLPs and chitins, are recognized by plants to induce the expression of CBP60g and SARD1, which subsequently regulate the expression of a number of immune regulators to defend against pathogen infection (Figure 8A). VdSCP41 secreted by V. dahliae functions as an intracellular effector that targets the transcription activator domain of CBPs, interrupting the function of this domain, to interfere with their transcription factor activity, and thus modulates both SA-dependent and SA-independent regulators to inhibit plant immunity against V. dahliae (Figure 8B).
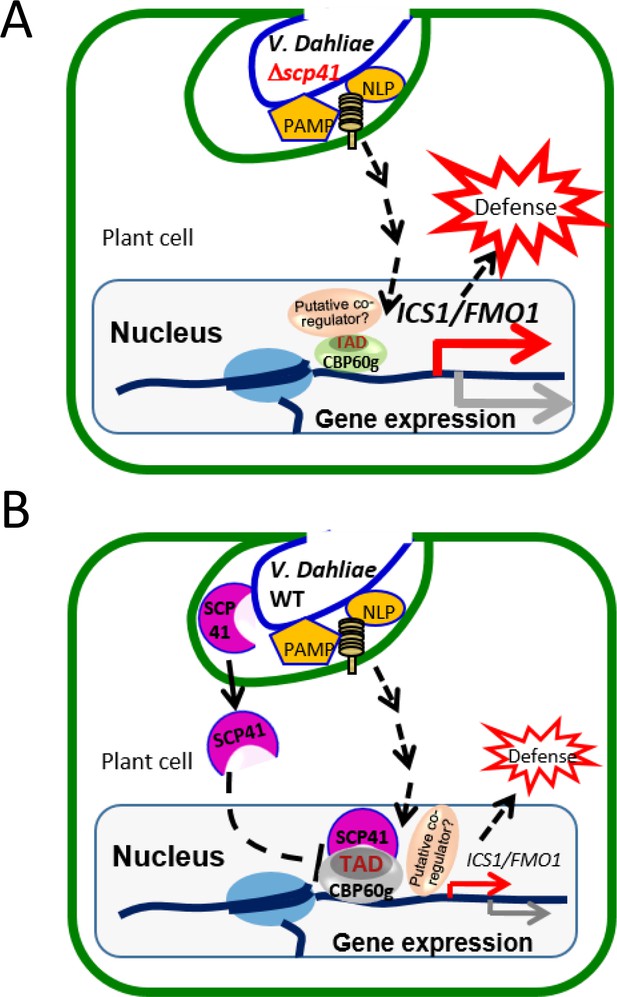
Model for VdSCP41–mediated suppression of defense in Arabidopsis during V. dahliae infection.
(A) In the absence of VdSCP41, PAMPs derived from V. dahliae, such as NLPs and chitins, induce the expression of CBP60g, which in turn upregulates the expression of a number of immune regulators to activate defense. (B) In the presence of VdSCP41, VdSCP41 secreted from V. dahliae translocates into the nucleus of plant cells. VdSCP41 targets the transcription activation domain (TAD) of CBP60g, interrupting either the activity of this domain or the recruitment of associated co-activators via this domain, to interfere with their activity and with plant immunity against V. dahliae. VdSCP41-mediated over-accumulation could provide an additional strategy to further interfere with the transcription factor activity of CBP60g.
Materials and methods
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Verticillium dahliae) | VdSCP41 | PMID:21829347 | NCBI Gene ID:20709665 | |
| Gene (Gossypium hirsutum) | GhCBP60b | NCBI Gene ID:107899061 | ||
| Strain, strain background (Verticillium dahliae) | V592 | PMID:21151869 | ||
| Other (cotton plant) | Xinluzao No. 16 | PMID:28282450 | ||
| Antibody | Anti-FALG (mouse monoclonal) | Sigma | RRID:AB_259529 | 1:10000 dilution |
| Antibody | Anti-HA (mouse monoclonal) | Roche | RRID:AB_514506 | 1:5000 dilution |
| Antibody | Anti-GFP (mouse monoclonal) | Roche | RRID:AB_390913 | 1:3000 dilution |
| Antibody | Anti-CLuc (mouse monoclonal) | Sigma | RRID:AB_439707 | 1:3000 dilution |
| Antibody | Anti-mCherry | Easybio | 1:3000 dilution | |
| Peptide | nlp20Vd2 | PMID:28755291 | ||
| Recombinant DNA reagent | pTRV1 (VIGS vector) | PMID:12220268 | ||
| Recombinant DNA reagent | pTRV2 (VIGS vector) | PMID:12220268 |
Fungal strains, plant materials and antibodies
Request a detailed protocolThe Verticillium dahliae strain V592 (Gao et al., 2010) was used in this study. Verticillium dahliae strains were grown on potato dextrose agar (PDA) medium at 25°C in the dark. To collect conidia, the mycelial plugs were cultured in potato dextrose broth (PDB) liquid medium at 25°C with shaking at 200 rpm for 3–5 days. Cotton plants ('Xinluzao No. 16') were used for virulence assessment in this study (Zhou et al., 2017). Arabidopsis thaliana plants used in this study include Col-0 (wild-type) and the cbp60g-1/sard1-1 mutant (Zhang et al., 2010). VdSCP41 was amplified from the V592 cDNA and cloned into pCambia1300-35S-FLAG (Zhang et al., 2010) to construct Arabidopsis transgenic lines expressing VdSCP41. Antibodies used in this study include anti-FLAG (RRID:AB_259529), anti-HA (RRID:AB_514506), anti-CLuc (RRID:AB_439707), anti-GFP (RRID:AB_390913), and anti-mCherry (Easybio, BE2026).
Generation of deletion and complementation fungal strains
Request a detailed protocolThe upstream and downstream flanking sequences were PCR amplified from V592 genomic DNA and cloned into a pGKO-HPT vector (Wang et al., 2016). The resulting construct was transformed into Agrobacterium tumefaciens EHA105, and used for A. tumefaciens-mediated transformation (ATMT) to generate the VdΔscp41 mutant strain (Wang et al., 2016). The genomic region of VdSCP41, including 1.5 kb upstream from the start codon, was amplified and cloned into a pNat-Tef-TrpC vector (Zhou et al., 2017) to generate a construct for complementation. The resulting construct was transformed into Agrobacterium tumefaciens EHA105, and used for ATMT to generate a VdΔscp41/VdSCP41-GFP strain. Primers used in this study are listed in Supplementary file 2.
Protein complex purification and mass spectrum analysis
Request a detailed protocolArabidopsis protoplasts isolated from 10 gram leaves were transfected with ΔspVdSCP41-FLAG, ΔspVdSCP45-FLAG, or empty vector for protein expression. Transfected protoplasts were collected and total protein was extracted with extraction buffer containing 50 mM HEPES (pH 7.5), 150 mM KCl, 1 mM EDTA, 1 mM DTT, 0.2% Triton X-100, and 1 × proteinase inhibitor cocktail. Total protein was incubated with 50 μl anti-FLAG agarose beads (Sigma) for 12 hr at 4°C. The immunocomplex was washed three times using the buffer described above and eluted with 100 μl 1 μg/μl 3 × FLAG peptide. The eluted proteins were run 10 mm into the separating gel and stained with Proteo Silver stain kit (Sigma). Total protein was destained and digested in-gel with sequencing grade trypsin (10 ng/mL trypsin, 50 mM ammonium bicarbonate [pH 8.0]) overnight. Peptides were sequentially extracted with 5% formic acid/50% acetonitrile and 0.1% formic acid/75% acetonitrile and concentrated to 20 μl. The extracted peptides were separated by an analytical capillary column packed with 5 mm spherical C18 reversed-phase material. The eluted peptides were sprayed into a LTQ mass spectrometer (Thermo Fisher Scientific) equipped with a nano-ESI ion source. The mass spectrometer was operated in data-dependent mode with one MS scan followed by five MS/MS scans for each cycle. Database searches were performed on an in-house Mascot server (Matrix Science Ltd., London, UK) against the IPI (International Protein Index) Arabidopsis protein database.
Co-immunoprecipitation assay in protoplasts
Request a detailed protocolFive-week-old Arabidopsis plants were used for protoplast isolation. pUC-35S-VdSCP41-FLAG, or its variant constructs, was co-transfected with pUC-35S-CBP60g-HA, or its variant constructs, pUC-35S-SARD1-HA or pUC-35S-GhCBP60b-HA, into Arabidopsis protoplasts. Total protein was extracted with extraction buffer. For anti-HA IP, total protein was incubated with 2 μg of anti-HA antibody together with protein A agarose at 4°C for 4 hr. The agarose beads were collected and boiled for 5 min with 1 × protein loading buffer. Immunoprecipitates were separated by 10% SDS-PAGE, and the presence of VdSCP41-FLAG or its variants, CBP60g-HA or its variants, SARD1-HA, or GhCBP60b-HA was detected by anti-FLAG or anti-HA immunoblot.
SA measurement
Request a detailed protocolSA was extracted and measured following the method described previously (Sun et al., 2015). Around 0.3 grams of leaf tissue, collected from 4-week-old plants, was ground into powder in liquid nitrogen. Plant leaves were infiltrated with or without Pst DC3000 hrcC– (OD600 = 0.1) 12 hr before sample collection. Three samples were analysed for each treatment. The samples were extracted with 0.8 mL 90% methanol and sonicated for 15 min, and the supernatant was transferred into a new tube. The pellet was re-extracted with 0.5 mL of 100% methanol, and the supernatant was combined with the first-step supernatant and dried by vacuum. The pellet was resuspended in 500 μL 0.1 M sodium acetate (pH 5.5) in 10% methanol. An equal volume of 10% TCA was added and the samples were vortexed and sonicated for 5 min. After centrifugation, the supernatant was extracted three times with 0.5 ml of extraction buffer (ethylacetate/cyclopentane/isopropanol:100/99/1 by volume). After spinning, the organic phases were collected and dried by vacuum. The samples were then dissolved in 250 μL 100% methanol and filtered through a 0.22 μm filter. The samples were then assayed by HPLC-MS/MS analysis on a AB SCIEX QTRAP 4500 system (AB SCIEX, Foster, CA, USA).
Luciferase complementation imaging assay
Request a detailed protocolAgrobacterium tumefaciens GV3101 strain carrying CLuc- or NLuc-tagged consctruct were infiltrated into leaves of 4-week-old N. b.. LUC activity in leaves was examined 2 days post infiltration. N. b. leaves were treated with 1 μM luciferin and kept in the dark for 5 min to quench the fluorescence. LUC image was captured by CCD imaging apparatus, and the quantitative LUC activity was determined by microplate luminometer. Expression of CLuc-tagged proteins or NLuc-tagged proteins was detected by anti-CLuc or anti-HA western blot, respectively.
Electrophoretic mobility shift assays (EMSAs)
Request a detailed protocolThe full-length CBP60g was cloned into the pGEX-6p-1 vector. VdSCP41C, VdSCP41100-163 or VDAG_01962 was cloned into the pET-28a vector. The resulting constructs were transformed into E. coli BL21 (DE3) competent cells. BL21 (DE3) strains containing the expression vectors were cultured and induced by isopropylthio-galactoside (IPTG) at 16°C for 16 hr. GST-tagged full-length CBP60g, His-tagged VdSCP41C or VdSCP41100-163, and His-tagged VDAG_01962 recombinant protein were affinity purified and used for EMSAs. A 60-bp probe within the DNA fragment 7 (Zhang et al., 2010) in the ICS1 promoter were labeled with [γ-32P]ATP using T4 polynucleotide kinase. Binding reactions were carried out in a 20 μl volume of reaction buffer (10 mM Tris-HCl [pH 7.5], 50 mM KCl, 1 mM DTT, 1 μl 50 ng/μl poly[dI-dC]) for 30 min at room temperature. Labeled DNA probe (2 fmol) was incubated with 4 μg; GST-CBP60g. 150 × unlabeled DNA probe was used for competition. 1.5 μg; His-tagged VdSCP41C163-end, VdSCP41100-163, or VDAG_01962 was preincubated with GST-CBP60g for 30 min at room temperature before DNA binding. The reaction was stopped by adding DNA loading buffer and the samples were separated by a 5% native PAGE gel. After electrophoresis, the gel was autoradiographed.
RNA isolation and qRT-PCR
Request a detailed protocolTotal RNA was isolated with the TRIzol reagent (Invitrogen) and used for cDNA synthesis with SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) following the instructions provided by the manufacturer. The quantitative PCR was performed with the SYBR Premix Ex Taq kit (TaKaRa) following standard protocols. Arabidopsis Col-0 or transgenic plants were treated with H2O, 1 μM flg22 or nlp20Vd2 (Du et al., 2017) as indicated for 3 hr. RNA was isolated and used for RT-PCR analysis for the expression of ICS1 and FMO1. AtTUB4 was used as internal control. To detect gene expression in cotton plants, leaves from the cotton plants were collected 14 days post Agrobacterium infiltration. Quantitative RT-PCR was performed as described above. Gossypium hirsutum HISTONE3 was used as internal control. For RT-PCR analyses of VdSCP41, the V592 or VdΔscp41 strain was incubated with the roots of 7-day-old Arabidopsis Col-0 plants for 2 days. Conidia were collected for RNA isolation and RT-PCR analysis to analyze the expression of VdSCP41. For RT-PCR analyses of V. dahliae-infected Arabidopsis plants, Arabidopsis plants infected with or without the V592 or VdΔscp41 strain were collected for RNA isolation and RT-PCR analysis for the expression of ICS1 and FMO1. VdELF1 was used as internal control for VdSCP41.
Fluorescence microscopy
Request a detailed protocolAgrobacterium tumefaciens EHA105 strain carrying pCambia1300-35S-CBP60g-GFP, pCambia1300-35S-SARD1-GFP, or pCambia1300-35S-GhCBP60b-GFP was infiltrated alone, or together with the A. tumefaciens EHA105 strain carrying pCambia1300-35S-VdSCP41-mCherry (or its variant mutants) into leaves of 4-week-old N. b. GFP and mCherry fluorescence were observed with Leica SP8 confocal microscopy 3 days post infiltration. The intensity of fluorescent signals was determined by Image J software. For fluorescence microscopy in Arabidopsis, Arabidopsis protoplasts were transfected with 35S-CBP60g-GFP alone or together with 35S-VdSCP41-mCherry (or its variant mutants). The protoplasts were incubated overnight under faint light before GFP and mCherry fluorescence were observed. To examine the subcellular localization of VdSCP41, conidia were cultured on cellophane and incubated for 3–9 days before observation by microscopy. The pieces of cellophane with mycelium were collected and observed as described (Zhou et al., 2017). The plasma membrane of the fungi was stained with FM4-64 (red).
Reporter assay in Arabidopsis protoplast
Request a detailed protocolArabidopsis protoplasts were co-transfected with ICS1::LUC or FMO1::LUC and 35S::RLUC (Renilla luciferase) alone, or together with VdSCP41, CBP60g, or their variants. 12 hr after transfection, the protein of transfected protoplasts was isolated, and the LUC activity was determined by using a Dual-Luciferase Reporter system (Promega) according to the manufacture’s instructions.
Virus-induced gene silencing in cotton plants
Request a detailed protocolThe VIGS was performed as described previously (Gao and Shan, 2013). Cotton plants were grown at 23–25°C in the growth room until two cotyledons had emerged. A 465-bp fragment of GhCBP60b cDNA was PCR amplified from G. hirsutum and cloned into pTRV2 plasmid (Liu et al., 2002). The Agrobacterium strain carrying pTRV1, together with the Agrobacterium strain carrying pTRV2 or pTRV2-GhCBP60b, was infiltrated into the cotyledons of the cotton plants. The cotton plants were root-dip-inoculated with V. dahliae V592 2 weeks post Agrobacterium infiltration.
Infection assay
Request a detailed protocolCotton or Arabidopsis plants were infected by the root-dip inoculation method (Gao et al., 2010). A conidial suspension of 107/ml from the indicated strain was used as the inoculum. The disease grade was classified as follows: Grade 0 (no symptoms), 1 (0–25% wilted leaves), 2 (25–50%), 3 (50–75%) and 4 (75–100%). The disease index was calculated as 100 × (sum [number of plants × disease grade])/ ([total number of plants] × [maximal disease grade]) (Xu et al., 2014). The onion epidermis infection assay was performed as described (Zhang et al., 2017). A conidial suspension of 107/ml from the V592-GFP, VdΔscp41/SCP41-GFP or VdΔscp41/SCP41-nls-GFP strain was inoculated onto the inner layer of onion epidermal cells and incubated on 1% water agar plates for 3–5 days before confocal imaging.
Data availability
All data generated or analysed during this study are included in the manuscript and supporting files. Source data files have been provided.
References
-
Plant-specific calmodulin-binding proteinsAnnual Review of Plant Biology 56:435–466.https://doi.org/10.1146/annurev.arplant.56.032604.144224
-
Plant immunity: a lesson from pathogenic bacterial effector proteinsCellular Microbiology 11:1453–1461.https://doi.org/10.1111/j.1462-5822.2009.01359.x
-
MYC2 differentially modulates diverse jasmonate-dependent functions in ArabidopsisThe Plant Cell Online 19:2225–2245.https://doi.org/10.1105/tpc.106.048017
-
NPR1, all things consideredCurrent Opinion in Plant Biology 7:547–552.https://doi.org/10.1016/j.pbi.2004.07.005
-
Expression of pathogenesis-related genes in cotton roots in response to Verticillium dahliae PAMP moleculesScience China Life Sciences 60:852–860.https://doi.org/10.1007/s11427-017-9071-9
-
Interfamily transfer of tomato Ve1 mediates Verticillium resistance in ArabidopsisPlant Physiology 156:2255–2265.https://doi.org/10.1104/pp.111.180067
-
Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1Plant Physiology 150:320–332.https://doi.org/10.1104/pp.109.136762
-
Cotton GhBAK1 mediates verticillium wilt resistance and cell deathJournal of Integrative Plant Biology 55:586–596.https://doi.org/10.1111/jipb.12064
-
Functional genomic analysis of cotton genes with agrobacterium-mediated virus-induced gene silencingMethods in Molecular Biology 975:157–165.https://doi.org/10.1007/978-1-62703-278-0_12
-
Virus-induced gene silencing in tomatoThe Plant Journal 31:777–786.https://doi.org/10.1046/j.1365-313X.2002.01394.x
-
Fungal effectors and plant susceptibilityAnnual Review of Plant Biology 66:513–545.https://doi.org/10.1146/annurev-arplant-043014-114623
-
Verticillium dahliae Sge1 differentially regulates expression of candidate effector genesMolecular Plant-Microbe Interactions 26:249–256.https://doi.org/10.1094/MPMI-08-12-0198-R
-
Evidence for functional diversification within a fungal NEP1-like protein familyMolecular Plant-Microbe Interactions 26:278–286.https://doi.org/10.1094/MPMI-09-12-0222-R
-
Inheritance of resistance to Verticillium wilt in a tomato crossPhytopathology 41:986–990.
-
Life Cycle and Epidemiology of Verticillium81–111, Life Cycle and Epidemiology of Verticillium, 10.1016/B978-0-12-464450-2.50009-7.
-
ChIP-seq reveals broad roles of SARD1 and CBP60g in regulating plant immunityNature Communications 6:10159.https://doi.org/10.1038/ncomms10159
-
Functional characterization of cotton genes responsive to verticillium dahliae through bioinformatics and reverse genetics strategiesJournal of Experimental Botany 65:6679–6692.https://doi.org/10.1093/jxb/eru393
Article and author information
Author details
Funding
The Strategic Priority Research Program of the Chinese Academy of Sciences (XDB11020600)
- Jie Zhang
National Natural Science Foundation of China (31730078)
- Hui-Shan Guo
- Jie Zhang
The Youth Innovation Promotion Association of Chinese Academy of Sciences
- Jie Zhang
The Strategic Priority Research Program of the Chinese Academy of Sciences (XDB11040500)
- Hui-Shan Guo
National Natural Science Foundation of China (31571968)
- Jie Zhang
National Natural Science Foundation of China (31501593)
- Jun Qin
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
The authors are grateful to Prof. Yuelin Zhang for providing the Arabidopsis cbp60g-1/sard1-1 mutant, to Prof. Yule Liu for providing the pTRV1 and pTRV2 plasmids, to Prof. Jian Ye for providing the TPS10::LUC plasmid, and to Yao Wu for help in SA measurement. This work was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB11020600) to JZ, the National Natural Science Foundation of China (31730078) to H-SG, the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB11040500) to H-SG, the Youth Innovation Promotion Association of the Chinese Academy of Sciences, the National Natural Science Foundation of China (31571968, 31501593), and the State Key Laboratory of Plant Genomics, Chinese Academy of Sciences.
Copyright
© 2018, Qin et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 4,660
- views
-
- 1,211
- downloads
-
- 141
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Citations by DOI
-
- 141
- citations for umbrella DOI https://doi.org/10.7554/eLife.34902






