Human LINE-1 retrotransposition requires a metastable coiled coil and a positively charged N-terminus in L1ORF1p
Figures
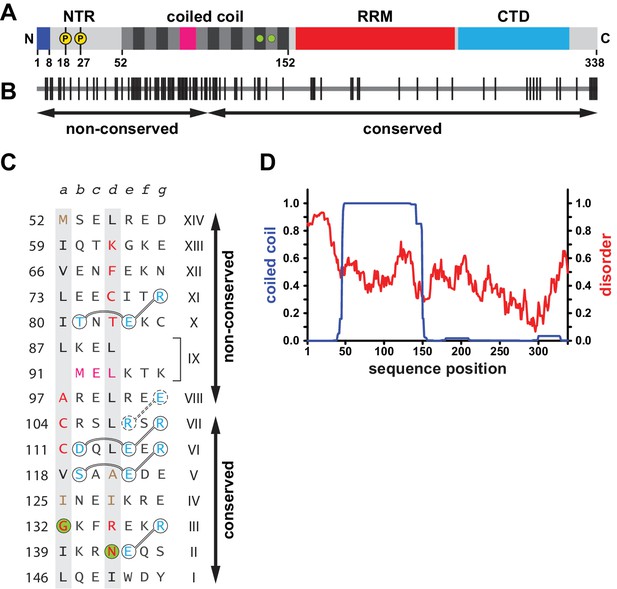
Sequence properties of human L1ORF1p.
(A) Domain organization of human L1ORF1p, including findings from this work. See also Figure 1—figure supplement 1. The bar diagram shows the NTR in light grey with phosphorylation sites marked in yellow (Cook et al., 2015) and with the positively charged N-terminal residues in blue (this work). The coiled coil domain shows heptads alternating grey and black. The irregular heptad IX harbors a stammer and is in magenta. Ion-coordinating heptads are marked by green circles. The RRM domain is in red and the CTD is in cyan. (B) Conservation and evolution of human L1ORF1p. Non-conserved amino acid positions from an alignment of primate sequences (Figure 1—figure supplement 2) are marked by vertical lines. Arrows indicate the previously crystallized conserved portion of L1ORF1p (Khazina et al., 2011) and the rapidly evolving, non-conserved portion. In contrast to the non-conserved portion, the conserved portion of L1ORF1p can be easily aligned in all mammals (Boissinot and Sookdeo, 2016; Khazina et al., 2011; Yang et al., 2014) (Figure 1—figure supplement 1B, Supplementary file 1). (C) Sequence of the crystallized coiled coil construct (hL1ORF1p-cc), listed as heptads. Heptad positions a and d are shaded in grey with non-canonical residues in red. Ion-coordinating layers are marked by green circles. In heptad IX, the three-residue insertion corresponding to the stammer is in magenta (this work). Polar residues involved in trimerization motifs and forming stabilizing salt bridges are in blue and circled with interactions shown as connecting lines. Residues in hL1ORF1p-cc that deviate from the human sequence are in brown. See the Materials and methods and Supplementary file 3 for further construct details. (D) Coiled coil propensity and probability of disorder. Coiled coil propensity (blue) was calculated from the alignment of primate sequences using PCoils (Alva et al., 2016; Lupas, 1996). The probability of disorder (red) was obtained from the human sequence using IUPred (Dosztányi et al., 2005).
-
Figure 1—source data 1
Output from Pcoils and IUpred.
- https://doi.org/10.7554/eLife.34960.006
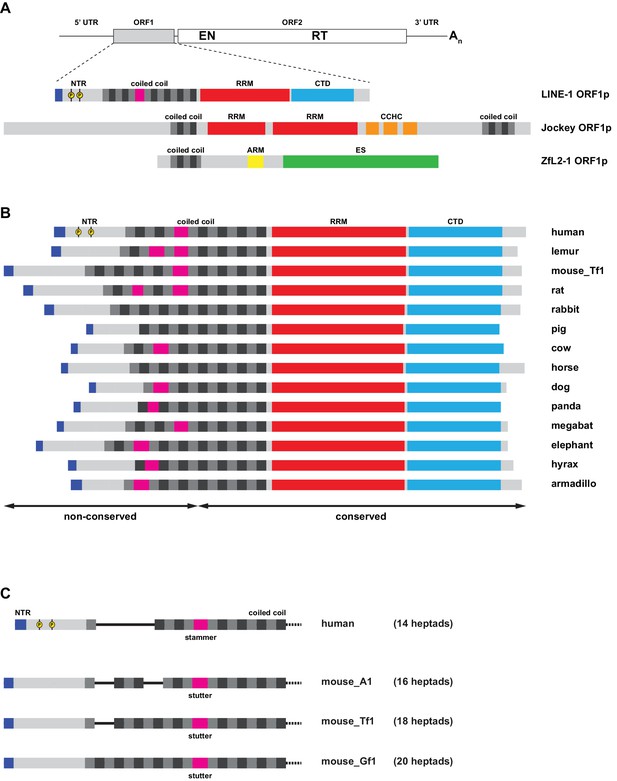
Organization of ORF1 proteins in L1 and other non-LTR retrotransposons.
(A) Distinct types of protein architecture among ORF1 proteins from non-LTR retrotransposons. Top: General organization of an RNA from a non-LTR retrotransposon, encoding a first, accessory ORF1 and a second, catalytic ORF2 that harbors both endonuclease (EN) and reverse transcriptase (RT) functions required for target-primed reverse transcription. Bottom: Commonly found types of ORF1p architecture, represented by the human L1 element (see Figure 1A for details), the Drosophila melanogaster Jockey element, and the Danio rerio ZfL2-1 element. All three proteins harbor coiled coil sequences with heptads colored in grey and black. Furthermore, all three proteins contain RNA binding elements: In the human L1ORF1p, a distinct RRM domain cooperates with a C-terminal domain (CTD) and the coiled coil. In Jockey-like ORF1ps, single or tandem RRMs are followed by one or more Gag-like CCHC zinc knuckles, and the ZfL2-1 ORF1p contains an arginine-rich motif (ARM). Finally, the ZfL2-1 ORF1p hosts a lipid binding SGNH esterase domain (ES) (Schneider et al., 2013). (B) Variability in mammalian L1ORF1p sequences. Bar diagrams are aligned via the C-terminal end of the coiled coil domain. Only the conserved portions of the L1ORF1ps can be unambiguously aligned (Boissinot and Sookdeo, 2016; Khazina et al., 2011; Yang et al., 2014). The non-conserved part of the coiled coil harbors a variable number of heptad repeats (magenta for non-heptad interruptions). The NTRs are of different length, but all show an accumulation of positive charges at the N-terminus (blue). Phosphorylation sites identified in the human NTR are marked in yellow (Cook et al., 2015). See Supplemental file 1 for the individual sequences. (C) Coiled coil heptad expansion and deletion in murine L1 elements. In the mouse, active L1 elements belong to different lineages (A1, Tf1, Gf1), where the non-conserved part of the L1ORF1p coiled coil has undergone heptad duplications and deletions (Sookdeo et al., 2013). Furthermore, murine L1ORF1ps contain a stutter as opposed to the stammer in human L1ORF1p, but the precise alignment of murine and human sequences remains ambiguous in the non-conserved part of the sequence. L1_Tf1 is also known as L1spa (Kingsmore et al., 1994; Naas et al., 1998). See Supplemental file 1 for the individual sequences.
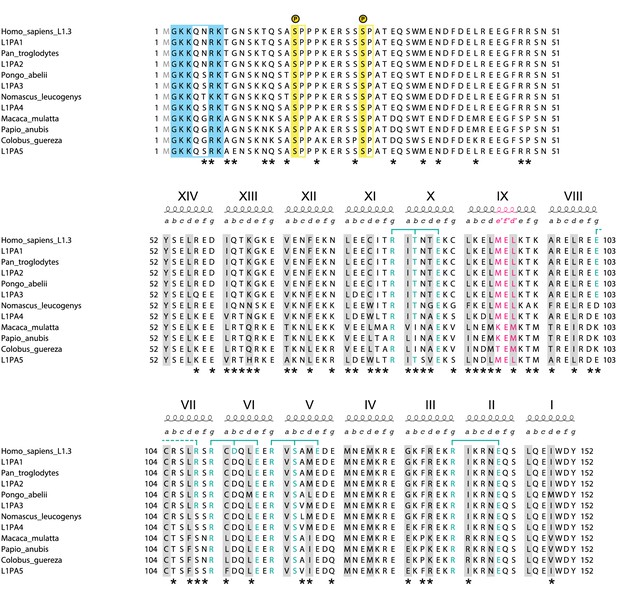
Alignment of closely related primate L1ORF1p sequences.
The human sequence is from the L1.3 element (NCBI accession L19088.1) used throughout this work (Dombroski et al., 1993; Sassaman et al., 1997). For the NTR and coiled coil domain of L1ORF1p, it is identical to the human L1PA1 consensus sequence from Khan et al., 2006. The most recent ancestral human sequences (L1PA2-L1PA5) are reconstructed consensus sequences and also taken from Khan et al., 2006. Sequences from the great apes and old world monkeys also are reconstructed consensus sequences (see Supplemental file 2 for individual accessions). Older and more distantly related primate sequences increasingly begin to show insertions, deletions and alignment ambiguities. As a consequence, the non-conserved portion of L1ORF1p cannot be confidently aligned anymore among mammalian orthologs. The alignment corresponds to the NTR (top), the non-conserved part of the coiled coil (middle) and the conserved part of the coiled coil (bottom). Heptad positions a and d, forming the core of the coiled coil are shaded in grey. Blue brackets indicate trimerization motifs and other stabilizing salt bridges (dashed) with participating residues in blue. In heptad IX, the three-residue insertion corresponding to the stammer is in magenta. Serine-proline phosphorylation sites in the NTR (Cook et al., 2015) are shaded in yellow. Positively charged residues at the N-terminus (after removal of the N-terminal methionine) are shaded in blue. Alignment positions where residues are not conserved are marked with an asterisk.
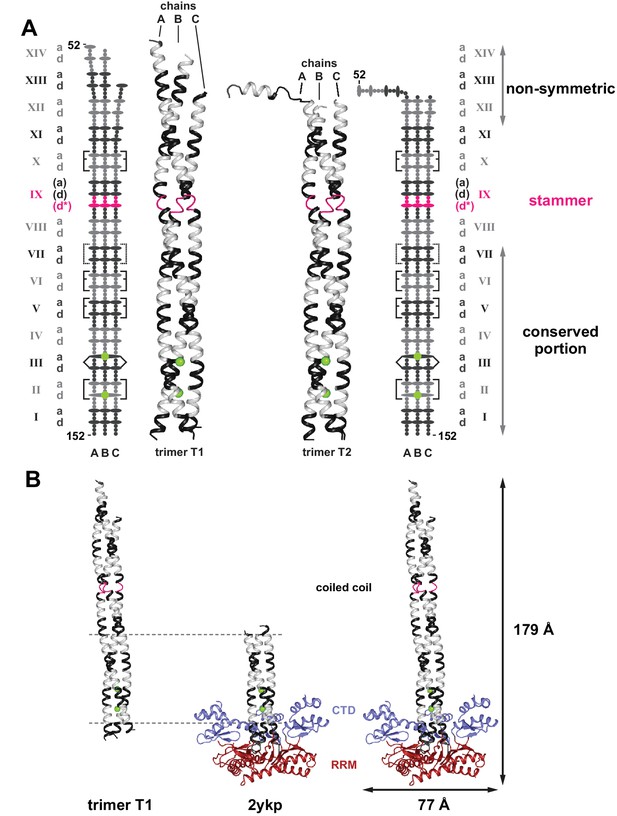
Crystal structure of the human L1ORF1p coiled coil domain.
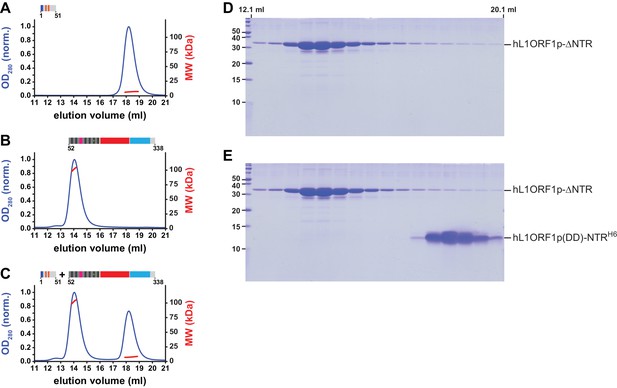
(A) The two crystallized trimers of the L1ORF1p coiled coil domain. See Table 1 for data collection and refinement statistics. Structures include the rapidly evolving N-terminal portion of the coiled coil. Structures are shown as three-dimensional ribbons or schematically, with colors as in Figure 1. Heptad positions a and d are symbolized by ovals, the remaining positions as small circles. Rectangular brackets symbolize trimerization motifs or other stabilizing salt bridges (dashed). The pointed bracket symbolizes R135 in position IIId and its coordination of a chloride ion in the preceding a-layer. (B) Composite structure model for the human L1ORF1p trimer. Superposition of the coiled coil trimer T1 (residues 52–152) via heptads II-VI (horizontal lines) with a crystal structure of the human L1ORF1p conserved portion (PDB-ID 2ykp, residues 107–223) (Khazina et al., 2011) results in a model comprising all parts of human L1ORF1p that are known to be structured. See also Figure 2—figure supplement 1.
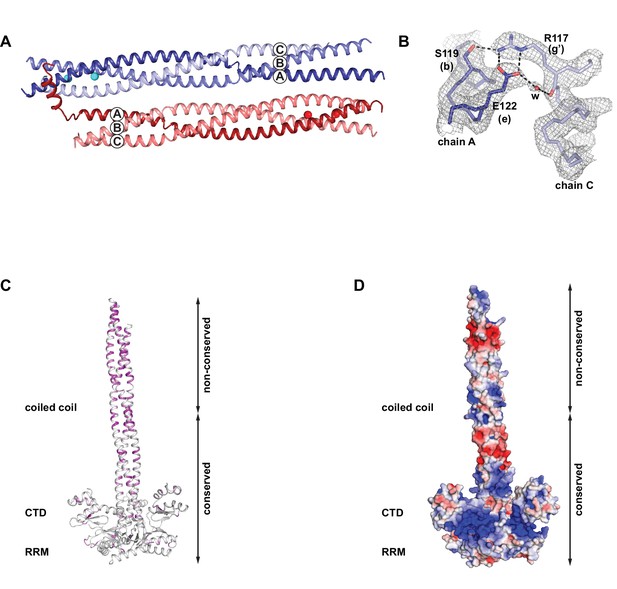
Crystallographic details and properties of the coiled coil in the context of the composite model of the L1ORF1p trimer.
(A) Crystal packing of the coiled coil trimer T1 (blue) and trimer T2 (red) in the asymmetric unit. (B) Electron density (2Fo-Fc), contoured at 1.0 sigma around the trimerization motif in heptad V of trimer T1. (C) Localization and distribution of non-conserved amino acids within the composite model of human L1ORF1p, using a color gradient from white (conserved) to magenta (non-conserved). (D) Electrostatic potential plotted onto the molecular surface of the composite model. The potential is contoured from –5 kT/e (red, acidic) to +5 kT/e (blue, basic).
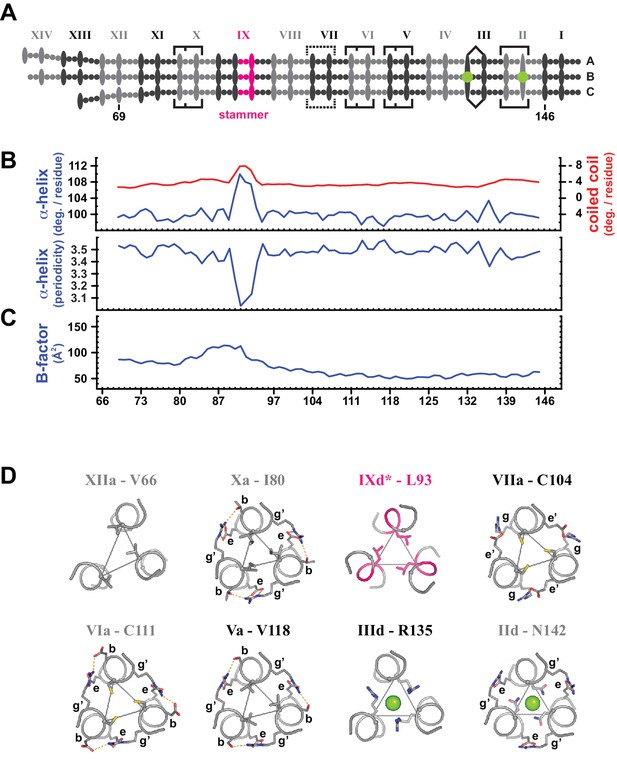
Coiled coil parameters and structural details (trimer T1).
(A) Schematic representation of trimer T1 (see Figure 2). (B) Coiled coil structural parameters as analyzed in TWISTER (Strelkov and Burkhard, 2002) and aligned with the scheme in (A). Top panel: Helical geometry of the α-helices (blue) and of the coiled coil bundle (red), expressed in degrees of right-handed rotation per residue. Negative values result from the left-handed geometry of the coiled coil. Bottom panel: Periodicity of the α-helices along the coiled coil axis, expressed in residues per turn. (C) Crystallographic B-factor, averaged over the main chain atoms of polypeptide chains A, B and C. (D) Individual selected core layers shown as sticks and viewed down the axis of the coiled coil. Heptad positions a or d of the respective layers are connected by thin lines, and chloride ions are shown as green spheres (heptads II and III). Peripheral heptad positions engaged in trimerization motifs (heptads II, V, VI, X) or other stabilizing salt bridges (heptad VII) are labeled, where g’ and e’ mark the positions from the preceding layer. Peripheral stabilization frequently compensates for non-canonical a- or d-layers. The stammer contributes an additional core layer (IXd*), geometrically closest to a d-type layer (Strelkov and Burkhard, 2002). N-terminal heptads are deformable with core layers deviating from three-fold symmetry (XIIa). For trimer T2, see Figure 3—figure supplement 1.
-
Figure 3—source data 1
Output from TWISTER and B-factor analysis.
- https://doi.org/10.7554/eLife.34960.012
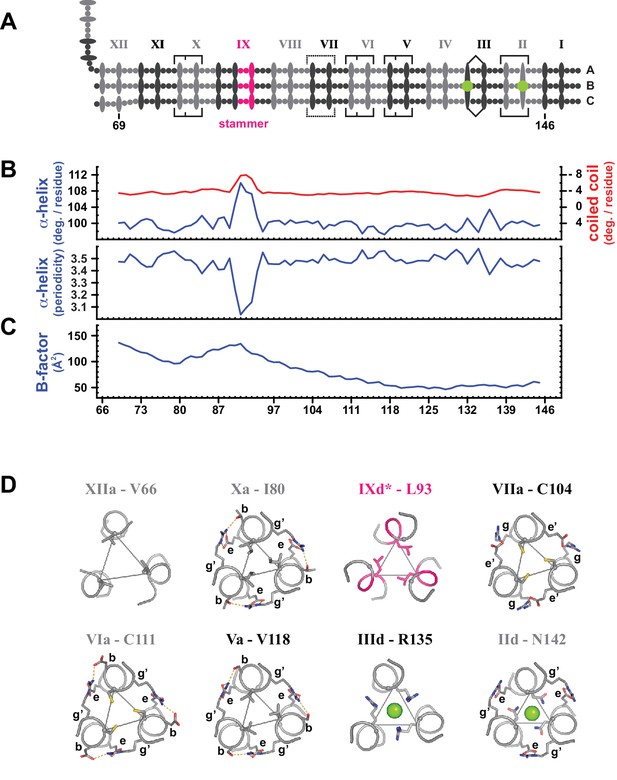
Coiled coil parameters and structural details (trimer T2).
(A) Schematic representation of trimer T2 (see Figure 2). (B) Coiled coil structural parameters as analyzed in TWISTER (Strelkov and Burkhard, 2002) and aligned with the scheme in (A). Top panel: Helical geometry of the α-helices (blue) and of the coiled coil bundle (red), expressed in degrees of right-handed rotation per residue. Negative values result from the left-handed geometry of the coiled coil bundle. Bottom panel: Periodicity of the α-helices along the coiled coil axis, expressed in residues per turn. (C) Crystallographic B-factor, averaged over the main chain atoms of polypeptide chains A, B and C. (D) Individual selected core layers shown as sticks and viewed down the axis of the coiled coil. Heptad positions a or d of the respective layers are connected by thin lines, and chloride ions are shown as green spheres. Apart from layer XIIa, the structures of the illustrated layers are highly similar to those of the corresponding layers in trimer T1.
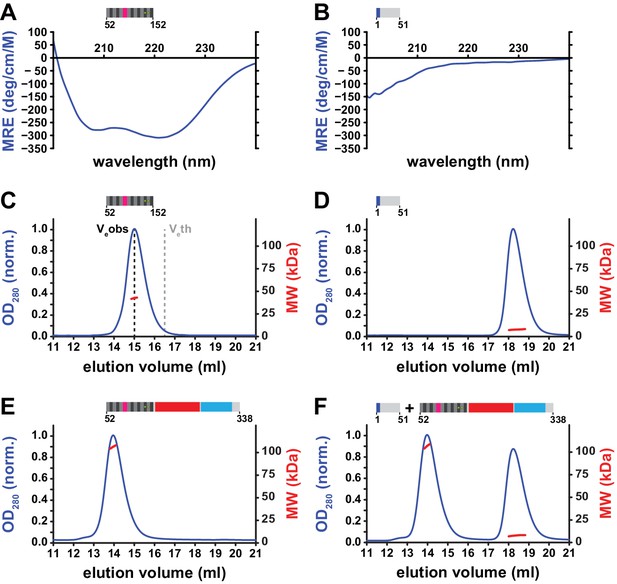
Structural properties of the N-terminal region (NTR) of L1ORF1p.
(A, B) Circular dichroism (CD) spectroscopy. The spectrum of the L1ORF1p coiled coil domain (A) is typical for an α-helix, whereas the spectrum of the NTR (B) indicates the absence of helices or strands. Ellipticity is calculated per residue as a mean residue ellipticity (MRE), expressed in degrees / (cm x M). (C, D) Size exclusion chromatography followed by multiangle static laser light scattering (MALLS). The molecular weights (MW) determined by MALLS from the elution peaks indicate the coiled coil domain to be trimeric in solution (C, 12.6 kDa per monomer), whereas the NTR remains monomeric (D, 6.7 kDa per monomer). The observed elution volume (Veobs) of the trimeric coiled coil domain is clearly larger than the theoretical elution volume (Veth) of a globular molecule with the same mass. Chromatography was done on a Superdex 200 column and the optical density at 280 nm (OD280) was normalized to 1.0 for the maximal absorption observed. (E, F) NTR-binding assay. L1ORF1p is trimeric in the absence of the NTR (E, 34.4 kDa per monomer). The NTR fails to interact with the remainder of L1ORF1p when mixed with the truncated trimer (F). See also Figure 4—figure supplement 1, Figure 4—figure supplement 2.
-
Figure 4—source data 1
Data from size exclusion chromatography, MALLS and CD spectroscopy.
- https://doi.org/10.7554/eLife.34960.016
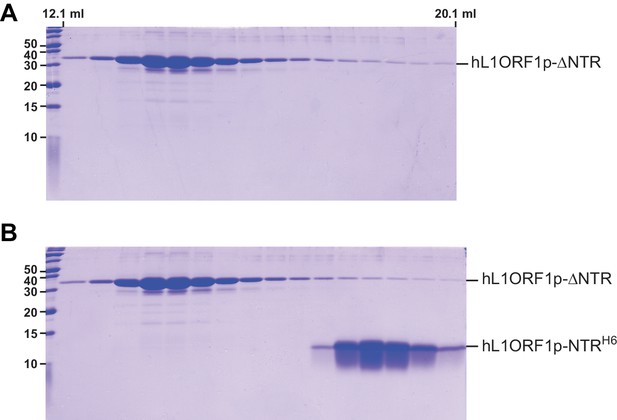
Gel analysis of the NTR-binding assay.
(A) Denaturing polyacrylamide gel (SDS PAGE) of size exclusion chromatography fractions corresponding to Figure 4E. (B) Denaturing polyacrylamide gel (SDS PAGE) of size exclusion chromatography fractions corresponding to Figure 4F.
MALLS and NTR-binding assay with an NTR peptide containing phospho-mimicking residues.
(A, B) Size exclusion chromatography followed by MALLS. The molecular weights (MW) determined by MALLS from the elution peaks indicate the NTR (A, 6.7 kDa per monomer) to remain monomeric in the presence (orange marks) of phospho-mimicking mutations S18D and S27D (Cook et al., 2015), whereas the L1ORF1p is trimeric in the absence of the NTR (B, 34.4 kDa per monomer). Chromatography was done on a Superdex 200 column and the optical density at 280 nm (OD280) was normalized to 1.0 for the maximal absorption observed. (C) NTR-binding assay. The mutated NTR fails to interact with the remainder of L1ORF1p when mixed with the truncated trimer. (D) Denaturing polyacrylamide gel (SDS PAGE) of size exclusion chromatography fractions corresponding to panel B. (E) Denaturing polyacrylamide gel (SDS PAGE) of size exclusion chromatography fractions corresponding to panel C.
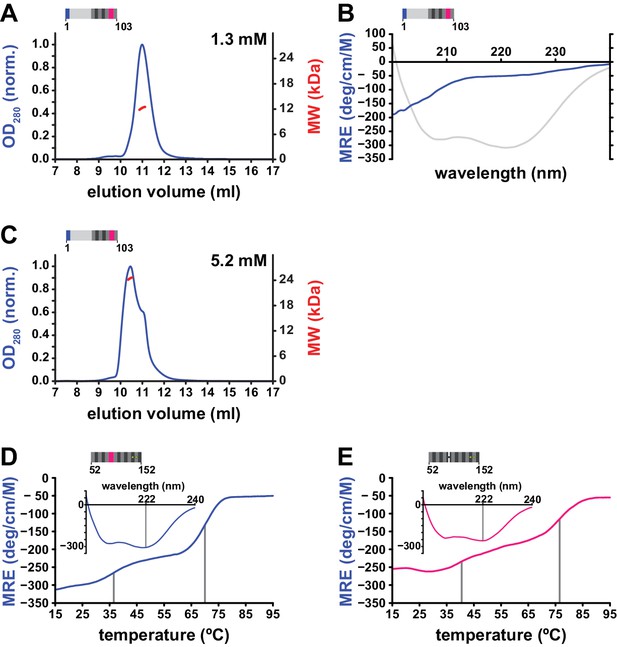
Trimerization properties and stability of the coiled coil domain.
(A, B) Loss of self-association and secondary structure in the non-conserved portion of human L1ORF1p upon deletion of the conserved portion. The non-conserved portion of the human L1ORF1p (12.4 kDa) fails to form oligomers at concentrations up to 1.3 mM, as revealed by size exclusion chromatography and MALLS (A). Chromatography was done on a Superdex 75 column and optical density (OD280) was normalized to 1.0 for the maximal absorption observed. Furthermore, CD spectroscopy (B) shows a total loss of α-helical structure (compare to the spectrum in grey from Figure 4A). Ellipticity is calculated per residue as a mean residue ellipticity (MRE), expressed in degrees / (cm x M). (C) Dimerization of the non-conserved portion at very high concentrations. At a protein concentration of 5.2 mM, size exclusion chromatography and MALLS reveal a partial dimerization. (D, E) Thermal melting curves of the coiled coil domain, monitored by CD spectroscopy at 222 nm. The coiled coil domain comes apart in two distinct steps, one of which occurs at physiological temperature (D). The removal of the stammer has a stabilizing effect on the structure and affects both transitions (E).
-
Figure 5—source data 1
Data from size exclusion chromatography, MALLS and CD spectroscopy.
- https://doi.org/10.7554/eLife.34960.018
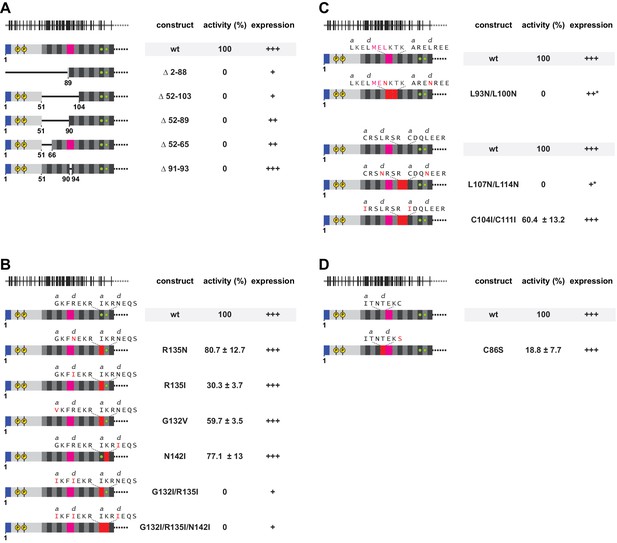
Functional importance of a non-ideal coiled coil.
(A) Deletion analysis of the N-terminal and poorly conserved heptads, monitored by L1 retrotransposition in HeLa cells. Deletions comprise the NTR and/or several complete heptads, or regularize heptad IX by a deletion of the stammer. Retrotransposition activity is calculated with respect to the wildtype (wt) protein, with the mean and standard deviations calculated from three independent replicates. Normal (+++), reduced (++) or poor (+) protein expression levels are marked, as well as an unusual gel migration (*). See also Figure 6—figure supplement 1. None of the deletions is tolerated. (B, C) Point mutational analysis of core layers. Mutations comprise non-canonical and presumably destabilizing residues in heptads II and III (B), as well as canonical and presumably stabilizing leucines in the d-layers of heptads VI/VII and VIII/IX (C). We also mutated potentially metal-coordinating cysteines (Ruckthong et al., 2016) in the a-layers of heptads VI/VII (C). Mutated amino acids and heptads are highlighted in red. (D) Point mutational analysis of a peripheral cysteine in heptad position Xg.
-
Figure 6—source data 1
Analysis of L1 retrotransposition.
- https://doi.org/10.7554/eLife.34960.021
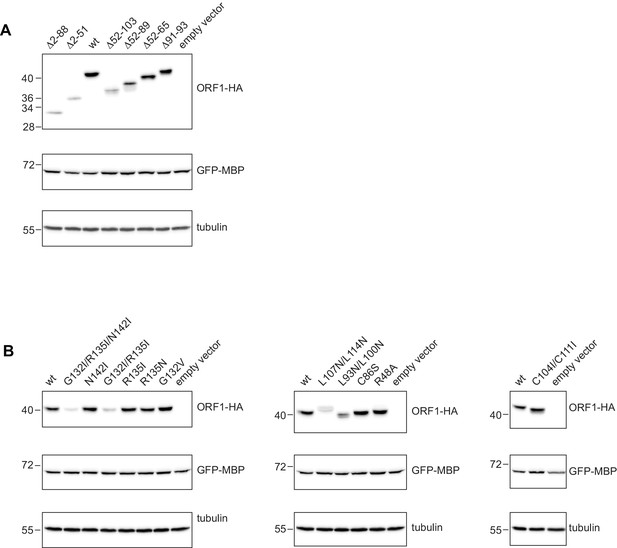
Western blots for the expression analysis of L1ORF1p coiled coil variants.
(A) Expression analysis of L1ORF1p deletion variants. Deletions comprise the NTR and/or several complete heptads, or regularize heptad IX by a deletion of the stammer. For expression analysis, HeLa cells were transfected with L1 retrotransposition plasmids encoding L1ORF1p variants with a C-terminal HA tag. Co-transfected GFP-MBP expression plasmids served as a transfection control. Empty vector (pcDNA3.1) served as a negative control and endogenous tubulin was detected as a gel loading control. Antibodies were against tubulin, GFP and HA. (B) Expression analysis of L1ORF1p point mutations including R48A in the NTR.
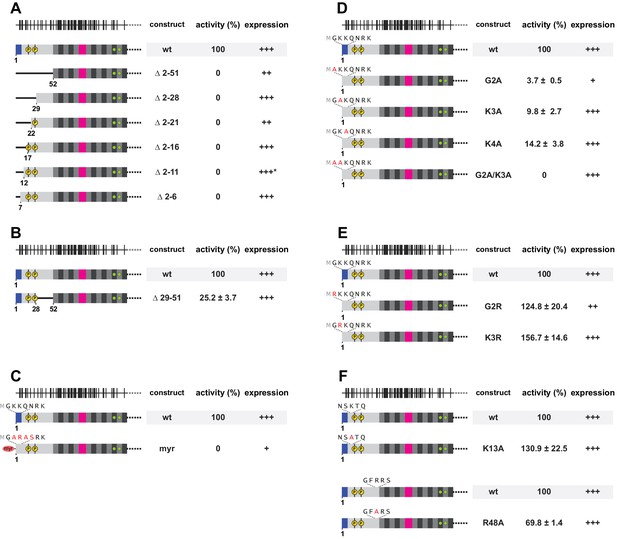
Requirement for a positively charged N-terminus on L1ORF1p.
(A) Systematic deletion analysis of the NTR, monitored by L1 retrotransposition in HeLa cells. Retrotransposition activity is calculated with respect to the wildtype (wt) protein, with the mean and standard deviations calculated from three independent replicates. Normal (+++), reduced (++) or poor (+) protein expression levels are marked. See also Figure 7—figure supplement 1. None of the deletions is tolerated. (B) Internal deletion in the NTR, permissive for retrotransposition. (C) Sequence conversion into an N-terminal myristoylation signal, abolishing retrotransposition. (D) Alanine scan of the three N-terminal residues, indicating the importance of these sequence positions. In the wildtype sequence, each of the three N-terminal residues carries a positive charge (after enzymatic removal of the N-terminal methionine). (E) Charge-preserving mutations, demonstrating the importance of electrostatics over sequence identity. (F) Point mutational analysis of internal NTR residues, demonstrating that internally located lysines or arginines are much less critical for retrotransposition.
-
Figure 7—source data 1
Analysis of L1 retrotransposition.
- https://doi.org/10.7554/eLife.34960.024
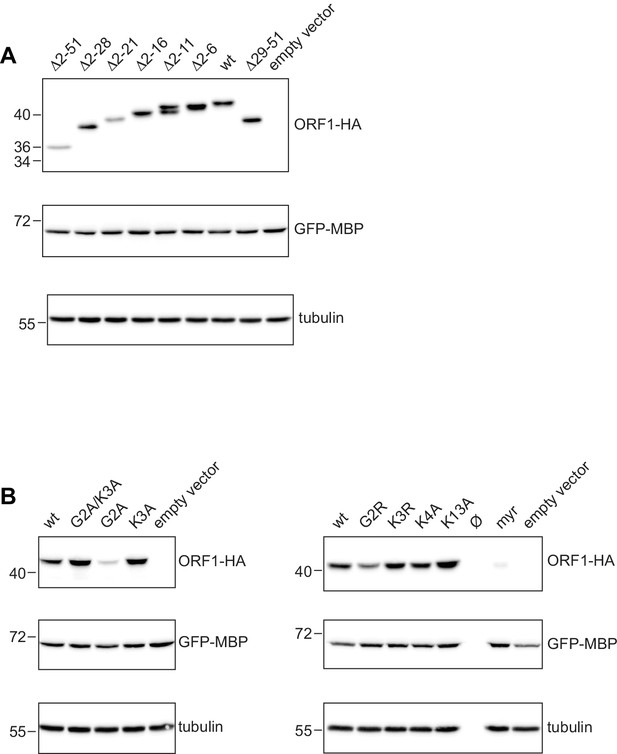
Western blots for the expression analysis of L1ORF1p NTR variants.
(A) Expression analysis of L1ORF1p NTR deletion variants. For expression analysis, HeLa cells were transfected with L1 retrotransposition plasmids encoding L1ORF1p variants with a C-terminal HA tag. Co-transfected GFP-MBP expression plasmids served as a transfection control. Empty vector (pcDNA3.1) served as a negative control and endogenous tubulin was detected as a gel loading control. Antibodies were against tubulin, GFP and HA. (B) Expression analysis of L1ORF1p point mutations, including a variant with an N-terminal myristoylation signal. As additional controls for the consistency of the protein expression results, several L1 reporter plasmids for the poorly expressed G2A mutant were independently generated and analyzed and also further mutagenized into normally expressing G2A/K3A variants (not shown here). For R48A, see Figure 6—figure supplement 1B.
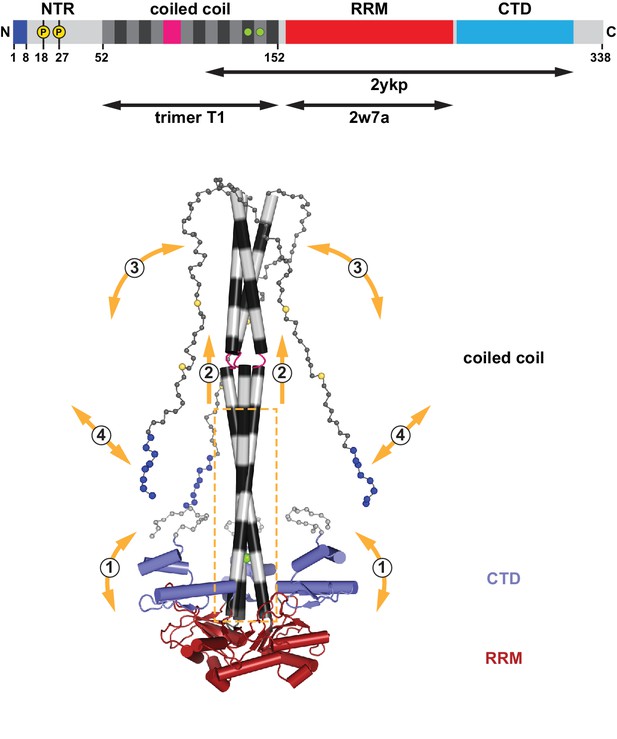
Assembly and flexibility of the human L1 ORF1p trimer.
A complete structural model for the human L1ORF1p trimer was generated by superimposing known crystal structures (ribbons representation with cylindrical helices) and by geometrically restrained free modeling of the unstructured regions as random coil (balls-and-sticks representation). Starting from the composite model in Figure 2B (generated from trimer T1, residues 52–152, and PDB-ID 2ykp, residues 107–223) (Khazina et al., 2011), the three RRM domains were replaced by the crystal structure of the isolated RRM domain (PDB-ID 2w7a, residues 157–252) (Khazina and Weichenrieder, 2009), where all of the connecting loops are defined. The presumably unstructured N-terminal and C-terminal regions were then added for size comparison, where each sphere represents a Cα atom. The conserved part of the coiled coil domain (boxed) is necessary and sufficient for the trimerization of L1ORF1p and serves as a scaffold for the flexible attachment of the CTDs, which cooperate with the RRM domains in L1RNA binding (Khazina et al., 2011) (1). The conserved part of the coiled coil domain is also required for the folding and trimeric assembly of the non-conserved part of the coiled coil (2), which can switch between structured and unstructured states (3). Finally, L1ORF1p functionally requires a strongly positively charged N-terminus (blues spheres) on the unstructured NTR. Interactions of the N-terminal amino acids might be regulated (4) by adjacent phosphorylation (yellow spheres), by the conformation of the coiled coil domain and/or by the bound L1RNA. For a video, see also Figure 8—video 1.
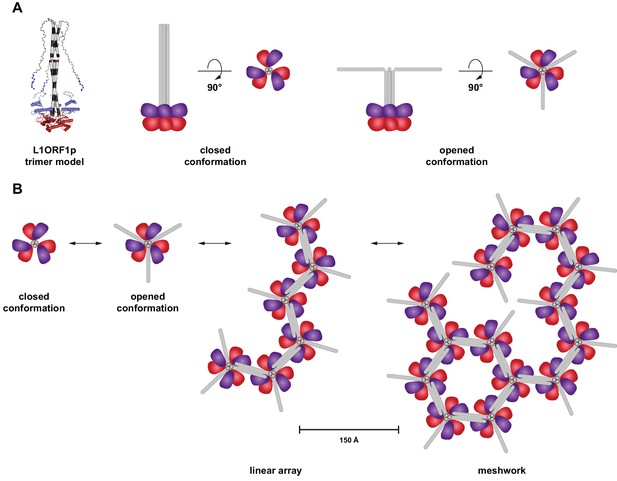
Hypothetical model for an oligomerization of L1ORF1p trimers into linear arrays and larger meshworks.
(A) Cartoon illustration of trimeric L1ORF1p in the closed and an opened conformation as indicated by thermal melting analysis. The NTR is omitted from the cartoon for simplicity. (B) Linear arrays and meshworks of trimeric L1ORF1p, enabled by the dimerization of the non-conserved portion of L1ORF1p. Dimerization might only occur upon assembly with the RNA substrate and also in geometrically distinct orientations.
Video of the complete structural model of the human L1ORF1p rotating around the threefold axis.
https://doi.org/10.7554/eLife.34960.027Tables
Data collection and refinement statistics.
https://doi.org/10.7554/eLife.34960.009| Data collection | |
| Space group | P21212 |
| Cell dimensions | |
| a, b, c (Å) | 92.2, 250.6, 33.8 |
| α, β, γ (°) | 90, 90, 90 |
| Wavelength (Å) | 1.000 |
| Resolution (Å)* | 125–2.65 (2.72–2.65) |
| Rsym* | 7.3 (81.3) |
| I/σI* | 14.9 (2.2) |
| Completeness (%)* | 99.8 (98.4) |
| Redundancy* | 5.4 (5.5) |
| Refinement | |
| Resolution (Å) | 125–2.65 |
| No. of reflections | 23830 |
| Rwork/Rfree (%) | 20.8/23.8 |
| No. of atoms | |
| Protein | 4851 |
| Ligand/ion | 4 |
| Water | 64 |
| B-factors (Å2) | |
| Protein | 87.6 |
| Ligand/ion | 69.3 |
| Water | 58.3 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 0.82 |
-
*Highest resolution shell is shown in parenthesis.
Additional files
-
Supplementary file 1
Mammalian L1ORF1p sequences used in Figure 1—figure supplement 1B
Mammalian L1ORF1p sequences are from Boissinot and Sookdeo (Boissinot and Sookdeo, 2016) apart from the human sequence (NCBI accession L19088.1) (Dombroski et al., 1993), the mouse sequences (Sookdeo et al., 2013) and the megabat sequence (NCBI accession KF796623.1) (Yang et al., 2014). The conserved and alignable portions of the proteins are in bold letters (Khazina et al., 2011), and the coiled coil domains are listed as heptads as in Figure 1C. Regular heptads are shaded grey and black and non-heptad interruptions are shaded in magenta, corresponding to the colors in Figure 1—figure supplement 1B. Similarly, the NTRs are light grey with the positively charged N-terminal peptides in blue, the RRM domains are in red and the CTDs are in cyan. Coiled coil propensity was calculated using PCoils (Alva et al., 2016; Lupas, 1996) and used to assign heptads by manual inspection of results from PCoils. Remaining ambiguities in the precise assignment of non-heptad interruptions require finer sequence sampling or experimental validation to be resolved.
- https://doi.org/10.7554/eLife.34960.028
-
Supplementary file 2
Individual accessions for primate L1ORF1p.
Primate L1ORF1p sequences in Figure 1—figure supplement 2 are reconstructed consensus sequences with 60% residue identity in the alignments of the listed accessions.
- https://doi.org/10.7554/eLife.34960.029
-
Supplementary file 3
Plasmid constructs based on pJM101 (Moran et al., 1996; Sassaman et al., 1997).
- https://doi.org/10.7554/eLife.34960.030
-
Reporting standard 1
- https://doi.org/10.7554/eLife.34960.031
-
Transparent reporting form
- https://doi.org/10.7554/eLife.34960.032





