Engineering ER-stress dependent non-conventional mRNA splicing
Figures
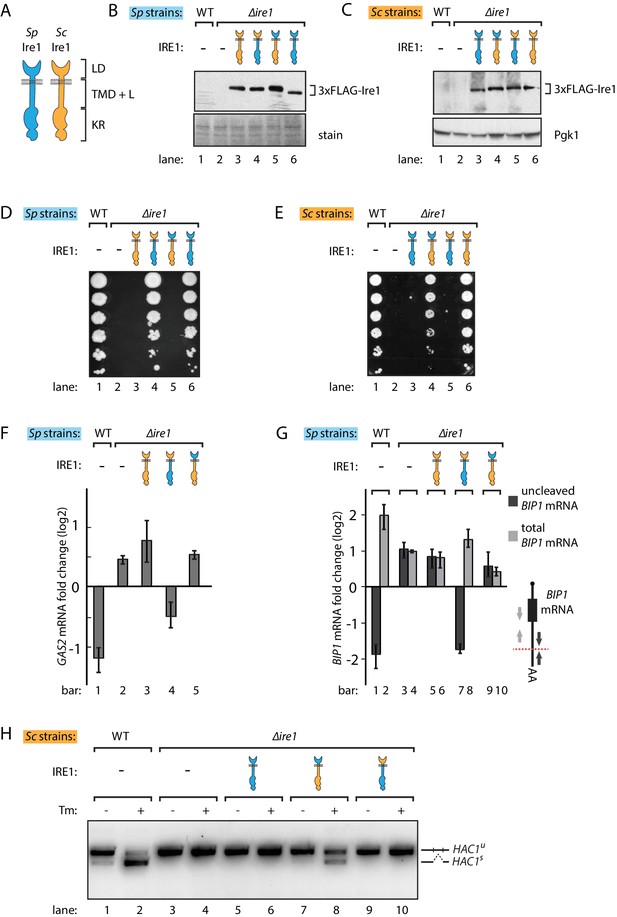
S. pombe and S. cerevisiae Ire1 have functionally conserved stress sensing ER-lumenal domains and divergent cytosolic domains.
(A) Cartoon illustration of lumenal domain (LD), transmembrane/cytosolic linker domain (TMD + L) and kinase/RNase domain (KR) for S. pombe (Sp) (blue) and S. cerevisiae (Sc) Ire1 (orange). (B, C) Expression levels of S. cerevisiae Ire1 (128 kD), S. cerevisiae lumenal S. pombe cytosolic Ire1 (126 kD), S. pombe lumenal S. cerevisiae cytosolic Ire1 (125 kD) and S. pombe Ire1 (122 kD) in S. pombe (B) and S. cerevisiae cells (C). Extracts were immunoblotted for 3xFLAG-Ire1. Ponceau stain (B) or Pgk1 (C) was used as loading control. (D, E) Cell growth assay on tunicamycin (Tm) plates. Serial dilutions of S. pombe (D) or S. cerevisiae (E) cells, which expressed the indicated Ire1 constructs, were spotted onto plates containing 0.05 μg/ml (D) or 0.1 μg/ml (E) of Tm. Plates were photographed after incubation at 30°C for 4 days. (F, G) qPCR assay for S. pombe GAS2 (F) or BIP1 (G) mRNA fold change upon 1 μg/ml Tm treatment for 1 hr. Experiments were done in triplicates. In (G), uncleaved (dark grey) or total (light grey) BIP1 mRNA was detected using the corresponding PCR primers illustrated as arrows in the schematic insert. The red dashed line indicates the Ire1 cleavage position on BIP1 mRNA. (H) Detection of S. cerevisiae HAC1 mRNA splicing by RT-PCR across the splice junction. Cells were treated with or without 1 μg/ml of Tm for 1 hr.
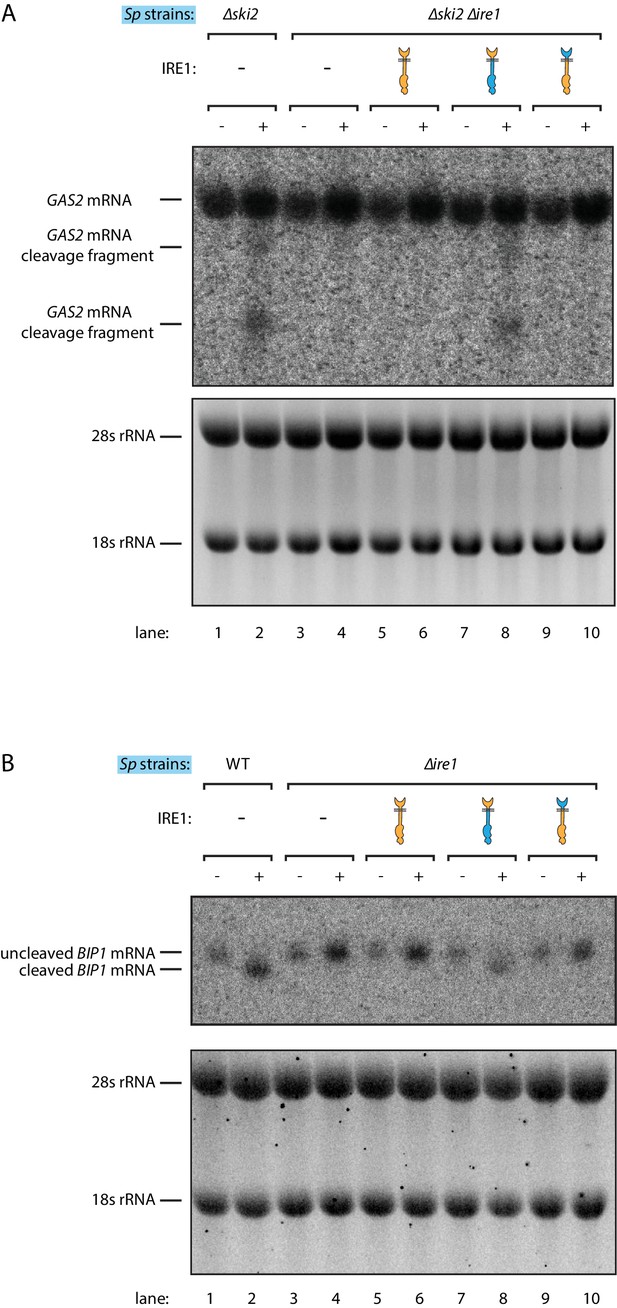
Ire1 chimeras with S. pombe cytosolic domain cleave BIP1 and GAS2 mRNA in S. pombe.
Northern blots of S. pombe GAS2 (A) and BIP1 (B) mRNA. Cells were treated with 1 μg/ml of Tm for 1 hr.
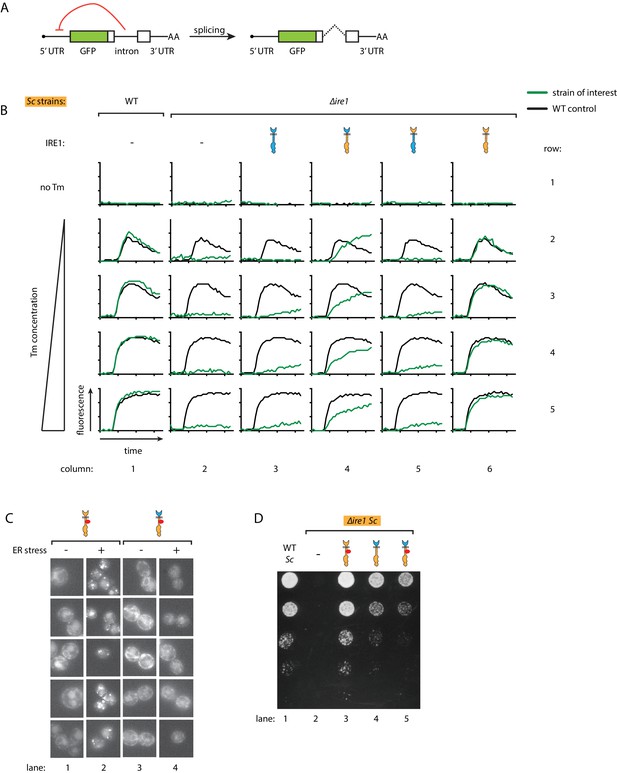
Ire1 oligomeric state determines the HAC1 mRNA splicing dynamics in S. cerevisiae cells.
(A) Illustration of the HAC1 mRNA derived splicing reporter. The splicing reporter contains a part of the HAC1 mRNA 5’ exon replaced by the green fluorescent protein (GFP) coding sequence. In the unspliced reporter mRNA, translation is inhibited by a translation block formed by the intron and 5'UTR (indicated by the red arrow). Upon splicing, intron is removed and translation begins. (B) Measuring the HAC1 mRNA splicing dynamics in S. cerevisiae using automated flow cytometry. After 1.5 hr of incubation, either no Tm or 0.25 μg/ml, 0.5 μg/ml, 1 μg/ml, 2 μg/ml of Tm was added. Then, we monitored the splicing dynamics for 10 hr. The splicing dynamics under various conditions is plotted. Green lines represent the strains of interest, which expressed indicated Ire1 variants, and the black line represents WT control strain under the same condition. (C) Examining Ire1 foci formation in S. cerevisiae cells via fluorescence microscopy with or without 1 μg/ml Tm treatment for 20 min. (D) Growth assay on Tm plate for S. cerevisiae cells expressing Ire1 constructs with or without mCherry inserted into the cytosolic linker. The inserted mCherry does not affect Ire1's ability to alleviate ER stress.
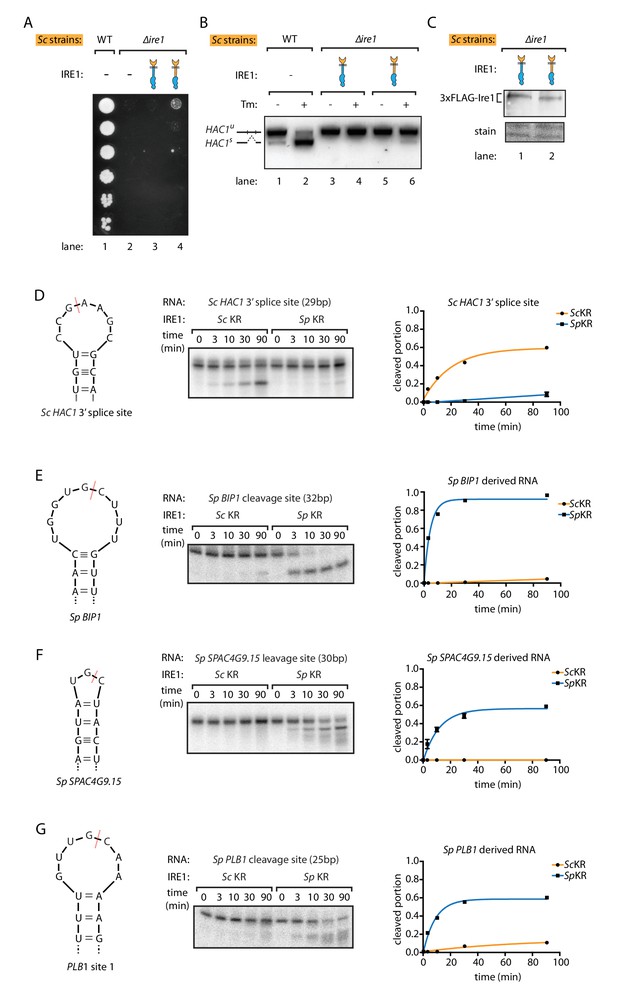
S. pombe and S. cerevisiae Ire1 have distinct RNase specificity.
(A) Growth assay for S. cerevisiae cells expressing indicated Ire1 constructs on Tm plates, as Figure 1E. (B) Measuring HAC1 mRNA splicing, as Figure 1H. (C) Comparing the expression levels of the indicated 3xFLAG-tagged Ire1 chimeras using immunoblotting. Ponceau stain was used as loading control. (D, E, F, G) In vitro RNA cleavage assays. 5'-radiolabeled hairpin RNA substrates were incubated with 12.5 μM S. cerevisiae or S. pombe Ire1 kinase/RNase domains (KR) at 30°C for the indicated time. (D) Hairpin RNA substrate derived from the 3' splice site of S. cerevisiae HAC1 mRNA. The calculated kobs is 9.4 ± 0.9 × 10−4 s−1 for S. cerevisiae Ire1 KR and 0.15 ± 0.01 × 10−4 s−1 for S. pombe Ire1 KR. (E) Hairpin RNA substrate derived from the Ire1 cleavage site on S. pombe BIP1 mRNA. The calculated kobs is 0.079 ± 0.0006 × 10−4 s−1 for S. cerevisiae Ire1 KR and 37.3 ± 4.4 × 10−4 s−1 for S. pombe Ire1 KR. (F) Hairpin RNA substrate derived from the Ire1 cleavage site on S. pombe SPAC4G9.15 mRNA, encoding a gene of unknown function. The calculated kobs was below our detection limit for S. cerevisiae Ire1 KR and 15.6 ± 2.2 × 10−4 s−1 for S. pombe Ire1 KR. (G) Hairpin RNA substrate derived from the Ire1 cleavage site on S. pombe PLB1 mRNA. The calculated kobs is 0.2 ± 0.003 × 10−4 s−1 for S. cerevisiae Ire1 KR and 19.0 ± 2.5 × 10−4 s−1 for S. pombe Ire1 KR.
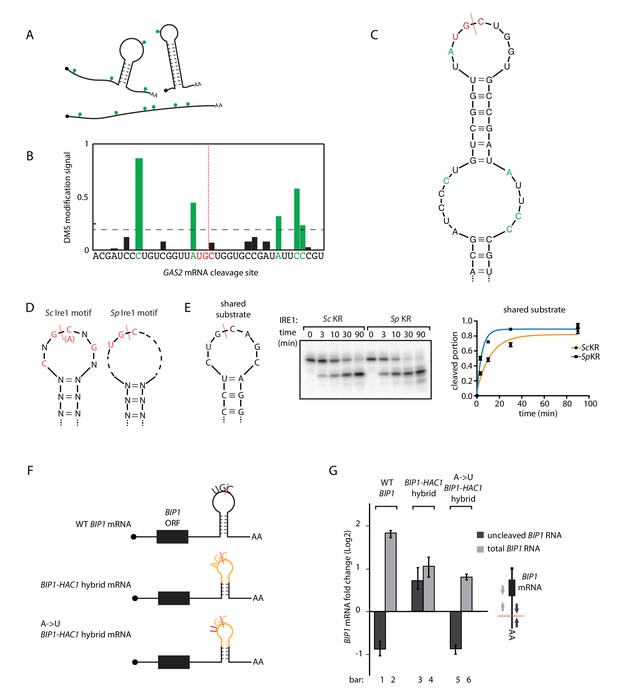
S.pombe and S. cerevisiae Ire1 recognize distinct RNA sequence and structural features.
(A) Illustration of RNA structural mapping by DMS modifications. Dimethyl sulfate (DMS) allows detection of unpaired adenine and cytosine RNA bases (green dots). (B) The normalized DMS modification signals near the Ire1 cleavage site on S. pombe GAS2 mRNA (cleavage site is indicated by the red dashed line). The positions with high DMS modification signals are labeled in green and the previously identified S. pombe Ire1 UG|C motif is labeled in red. (C) In sillico RNA secondary structure prediction of the Ire1 cleavage site on GAS2 mRNA. Structure prediction was constrained by forcing the positions with high DMS modification signals (green) to be unpaired. (D) RNA sequence and structural motifs recognized by the S. cerevisiae and S. pombe Ire1. (E) In vitro cleavage assay using an RNA hairpin derived from human XBP1 mRNA 3' splice site, which is predicted to be a shared substrate for S. cerevisiae and S. pombe Ire1 KR. The calculated kobs is 16.7 ± 2.3 × 10−4 s−1 for S. cerevisiae Ire1 KR and 38.9 ± 4.0 × 10−4 s−1 for S. pombe Ire1 KR. (F) Illustrations of the S. pombe BIP1 mRNA variants and (G) their uncleaved (dark grey) or total (light grey) mRNA fold change upon ER stress in S. pombe cells. Experiments were done in triplicates.
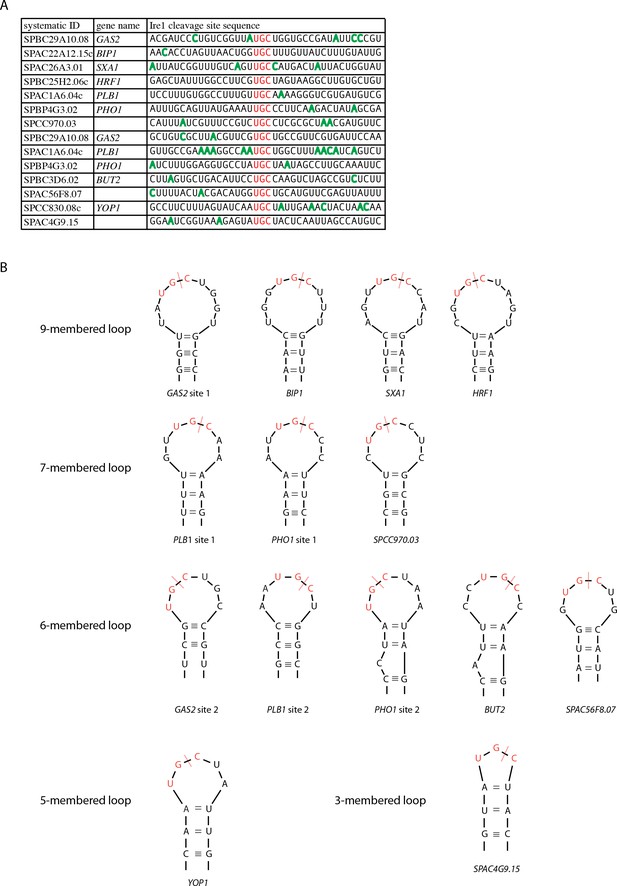
S. pombe Ire1 cleaves at UG|C positioned near the center of loops in RNA stem-loop structures.
(A) A list of all 14 S. pombe Ire1 mRNA cleavage sites, which were independently identified by both Kimmig, Diaz et al. and Guydosh et al. (Guydosh et al., 2017; Kimmig et al., 2012). The UG|C motifs are labeled in red. The positions with high DMS modification signals are labeled in green. (B) Predicted RNA secondary structures of S. pombe Ire1 cleavage sites. DMS modification signals were used to guide the secondary structure prediction of S. pombe Ire1 mRNA cleavage sites. The red dashed lines indicate the Ire1 cleavage sites.
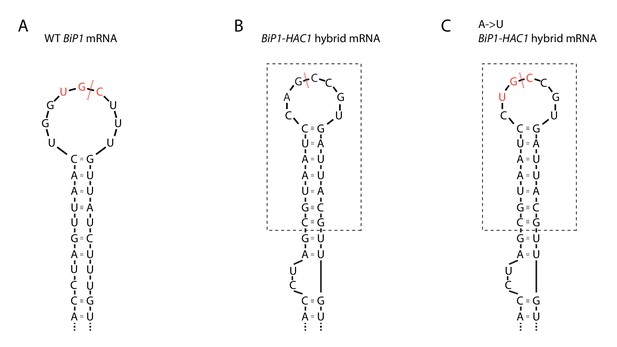
Ire1 cleavage sites on BIP1 mRNA variants.
Sequence and predicted RNA secondary structures of Ire1 cleavage sites on (A) S. pombe BIP1 mRNA, (B) BIP1-HAC1 hybrid mRNA and (C) BIP1-HAC1 hybrid mRNA with an A to U mutation. The part included in the dashed box is derived from S. cerevisiae HAC1 mRNA 5' splice site.
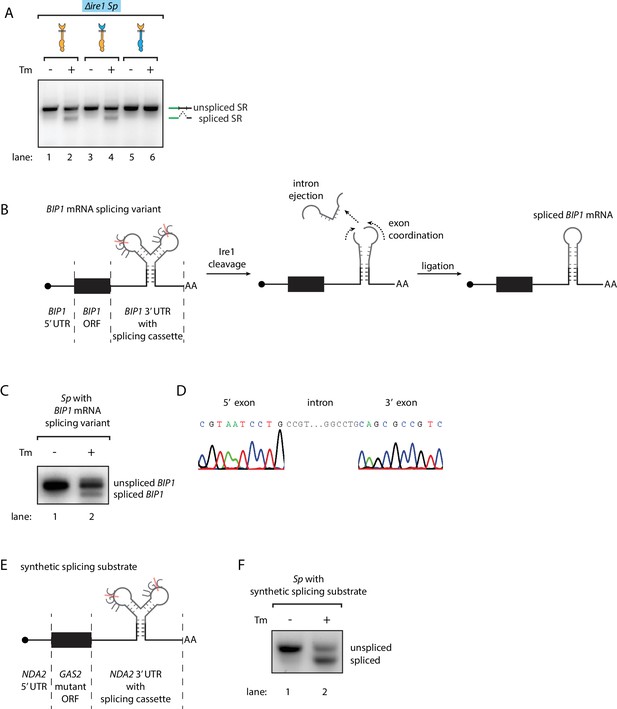
Engineering the Ire1-mediated non-conventional mRNA splicing in S. pombe cells.
(A) Measuring the non-conventional mRNA splicing in S. pombe cells, which were transformed with the S. cerevisiae HAC1 mRNA splicing reporter (SR) and the indicated Ire1 constructs. Cells were treated with 1 μg/ml Tm for 1 hr. (B) Illustration of the engineered S. pombe BIP1 mRNA splicing variant. (C) Measuring the non-conventional mRNA splicing of the engineered S. pombe BIP1 mRNA splicing variant. Experimental conditions are the same as those for Figure 4A. (D) Sequencing reads of the spliced BIP1 mRNA. The schematic illustration (E) and the splicing assay (F) of the synthetic splicing substrate in S. pombe. Cells were treated with 1 μg/ml Tm for 1 hr.
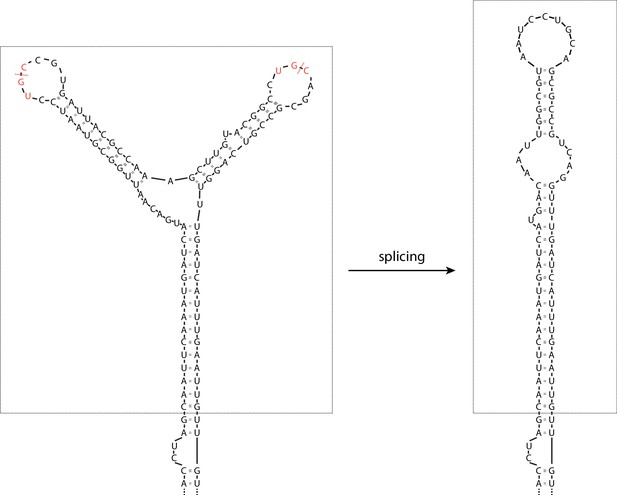
The splicing cassette in the engineered S. pombe BIP1 mRNA splicing variant.
The part included in the dashed box is the inserted synthetic splicing cassette. The red dashed lines indicate the Ire1 cleavage sites. The S. pombe Ire1 UG|C motifs are labeled in red.
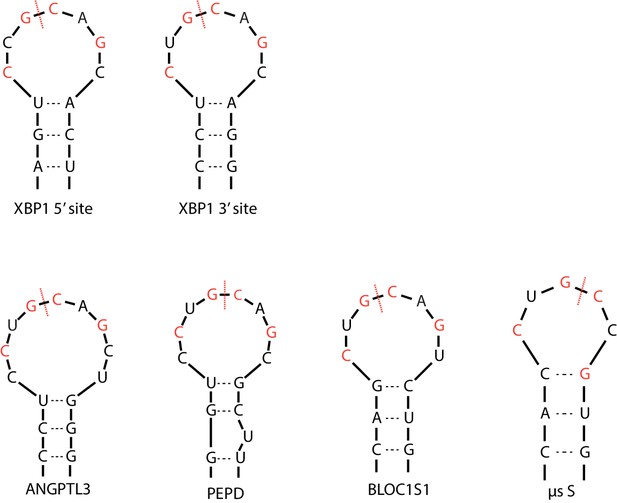
The Ire1α cleavage sites on XBP1 mRNA and RIDD targets.
Red dashed lines mark the cleavage sites and the red letters indicate the previously identified sequence motif.
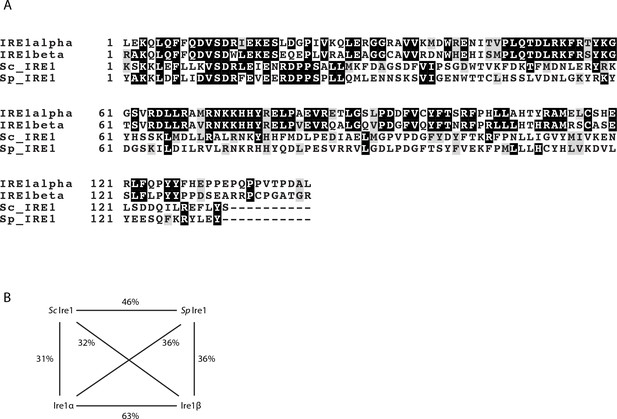
The sequence alignment of the kinase/RNase domains of Ire1α, Ire1β, the S. cerevisiae Ire1 and the S. pombe Ire1.
(A) The sequence alignment and colored with BoxShade Server. (B) The sequence identities between the indicated pairs of Ire1 constructs.
Tables
Yeast strains used in this study.
All strains are derived from WL001 and WL002. All Ire1 constructs listed below are 3x FLAG-tagged within their lumenal domains.
| Strain | Species | Description |
|---|---|---|
| yWL001 | Sc | WT, mat A, leu2-3,112 TRP1 can1-100 ura3-1 ADE2 his3-11,15 |
| yWL002 | Sc | ire1∆::NATR |
| yWL003 | Sc | ire1∆::NATR, HO::Sp IRE1 |
| yWL004 | Sc | ire1∆::NATR, HO::SpLumScCyto IRE1 |
| yWL005 | Sc | ire1∆::NATR, HO::ScLumSpCyto IRE1 |
| yWL006 | Sc | ire1∆::NATR, HO::Sc IRE1 |
| yWL007 | Sc | ire1∆::NATR, HO::ScLum/transmembrane/linkerSpKR IRE1 |
| yWL008 | Sc | WT, leu2::5'hac1-gfp-3'hac1 |
| yWL009 | Sc | ire1∆::NATR, leu2::5'hac1-gfp-3'hac1 |
| yWL010 | Sc | ire1∆::NATR, leu2::5'hac1-gfp-3'hac1, HO::Sp IRE1 |
| yWL011 | Sc | ire1∆::NATR, leu2::5'hac1-gfp-3'hac1, HO:: SpLumScCyto IRE1 |
| yWL012 | Sc | ire1∆::NATR, leu2::5'hac1-gfp-3'hac1, HO:: ScLumSpCyto IRE1 |
| yWL013 | Sc | ire1∆::NATR, leu2::5'hac1-gfp-3'hac1, HO::Sc IRE1 |
| yWL014 | Sc | leu2::5'hac1-gfp-3'hac1, his3::pTdh3-mCherry |
| yWL015 | Sp | WT, mat h+, ade6-M210, ura4-D18, leu1-32 |
| yWL016 | Sp | ire1∆::KANR |
| yWL017 | Sp | ire1∆::KANR, leu1::Sp IRE1 |
| yWL018 | Sp | ire1∆::KANR, leu1::SpLumScCyto IRE1 |
| yWL019 | Sp | ire1∆::KANR, leu1::ScLumSpCyto IRE1 |
| yWL020 | Sp | ire1∆::KANR, leu1::Sc IRE1 |
| yWL021 | Sp | ire1∆::KANR, leu1::SpLumScCyto IRE1, ura4::5'hac1-gfp-3'hac1 |
| yWL022 | Sp | ire1∆::KANR, leu1::ScLumSpCyto IRE1, ura4::5'hac1-gfp-3'hac1 |
| yWL023 | Sp | ire1∆::KANR, leu1::Sc IRE1, ura4::5'hac1-gfp-3'hac1 |
| yWL024 | Sp | bip1::bip1-hac1 hybrid |
| yWL025 | Sp | bip1::bip1-hac1 hybrid A->U |
| yWL026 | Sp | bip1::bip1 splicing variant |
| yWL027 | Sp | ura4::synthetic splicing substrate |
qPCR primers used in this study.
https://doi.org/10.7554/eLife.35388.014| qPCR primers description | Sequence |
|---|---|
| uncleaved Sp BiP1 mRNA forward primer | GAATCGTGACTCTATAGCCATTAACA |
| uncleaved Sp BiP1 mRNA reverse primer | CAATTATTGTCAGTTCCACAAAGC |
| total Sp BiP1 mRNA forward primer | TGGTAAGGTTGATCCCGAAG |
| total Sp BiP1 mRNA reverse primer | CATCGAGTTTTTGACGCTGA |
| Sp GAS2 mRNA forward primer | GTTGTCAACAATGCCTCGAA |
| Sp GAS2 mRNA reverse primer | CGGTCTCAGAGTTGGTGTCA |
| Sp NDA2 mRNA forward primer | TCCATGAATCCAACAGCGTA |
| Sp NDA2 mRNA reverse primer | CTAGTAACGGCAGCCTGGAC |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.35388.015





