Dual roles for ATP in the regulation of phase separated protein aggregates in Xenopus oocyte nucleoli
Figures
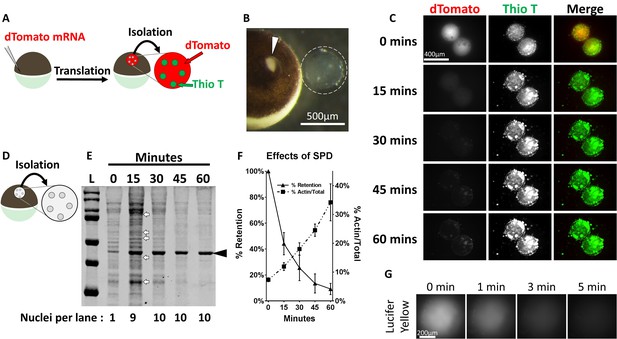
Protein aggregates are selectively retained in isolated oocyte nuclei.
(A) Synthetic mRNA encoding nuclear localized dTomato was injected into stage V-VI Xenopus laevis oocytes 1 day prior to isolation. Following isolation nuclei were incubated in OR2 buffer for the indicated times then assayed. (B) Stage VI oocyte with incision site (arrowhead) and manually isolated nucleus (dashed circle). (C) 1 hr time-lapse images of aqueously isolated and Thioflavin T (Thio T, green) stained nuclei from dTomato-NLS (red) expressing stage VI oocytes demonstrates loss of soluble dTomato, but retention of Thioflavin T positive aggregates. (D–F) Nuclei were isolated from un-manipulated oocytes, incubated in OR2, collected at 15 min intervals, and analyzed by SDS-PAGE (D). Coomassie staining (E) with quantitation (F) of soluble protein depleted nuclei demonstrates rapid loss of soluble endogenous proteins and retention of aggregate associated proteins. The number of nuclear equivalents per lane is indicated at the bottom of (E). Arrows highlight the subset of proteins enriched following depletion of soluble proteins. Arrowhead highlights 42 kDa actin, which is enriched following soluble protein depletion (F). (G) Time-lapse images of an isolated stage VI oocyte nucleus immediately following nuclear injection of Lucifer Yellow, a fluorescent ATP surrogate. Images in (C) and (G) are representative from two independent experiments encompassing at least six nuclei. Data in (F) contains three biological replicates (3–10 nuclei per replicate) representing material from two separate frogs.
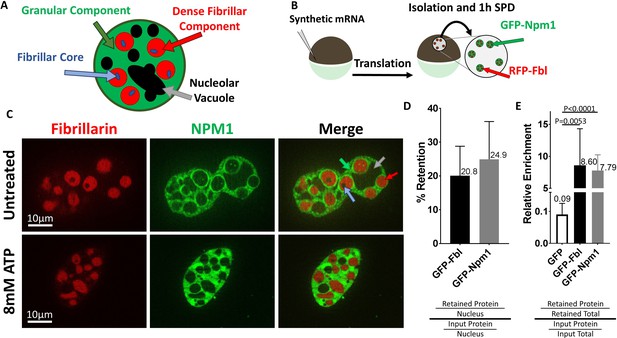
Nucleoli are resistant to ATP mediated hydrotropic solubilization.
(A) Schematic diagram of tri-laminar nucleolar architecture, with granular component in green, dense fibrillar component in red, and fibrillar core in blue. Nucleolar vacuoles are unlabeled and will appear black. (B) Synthetic mRNA encoding GFP-NLS or fluorescently labeled Npm1 and/or Fbl were injected into stage V/VI oocytes. Following overnight incubation nuclei were isolated, depleted of soluble proteins for 1 hr in OR2, then analyzed by fluorescent microscopy (C) or anti-GFP immunoblotting (D–E). (C) Npm1 (green) or Fbl (red) are retained (top) and resistant to solubilization by 8 mM ATP (bottom). (D–E and Figure 2—figure supplement 1) Nuclei from GFP-Fbl/GFP-NLS or GFP-Npm1/GFP-NLS co-expressing oocytes were isolated and depleted of soluble proteins (retained) and compared to GFP-Fbl or GFP-Npm1 levels in immediately harvested (input) nuclei (D) or total protein (E). Labeled proteins were analyzed via anti-GFP Western Blot, and total protein was determined via REVERT protein staining (LI-Cor). Schematic of methodology and representative raw data for (D–E) can be found in Figure 2—figure supplement 1. Images in (C) are representative optical sections captured with an Apotome equipped Zeiss Axioplan2 microscope, and do not have identical exposure times. Data in (D–E) is derived from 4 (GFP-Npm1 and GFP-FBL), or 6 (GFP) biological replicates (groups of 2–20 nuclei).
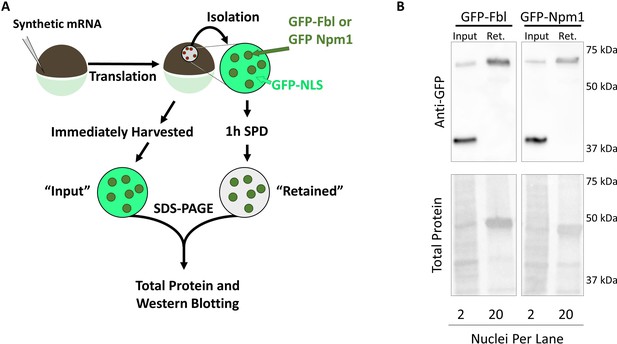
GFP-Fbl and GFP-Npm1, but not GFP-NLS, are Retained Following Soluble Protein Depletion.
(A) GFP-Fbl/GFP-NLS or GFP-Npm1/GFP-NLS synthetic mRNA was co-injected into stage V-VI oocytes and allowed to translate overnight. Nuclei from co-expressing oocytes were isolated and depleted of soluble proteins (retained), or immediately harvested (input), and resolved with SDS-PAGE. Following transfer to a nitrocellulose membrane, total protein was determined via REVERT protein staining (LI-Cor), and labeled proteins were analyzed via anti-GFP Western Blot. (B) Representative raw data from Figure 2E–D. Anti-GFP Western blotting (Top) demonstrates enrichment of GFP-Fbl and GFP-Npm1 as compared to soluble GFP-NLS internal control, as well as total protein (bottom). Total number of nuclei per lane is as indicated (bottom).
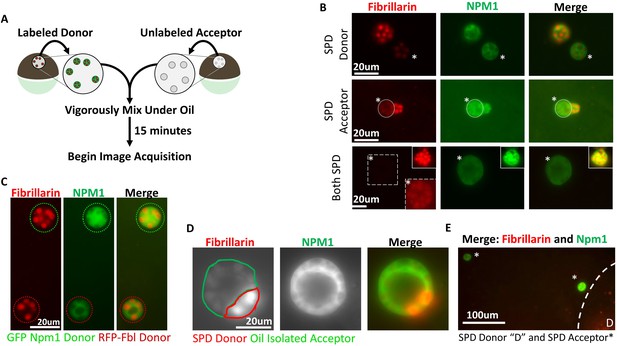
Aggregated nucleolar proteins retain intrinsic capacity for dynamic proteins exchange, but that capacity is enhanced by soluble nuclear components.
(A) Donor nuclei were isolated from oocytes co-expressing GFP-Npm1 and RFP-Fbl, while acceptor nuclei were isolated from un-injected oocytes. (B) Top, soluble protein depleted (SPD) donor nucleus was mixed with oil isolated acceptor nucleus, demonstrating soluble protein depletion does not irreversibly alter nucleoli, and suggests that a soluble nuclear factor(s) is responsible for the dynamic nature of nucleoli. Middle, transfer of labeled protein from oil isolated donor to soluble protein depleted acceptor indicates that soluble protein depletion does not prevent recruitment nor aggregation of soluble proteins. Bottom, aggregated proteins exchange between soluble protein depleted nucleoli. Exposure was adjusted in bottom images to account for decreased signal, and donor nucleolus was included in top inset for comparison. Brightness and contrast were adjusted in dashed inset to aid in RFP-Fbl visualization. Asterisks indicates the acceptor nucleolus. (C) Exchange of fluorescently labeled proteins from oil isolated and merged nuclei singly expressing GFP-Npm1 or RFP-Fbl. (D) Trans-nucleolar fusion following mixing of soluble protein depleted RFP-Fbl expressing donor (red) and oil isolated GFP-Npm1 (green) under oil demonstrates rescue more normal rounded phenotype. (E) Exchange of GFP-Npm1 and RFP-Fbl is diffusion limited. As distance from donor ‘D’ increases fluorescence intensity of acceptor decreases. Images in (B–C) are representative of 3 independent experiments and at least nine biological replicates (merged nuclei). Images acquisition began 15 min after trans nuclear mixing. The fusion event depicted in (D) is the most complete event we observed. Partial fusions (e.g. middle panel B) are commonly observed.
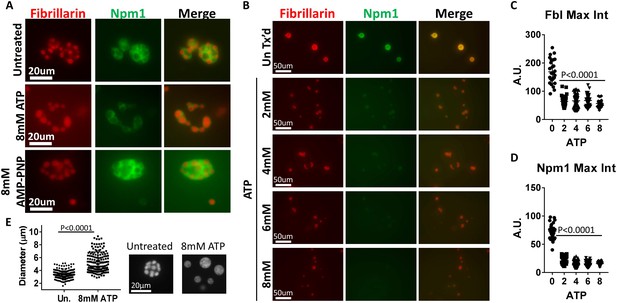
ATP hydrolysis and a soluble factor(s) are required for nucleolar remodeling.
(A) Nuclei were isolated from RFP-Fbl and GFP-Npm1 expressing oocytes, immediately transferred into ISB alone (top), ISB supplemented with 8 mM ATP (middle), or ISB supplemented with 8 mM AMP-PNP (bottom), and imaged. (B) Freshly isolated nuclei were transferred into ISB supplemented with increasing amounts of ATP. (C–D) Relative maximum fluorescence intensity of GFP-NPM1 and RFP-FBL co-expressing nuclei incubated in ISB (untreated) or ISB supplemented with 0–8 mM ATP. (E) Diameter of RFP-Fbl positive Dense Fibrillar Component following incubation of freshly isolated nuclei in increasing concentrations of ATP. To test the effect of soluble nuclear proteins, nuclei in (A–E) were isolated and immediately placed into ISB with or without indicated supplementation. Images in (A–D) are representative of at least three independent experiments. Data in (E) represents 298 independent untreated and 144 independent 8 mM ATP treated RFP-Fbl foci. Data in (C–D) is from a single experiment with at least 25 nucleoli per data point.
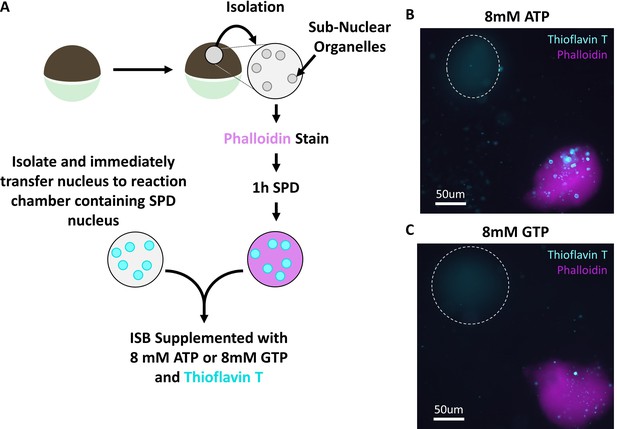
Treatment with 8 mM GTP replicates ATP results.
(A) Nuclei were stained with stained with Alexa Fluor 568 conjugated phalloidin (Molecular Probes), depleted of soluble proteins for 1 hr and then placed into a reaction chamber with ISB supplemented with thioflavin T and 8 mM ATP or 8 mM GTP. A second nucleus was subsequently isolated and immediately added to the same reaction chamber. Isolation and immediate addition to ISB ensures transfer of soluble proteins. Images were acquired 15 min after addition of freshly isolated nucleus.
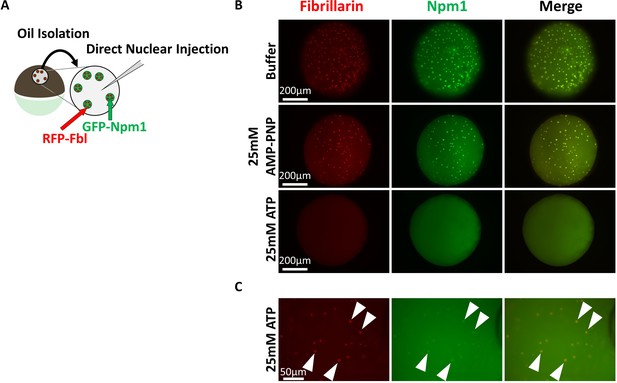
Super physiologic ATP promotes Nucleolar Disassembly Ex Vivo but AMPPNP does not.
(A) Nuclei from GFP-Npm1 and RFP-Fbl expressing oocytes were isolated under oil to maintain nuclear contents and were injected with an ~equal vol (9.2 nL) of buffer (Top), 50 mM AMP-PNP (middle), or 50 mM ATP (Bottom). (B) High magnification image of 50 mM ATP injected nucleus. Arrowheads indicate resistant RFP-Fbl positive puncta. Images in (B–C) are representative of 9 (buffer), 12 (AMP-PNP), or 18 (ATP) biological replicates.
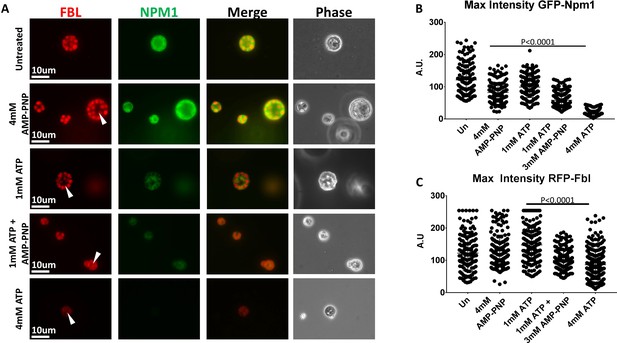
Hydrotropic Solubilization Contributes to Nucleolar Protein Dynamics.
(A) Oocyte nuclei were isolated from RFP-Fbl and GFP-Npm1 expressing oocytes and immediately transferred into ISB alone (top) or ISB supplemented with ATP, AMP-PNP, or both nucleotides and imaged. (B) When compared to untreated controls, maximum fluorescence intensity measurements for GFP-Npm1 and C) RFP-Fbl reveals loss of signal intensity after 1 mM ATP + 3 mM AMP-PNP or 4 mM ATP supplementation. Images in (A) are representative of 3 independent experiments in which maximum fluorescence intensity of GFP-Npm1 (B) and RFP-Fbl (C) from 155 to 264 nucleoli was quantified, pooled, and analyzed.
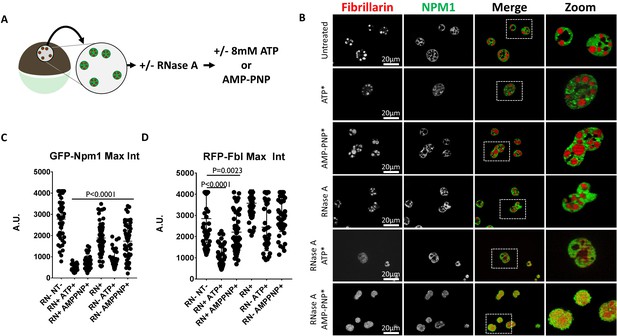
RNase Treatment Sensitizes Nucleoli to Hydrotropic Solubilization.
(A) Nuclei from GFP-Npm1 and RFP-Fbl expressing oocytes were isolated, transferred into OR2 or OR2 supplemented with 1 mg/mL RNase A, and incubated for 1 hr prior to nucleotide treatment. (B) Optical sections through soluble protein depletion and RNase A treated soluble protein depleted nuclei left untreated (left), treated with 8 mM ATP (middle), or 8 mM AMP-PNP (right). (C–D) Intensity of GFP-Npm1 and RFP-Fbl. Data in (B–D) is representative of three independent experiments and three independent frogs. (C–D) represent a single experiment containing 3–6 biological replicates per treatment group and 129–205 individual nucleoli.
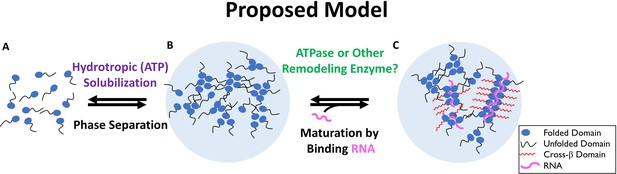
Two-Step Model of Nucleolar Aggregation and Disaggregation.
Monomeric proteins self-associate driving phase separation. RNA binding stabilizes and matures aggregates, conferring resistance to hydrotropic solubilization. In contrast, ribonucleoprotein particles are destabilized by a diffusible ATPase or RNA depletion, sensitizing them to ATP mediated hydrotropic solubilization.
Tables
Representative listing of aggregate and aggregate associated proteins in oocyt nuclei.
https://doi.org/10.7554/eLife.35224.003| Process | Gene name | Function (summarized from www.genecards.org) |
|---|---|---|
| DNA Replication | ||
| TOP2A | Topoisomerase II Alpha, DNA topoisomerase | |
| MCM2 | Minichromosome Maintenance Complex Component 2, involved in replication initiation | |
| POLE | DNA Polymerase Epsilon, Contributes to DNA repair and replication | |
| WRN | Werner Syndrome RecQ Like Helicase, DEAH 5' to 3' DNA with 3' to 5' exonuclease activity | |
| PRIM2 | Primase Subunit 2, DNA directed RNA polymerase that generates lagging | |
| RNA Processing | ||
| GTF2H2 | General Transcription Factor IIH Subunit 2, member of RNA pol II transcription initiation factor IIH complex | |
| SRRM1 | Serine and Arginine Repetitive Matrix 1, Part of pre- and post-splicing multiprotein mRNP complexes | |
| XAB2 | XPA Binding Protein 2, participates in mRNA splicing and nucleotide excision repair | |
| NOLC1 | Nucleolar and Coiled-Body Phosphoprotein, heavily modified regulator of RNA pol I | |
| SRSF5 | Serine Arginine Rich Splicing Factor 5, spliceosome component with an RRM and RS domains | |
| Transcription, DNA-templated | ||
| POLR1C | RNA Pol I subunit C, component of RNA Pol I and III complexes | |
| HSPA8 | Heat Shock Protein Family A Member 8, constitutively expressed member of HSP70 family | |
| NCL | Nucleolin, binds histone H1 to decondense DNA | |
| POLR1A | RNA pol I subunit A, catalytic and largest subunit or RNA Pol I complex | |
| POLR2E | RNA Pol II subunit E, subunit shared between RNA Pol I, II, and III | |
| Ribonucleoprotein Complex Biogenesis | ||
| PES1 | pescadillo, member of PeBoW complex and contributes to ribosome biogenesis, binds BRCA1 | |
| BOP1 | Block of Proliferation 1, participates in rRNA processing and is a member of PeBoW complex | |
| NPM1 | Nucleophosmin, multi-functional nucleolar phosphoprotein | |
| DDX21 | DEAD-Box Helicase 21, facilitates processing or RNA Pol I and II transcripts | |
| FBL | Fibrillarin, rRNA and protein methyltransferase | |
-
Table 1—source data 1
Retained proteins following depletion of soluble nuclear components.
- https://doi.org/10.7554/eLife.35224.004
-
Table 1—source data 2
Key with brief descriptions of the data sets.
- https://doi.org/10.7554/eLife.35224.005
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.35224.015





