A genetic program mediates cold-warming response and promotes stress-induced phenoptosis in C. elegans
Figures
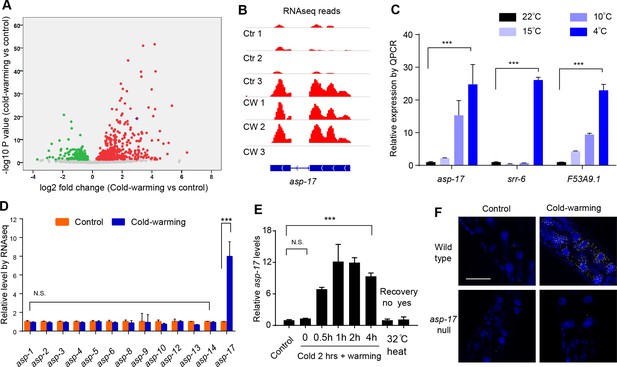
asp-17 is robustly and specifically induced by cold-warming.
(A) Volcano plot of RNA-seq showing differentially regulated genes (up-regulated genes in red; down-regulated genes in green; asp-17 was indicated by purple dot) by cold-warming in wild type C. elegans. (B) RNA-seq reads at the asp-17 locus showing consistent up-regulation of asp-17 transcript levels in triplicate samples after cold-warming. (C) QPCR measurements of gene expression levels showing up-regulation of three representative CW inducible genes, including asp-17, after shifting from 25°C to different degrees of hypothermia (4°C, 10°C, 15°C and 22°C) lasting 2 hrs followed by recovery at 25°C for 0.5 hr. (D) Quantification of RNA-seq reads indicating specific up-regulation of asp-17 but not other members of the asp family genes (only expressed asp genes are shown). (E) QPCR measurements of asp-17 levels under conditions of indicated durations of cold and warming (with or without 25°C recovery for ‘yes/no’). (F) smFISH images showing asp-17 up-regulation (signals indicated by yellow) by CW predominantly in the intestine of wild type but not asp-17 null animals. n ≥ 20 total animals for each group with N ≥ 3 independent biological replicates; *** indicates p<0.001. Scale bar: 50 µm.
-
Figure 1—source data 1
Lists of genes up- and down-regulated by CW with adjusted p<0.05 and log2FoldChange from biological triplicate samples of wild-type C. elegans.
- https://doi.org/10.7554/eLife.35037.005
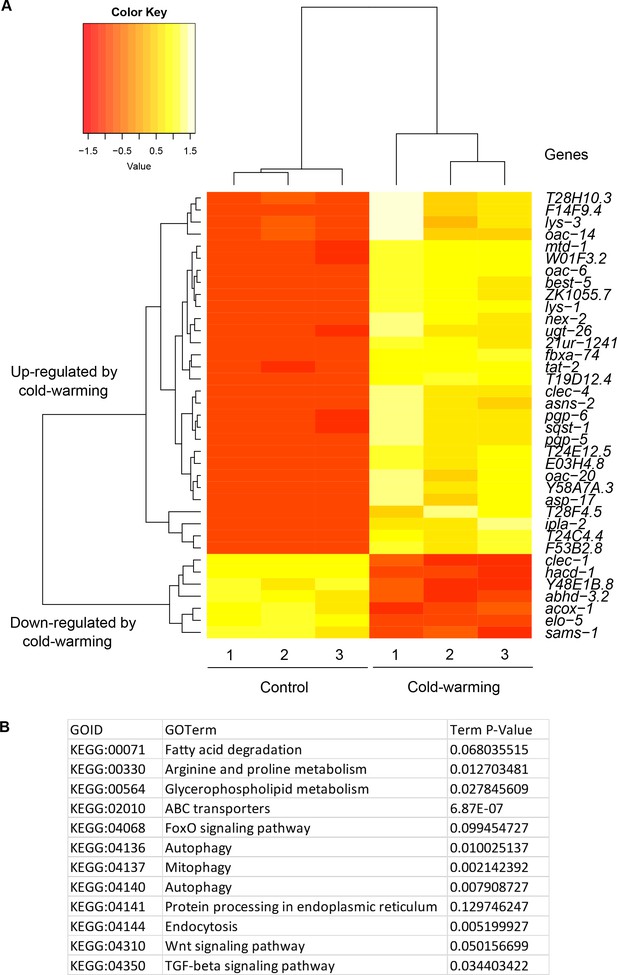
RNA-seq identified genes up-regulated by cold-warming.
(A) Hierarchical clustering analysis of genes up- and down-regulated genes by CW. (B) Gene ontology analysis of genes up- and down-regulated genes by CW.
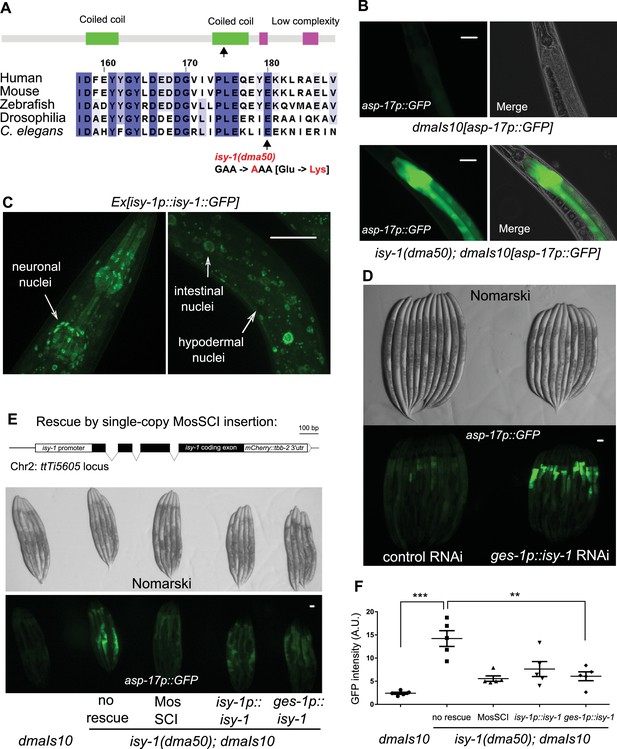
A forward genetic screen identifies C. elegans isy-1 as a causal regulator of asp-17::GFP expression.
(A) Schematic of the C. elegans ISY-1 protein showing the domain structure predicted by SMART (top) (http://smart.embl-heidelberg.de) and a multiple sequence alignment of ISY-1 homologues from major metazoans showing the conservation of the glutamate residue substituted to lysine by the dma50 mutation isolated from EMS screens. (B) Nomarski and fluorescence images showing the phenotype of intestinal asp-17p::GFP in wild type and dma50 mutants. (C) Fluorescence images showing the distribution of ISY-1::GFP driven by the endogenous isy-1 promoter. Arrows indicate neuronal, hypodermal and intestinal nuclei. (D) Nomarski and fluorescence images showing intestinal asp-17p::GFP in a transgenic strain expressing RNAi against isy-1 specifically in intestine. (E) Schematic of the C. elegans isy-1 gene with mCherry tagged at the C-terminus (top); Nomarski and fluorescence images showing rescue of dma50 by various transgenes (below). (F) Quantification of fluorescence intensities showing rescue of dma50 in asp-17p::GFP activation. N ≥ 3 independent biological replicates; *** indicates p<0.001. Scale bar: 20 µm.
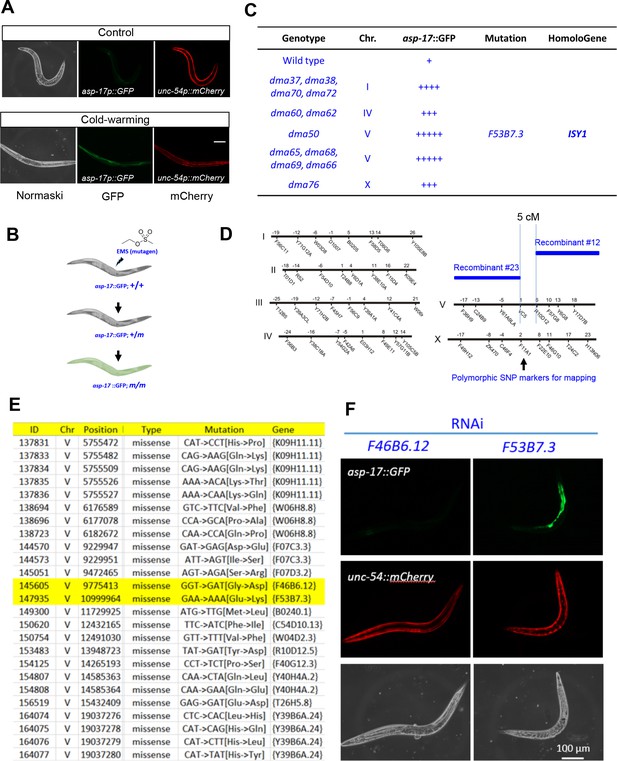
EMS screens identified isy-1 as a regulator of asp-17.
(A) Fluorescence images showing up-regulation of asp-17p::GFP but not the co-injection marker unc-54p::mCherry by CW. (B) Schematic illustrating the workflow of EMS mutagenesis to isolate mutants with abnormal asp-17p::GFP expression. (C) Table showing the mutants isolated from the screen, including dma50 that defines the gene F53B7.3, named as isy-1. (D) Chromosome genetic maps showing DraI SNP sites and the mapped interval for dma50. (E) Table listing candidate genes with mutations identified by WGS, with two genes highlighted in yellow within the mapped interval. (F) fluorescence images showing that RNAi of isy-1 phenocopies dma50. N ≥ 3 independent biological replicates; *** indicates p<0.001. Scale bar: 20 µm.
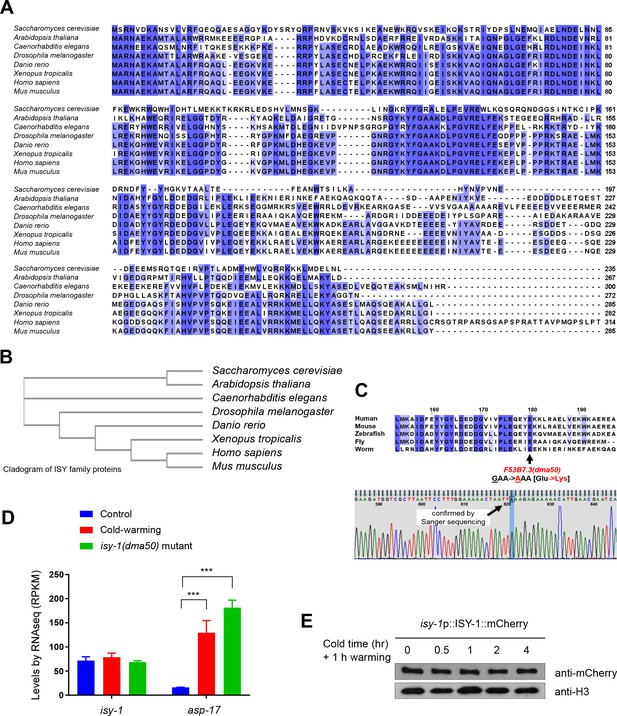
ISY-1 is a C. elegans member of evolutionarily conserved protein family and its level is not regulated by cold-warming.
(A) Multiple sequence alignment (Cluster Omega, visualized by Jalview) of ISY-1 family members in major metazoan species. (B) Cladogram of ISY family proteins. (C) Sanger sequencing confirmation of isy-1(dma50). (D) RNA-seq measurements of isy-1 and asp-17 levels under CW conditions and in isy-1(dma50) mutants. (E) Western blot of isy-1p::ISY-1::mCherry showing no regulation by CW. N ≥ 3 independent biological replicates; *** indicates p<0.001.
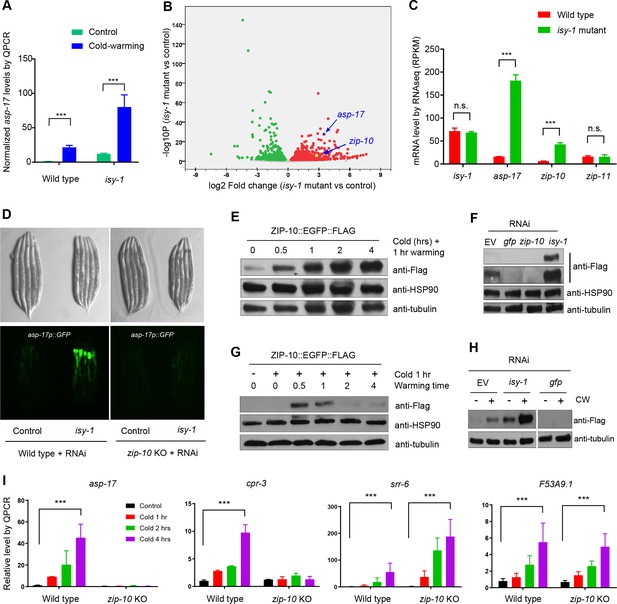
ZIP-10 acts downstream of ISY-1 and mediates transcriptional response to CW.
(A) QPCR measurements of asp-17 levels induced by CW in wild type and isy-1(dma50) mutants. (B) Volcano plot of RNA-seq showing differentially regulated genes (up-regulated genes in red; down-regulated genes in green) in isy-1 mutants compared with wild type. (C) RNA-seq measurements of expression levels for indicated genes in wild type and isy-1(dma50) mutants. (D) Nomarski and fluorescence images showing asp-17p::GFP induction by isy-1 RNAi was blocked in zip-10 mutants. (E) Western blots of the integrated zip-10p::zip-10::EGFP::FLAG strain showing time-dependent protein induction by CW. (F) Western blots of the integrated zip-10p::zip-10::EGFP::FLAG strain showing its up-regulation by isy-1 RNAi and down-regulation by GFP or zip-10 RNAi. Both short- and long-exposure blots are shown. (G) Western blots of the integrated zip-10p::zip-10::EGFP::FLAG strain showing its up-regulation strictly required warming after cold shock. (H) Western blots of the integrated zip-10p::zip-10::EGFP::FLAG strain showing its up-regulation by CW was further enhanced by isy-1 RNAi. (I) QPCR measurements of gene expression levels showing ZIP-10 dependent up-regulation of asp-17 and cpr-3 but not srr-6 or F53B9.1 after cold for indicated durations and 1 hr warming. n ≥ 20 total animals for each group with N ≥ 3 independent biological replicates; *** indicates p<0.001. Scale bar: 20 µm.
-
Figure 3—source data 1
Lists of genes up- and down-regulated by the isy-1(dma50) mutation with adjusted p<0.05 and log2FoldChange from biological triplicate samples of wild-type and isy-1(dma50) mutant C. elegans.
- https://doi.org/10.7554/eLife.35037.012
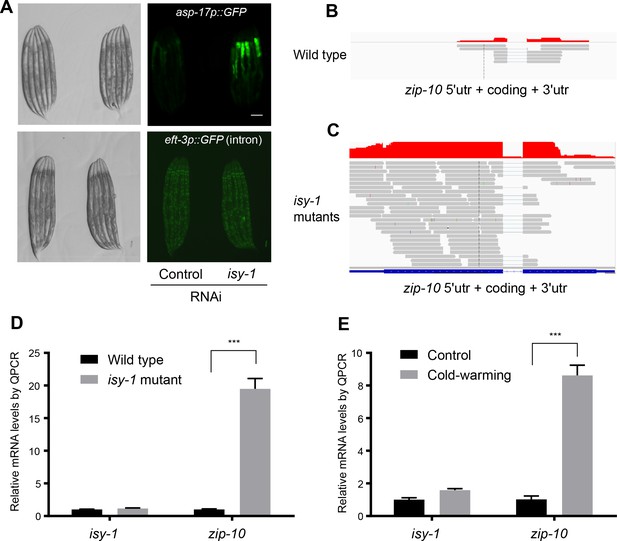
ISY-1 does not affect general splicing of a GFP reporter nor specific splicing of zip-10.
(A) Nomarski and fluorescence images showing that animals with isy-1 RNAi are not defective in general intron splicing of an eft-3p::GFP (intron-containing) reporter. (B) Read plot from RNA-seq of wild type at the zip-10 locus. (C) Read plot from RNA-seq of isy-1(dma50) mutants at the zip-10 locus indicating the intact splicing of the zip-10 intron despite increased mRNA levels. (D) Normalized QPCR measurements of mRNA expression levels of isy-1 and zip-10 from wild type and isy-1(dma50) mutants. (E) Normalized QPCR measurements of mRNA expression levels of isy-1 and zip-10 from wild type and CW-treated animals.
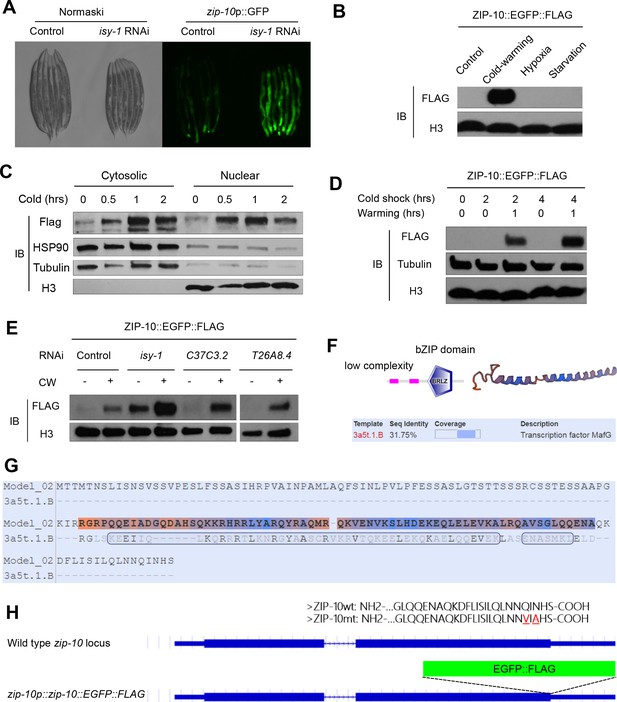
Mechanisms of regulation and function of zip-10.
(A) Nomarski and fluorescence images showing the zip-10p::GFP reporter activated by isy-1 RNAi. (B) Western blot of lysates from zip-10p::zip-10::EGFP::FLAG transgenic animals showing its increased abundance after CW but not by hypoxia (0.5% O2) or starvation for 24 hrs. (C) Western blot of fractionated lysates from zip-10p::zip-10::EGFP::FLAG transgenic animals after CW showing its increased abundance in cytosol and nucleus. (D) Western blot of lysates from zip-10p::zip-10::EGFP::FLAG transgenic animals showing its increased abundance after CW when warming was permitted and cold exposure was prolonged. (E) Western blot of lysates from zip-10p::zip-10::FLAG transgenic and various RNAi-treated animals showing its increased abundance after CW in isy-1 RNAi-treated animals but was unaffected by RNAi against C37C3.2, encoding the C. elegans orthologue of translation initiation factor 5 (eIF5), or T26A8.4, encoding the C. elegans orthologue of Saccharomyces cerevisiae Caf120, a component of the Ccr4-Not deadenylase RNA-degrading complex. (F) ZIP-10 domain organization based on analysis by SMART (smart.embl-heidelberg.de) and modeled structure of ZIP-10’s bZIP domain (swissmodel.expasy.org). (G) Sequence alignment of ZIP-10’s bZIP domain and the Maf transcription factor as a template of the modelled ZIP-10 structure. (H) Schematic of the zip-10 locus showing the wild type, EGFP::FLAG tagged allele, and the wild-type and C-terminal transactivation mutant ZIP-10 protein sequences. The mutated residues (from Q and N to V and A, respectively) at the ZIP-10 C-terminus are completely conserved in nematodes.
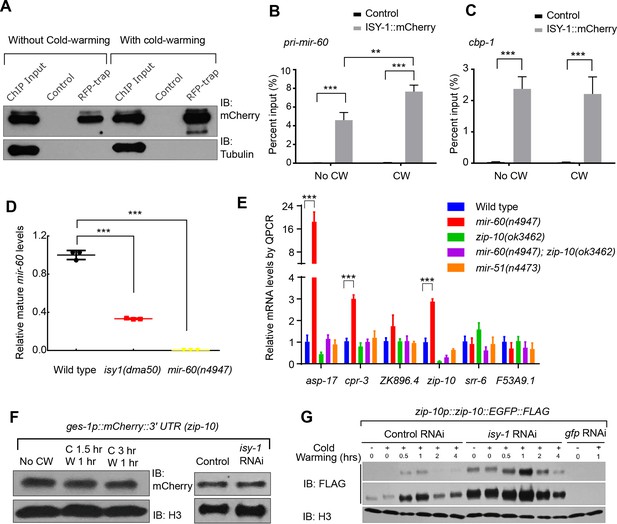
ISY-1 regulates zip-10 via mir-60.
(A) Western blot of mCherry/RFP-trapped RNA immunoprecipitates in animals treated with or without cold-warming. (B) QPCR measurements of the percent input for primary mir-60 transcripts from mCherry/RFP-trapped RNA immunoprecipitates in animals treated with or without cold-warming. (C) QPCR measurements of the percent input for cbp-1 transcripts from mCherry/RFP-trapped RNA immunoprecipitates in animals treated with or without cold-warming. (D) QPCR measurements of the mature mir-60 transcript levels from wild type, isy-1(dma50) and mir-60(n4947) deletion mutants. (E) QPCR measurements of the levels of CW-inducible gene transcripts in animals with indicated genotypes and conditions. (F) Western blot of lysates from animals carrying the array ges-1p::mCherry::3’utr(zip-10) reporters with CW or isy-1 RNAi. No change of reporter activity was observed. (G) Western blot of lysates from animals carrying zip-10p::zip-10::EGFP::FLAG reporters with various indicated CW and RNAi conditions. n ≥ 20 total animals for each group with N ≥ 3 independent biological replicates; *** indicates p<0.001; ** indicates p<0.01.
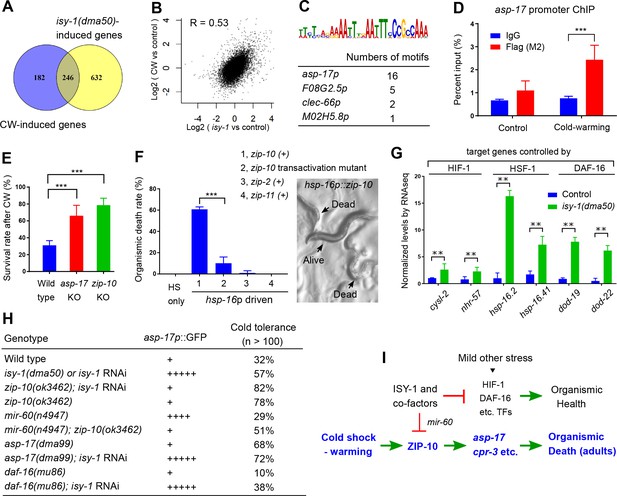
ISY-1 and ZIP-10 regulate a genetic program to promote organismic death.
(A) Venn diagram indicating numbers of genes commonly regulated by CW and isy-1(dma50) mutants. (B) Scatter plot depicting the correlation between the transcriptome response to cold (y-axis) and the transcriptome response to isy-1 mutation (x-axis). Shown are the log2 fold changes compared with corresponding controls. Pearson correlation coefficient and the associated P-value were calculated using R functions. (C) AT-rich motif identified by MEME enriched among the CW and isy-1(dma50) regulated genes, with a table listing numbers of the motif present in top-ranked four genes. (D) ChIP-QPCR measurements of ZIP-10::FLAG binding to the asp-17 promoter. (E) Survival rates of indicated genotypes after prolonged CW (4°C for 48 hrs followed by 4 hrs of warming). (F) Organismic death rates of indicated genotypes after heat shock (32°C) induction of zip-10 wild type, mutant with defective transactivation C-terminus, zip-2 and zip-11 (left), without CW. Nomarski image (right) indicates morphologies of normal and dead animals with induction of zip-10. (G) RNA-seq measurements of gene targets of indicated TFs. (H) Table showing the asp-17p::GFP and cold tolerance phenotypes of animals with indicated genotypes. (I) Model for the role and regulation of the ZIP-10 pathway. n ≥ 20 total animals for each group with N ≥ 3 independent biological replicates; *** indicates p<0.001.
-
Figure 5—source data 1
Lists of genes commonly regulated by CW and the isy-1(dma50) mutation with adjusted p<0.05 and log2FoldChange from biological triplicate samples of wild-type and isy-1(dma50) mutant C. elegans.
- https://doi.org/10.7554/eLife.35037.017
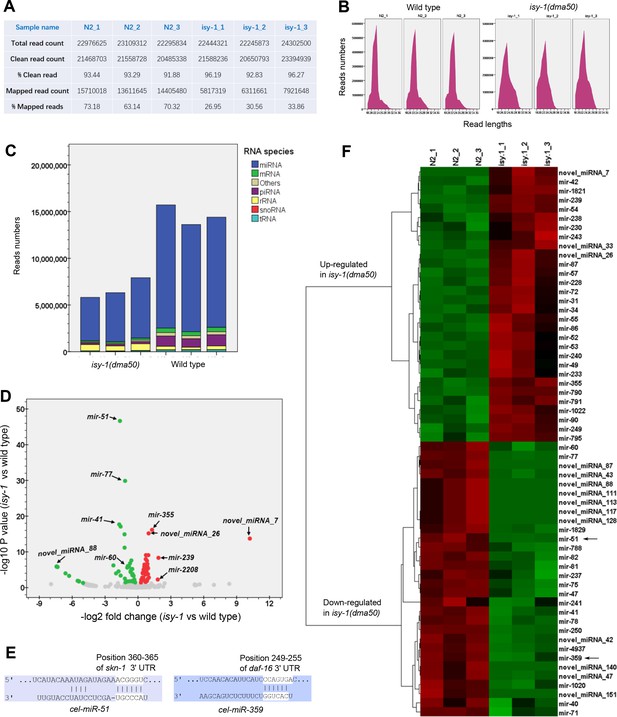
Identification of differentially regulated microRNAs in wild type and isy-1(dma50) mutants.
(A) Summary table showing the read statistics for the small RNA-seq experiments using three biologically triplicate samples in wild type and isy-1(dma50) mutants. (B) Small RNA length distribution plots for wild type and isy-1(dma50) mutants. (C) Distribution graphs for various types of small RNAs identified by small RNA sequencing for wild type and isy-1(dma50) mutants. (D) Volcano plot showing the differentially regulated microRNAs, including mir-60, in the isy-1(dma50) mutants (three replicates each). (E) Schematic of two examples illustrating the TF-encoding genes, skn-1 and daf-16 at 3’ UTRs, targeted by two microRNAs, mir-51 and mir-359, respectively, which are down-regulated in isy-1(dma50) mutants. (F) Hierarchical clustering analysis of microRNAs up- or down-regulated in isy-1(dma50) mutants. mir-51 and mir-359 down-regulation were indicated by arrows.
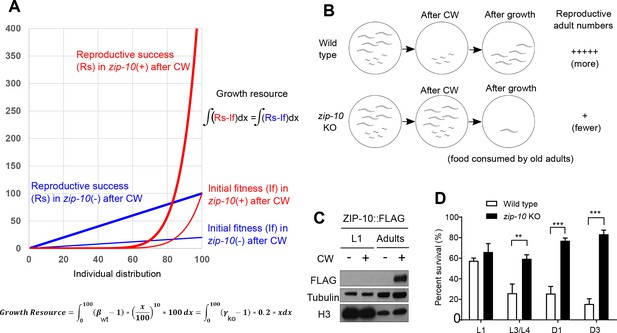
ZIP-10 constitutes a genetic program promoting kin selection of C. elegans under resource-limiting and stress conditions.
(A) Mathematical model illustrating the advantage of wild-type (red line) versus zip-10 KO animals (blue line) at the population level under growth resource-liming and thermal stress conditions. Individual fitness distribution (x-axis) after cold-warming thermal stress is set to be exponential (thin red line) and linear (thin blue line) to fit the phenotypic differences in death rates of wild type and zip-10 KO animals and to simplify comparison and calculation. Given the same period of time and the same amount of growth resources (integral differences between thin and thick lines), wild-type population yields more thermal stress-adapted reproducing animals than zip-10 deficient animals. The parameters for integrals were adjusted so that the ‘growth resources’ are the same despite that the population growth modes are different for wild type and zip-10 KO. (B) Schematic illustrating an exemplar situation of the mathematical model in which five post-reproduction adults and five larvae are living in a food-limiting condition. After cold-warming thermal stress, wild-type adults die out, leaving food for young larvae to grow into reproductive adults whereas zip-10 KO post-reproduction adults do not die, consuming the limited food and competing out the young larvae so that fewer reproductively active zip-10 KO animals emerge from such conditions. (C) Experimental evidence indicating the specific induction of zip-10p::zip-10::EGFP::FLAG by CW as measured by western blot in adults but not in larvae. (D) Survival rates of wild type and zip-10 KO (ok3462) animals at different stages after prolonged CW thermal stresses (96 hrs cold shock for L1 while 48 hrs for others) showing that ZIP-10’s pro-death effects are more prominent in old animals. n ≥ 20 total animals for each group with N ≥ 3 independent biological replicates; *** indicates p<0.001, ** indicates p<0.01.
Additional files
-
Supplementary file 1
A specific subset of CW-inducible genes is dependent on ZIP-10.
Shown is a table of fold induction for gene expression levels determined by QPCR measurements of top-ranked randomly selected CW-inducible genes in wild type and zip-10 mutant animals.
- https://doi.org/10.7554/eLife.35037.018
-
Transparent reporting form
- https://doi.org/10.7554/eLife.35037.019





