A stable mode of bookmarking by TBP recruits RNA polymerase II to mitotic chromosomes
Figures
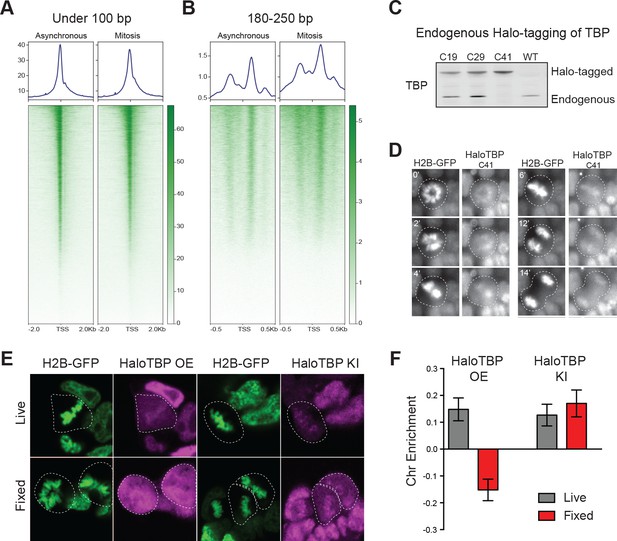
Endogenous TBP associates with mitotic chromosomes.
(A–B) Previous ATAC-seq data (Teves et al., 2016) were examined for TSS analysis. Heatmaps for all genes centered at the TSS (bottom) and aggregate plots of global average signal (top) surrounding the TSSs of all genes were generated for reads under 100 bp (A), and for mononucleosome-sized fragments (B). (C) Western blot analysis of whole cell extracts of clones derived from endogenous tagging of TBP to insert the HaloTag. (D) Time-lapse live-cell imaging of C41 HaloTBP cells stably expressing H2B-GFP as cells undergo mitosis. (E) Live imaging versus fixed immunofluorescence for cells over-expressing Halo-TBP or the endogenous C41 Halo-TBP knock-in. (F) Chromosome enrichment levels for either the over-expressing Halo-TBP cells under live or fixed conditions, or the C41 Halo-TBP knock-in cells under live or fixed conditions. n = 40 cells. Data are represented as mean ± SEM.
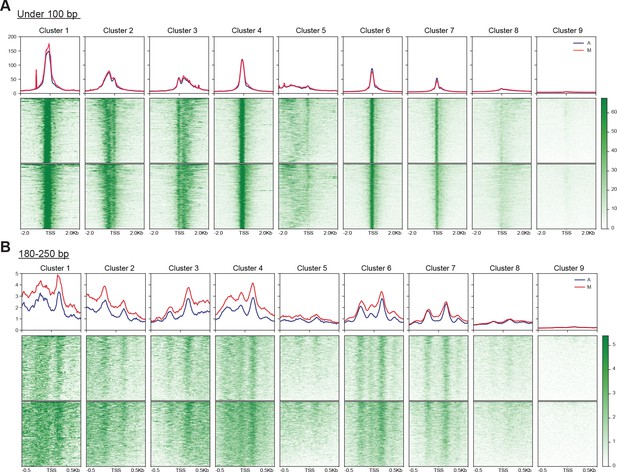
Accessibility at the TSS is maintained in mitosis.
We performed unbiased k-means clustering of ATAC-seq data for reads under 100 bp (A) and 180–250 bp (B) with k = 9. The heatmaps for each cluster are shown on the bottom, and the average signal surrounding the TSS for each cluster is plotted on top.
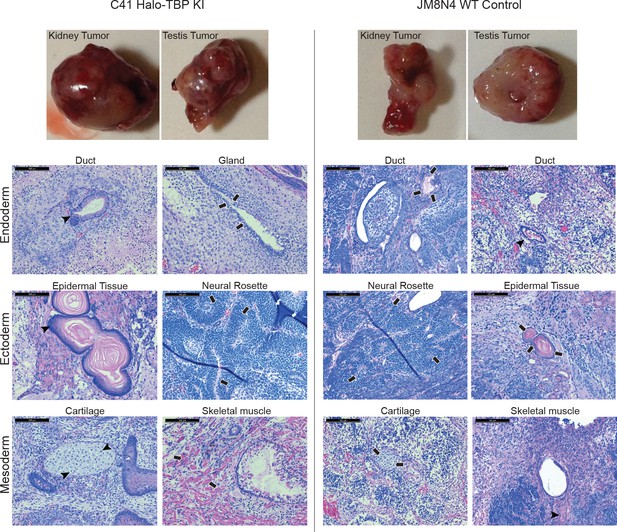
Halo-tagged TBP mouse ES cell line remains pluripotent.
The C94 Halo-TBP mES cells (left) or WT JM8.N4 mES cells (right) were injected into SCID-Beige mice. Thirty days post inoculation, kindey and testis tumors were harvested and a sample tumor is shown (top). Histological analyses of tumors show small areas of differentiated cells, representing endoderm, mesoderm, and ectoderm tissues (bottom).
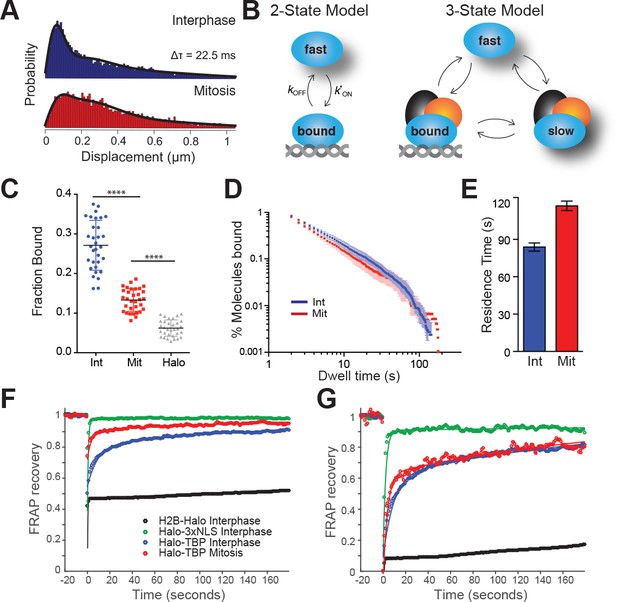
TBP dynamics within live cells.
(A) Cells were labeled with 25 nM pa-JF549 and individual molecules were tracked over 25,000 frames. Jump length histogram measured in displacements (μm) after three consecutive frames (Δτ = 22.5 ms) during spaSPT for cells in interphase (blue) or in mitosis (red). (B) Depiction of different states for the 2-state or the 3-state kinetic model. For the 2-state model, the molecules switch from fast diffusing mode to bound states. In the 3-state model, the freely diffusing molecules can be divided into two categories, fast and slow, which can switch to a DNA-bound state. (C) Scatter plot of fraction bound from individual cell model fits from interphase (Int) and mitotic cells (Mit). mESCs stably expressing HaloTag only (Halo) were imaged and the fraction bound for each individual cell was also extracted from model fitting to show non-specific binding population. n = 32 cells over four biological replicates. ****p-value<0.0001 (D) Dwell time histogram of the fraction of endogenously-tagged Halo-TBP molecules remaining bound for interphase (blue) and mitotic (red) cells. (E) Quantification of the residence time of Halo-TBP in interphase (blue) and mitotic (red) cells. n = 30 cells. (F) Quantification of fluorescence recovery at the bleach spot for the indicated Halo-tagged construct in interphase and mitosis. n = 30 cells. (G) Data from F, normalized for bleach depth. Data are represented as mean ± SEM.
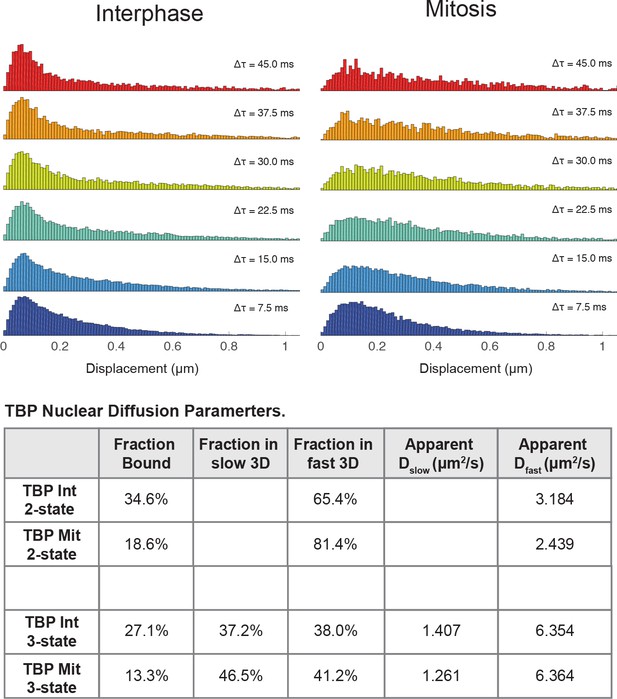
Halo-TBP shows increased movement in mitosis relative to interphase.
The jump lengths distribution is displayed for consecutive time frames, from Δτ = 7.5 ms to Δτ = 45.0 ms for interphase (left) and mitotic (right) cells. A summary table showing the observed fits for each parameter in a 2-state vs 3-state kinetic model is also presented.
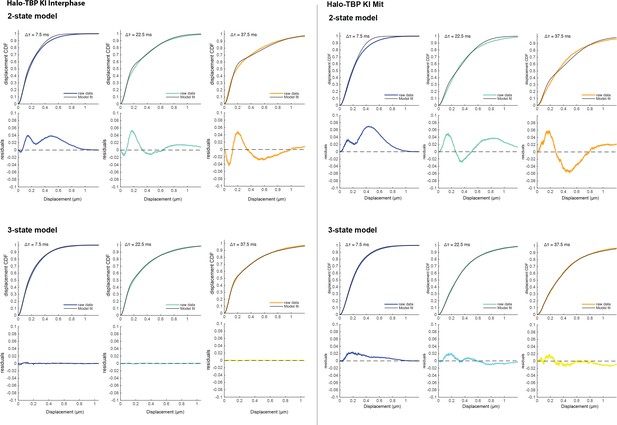
Model fitting of Halo-TBP fast tracking data for interphase and mitotic cells.
The displacement CDFs for consecutive time frames, from Δτ = 7.5 ms to Δτ = 45.0 ms, are plotted for Halo-TBP in interphase (left) and mitotic cells (right) as colored lines. The displacement CDF is then fitted with a 2-state (top) or a 3-state (bottom) model. The resulting fit is plotted as a black line, and the corresponding residuals from the fit is plotted below for each consecutive frame. The 2-state model resulted in large residuals after fitting, necessitating a 3-state model.
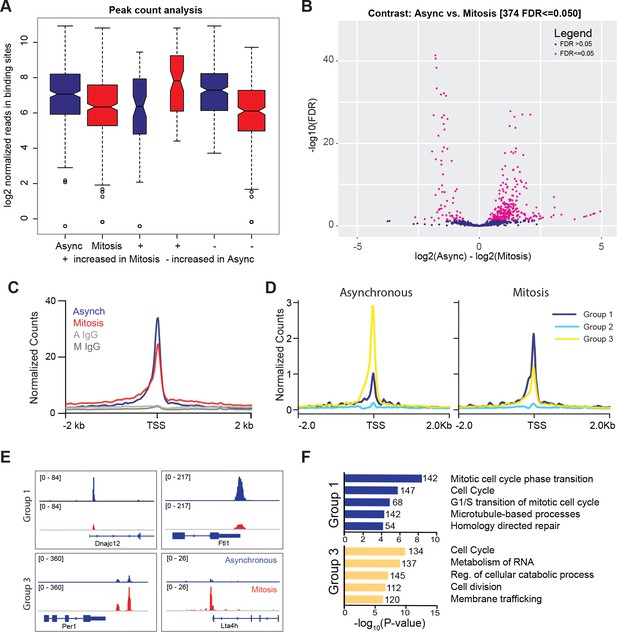
TBP maintains binding to promoters of active genes during mitosis.
(A) Box plot of read distributions for all identified peaks. Asynchronous and mitotic samples are shown in blue and red, respectively. The first two boxes represent all peaks, the middle two are peaks that have higher levels in mitosis, and the last two boxes are peaks that have higher reads in asynchronous samples. (B) Volcano plot of all peaks in asynchronous and mitotic samples. Peaks identified as differentially bound are shown in pink (n = 374). (C) Average ChIP-seq read density for all TSS and surrounding regions for asynchronous (blue) and mitotic (red) samples and corresponding IgG controls (grays). (D) Unbiased k-means clustering of data from B with k = 6. Clusters are grouped into three groups depending on changes in signal. (E) Genome browser snapshots of genes in Group one and genes in Group 3. TBP ChIP-seq from asynchronous and mitotic samples are shown in blue and red, respectively. (F) Gene Ontology term analysis of genes in Group one and Group three from (D). Numbers correspond to the number of genes within the group that is labeled with the specific GO term.
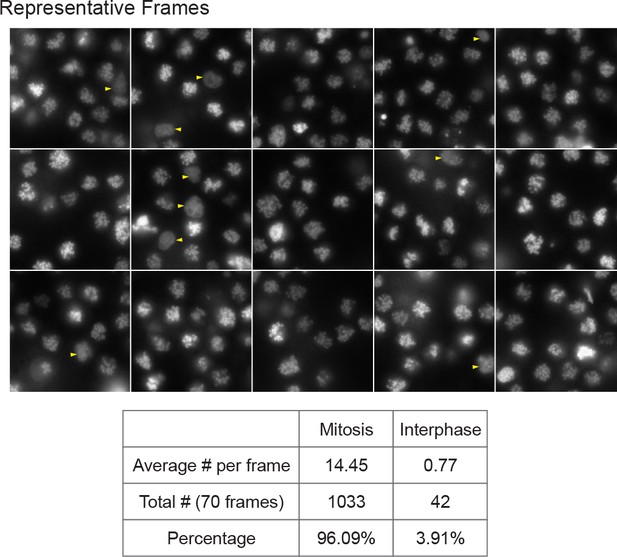
Synchronization for mitotic cells.
Cultured mESCs were treated with 100 ng/mL of Nocodazole for 6 hr, and mitotic cells were collected by shake off. The collected cells were imaged for H2B-GFP in 70 frames, with 15 representative frames shown. Each cell was classified as mitotic or interphase. Interphase cells are marked with yellow arrowheads. The average cell number per frame, the total cell count in 70 frames, and percentage for mitotic and interphase cells are shown in the table.
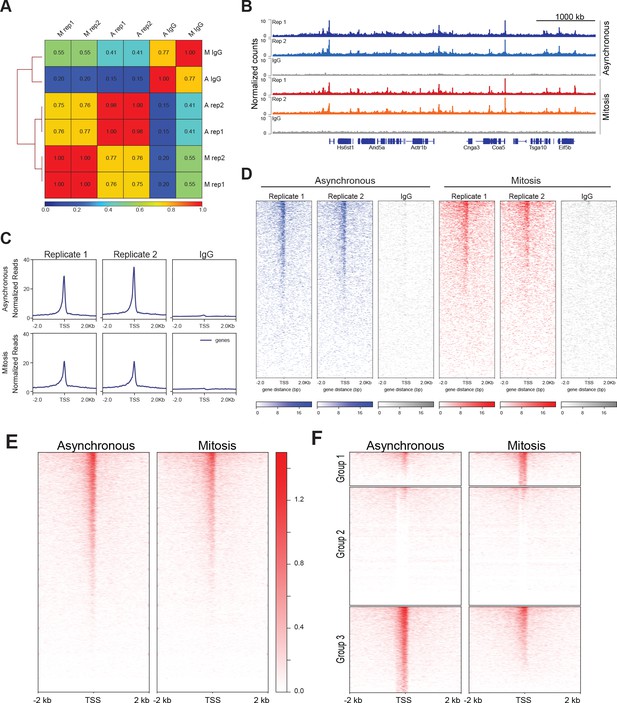
TBP ChIP-seq is highly reproducible.
(A) The Pearson correlation was calculated for reach replicate in Asynchronous (A) and Mitotic (M) samples as well as the corresponding IgG control, and the correlation values are plotted as a correlation heatmap. (B) Genome browser profiles for TBP ChIP-seq replicates in asynchronous and mitotic cells and the corresponding IgG ChIP control. (C) Average of all ChIP-seq reads centered at the TSS for each replicate in Asynchronous and Mitosis samples and the corresponding IgG controls. (D) ChIP-seq reads for each gene are plotted as heatmaps with regions centered at TSS for each replicate in Asynchronous and Mitosis samples and the corresponding IgG controls. (E) Heatmaps of the replicate-combined ChIP-seq signal surrounding the TSS of all genes, arranged by decreasing gene expression for asynchronous and mitotic samples. (F) Heatmap of E after k-means clustering with k = 3.
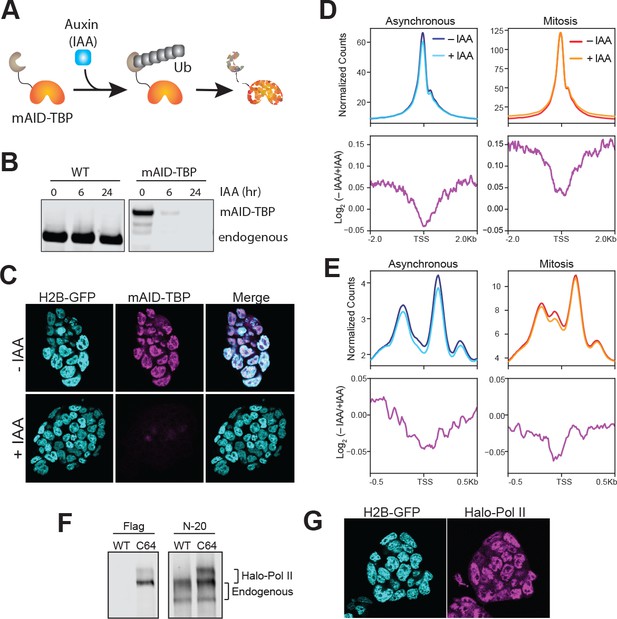
Drug-inducible degradation of endogenous TBP.
(A) Schematic for mechanism of auxin-inducible degradation. Ub, ubiquitin. (B) Western blot analysis for time-course of Auxin (IAA)-dependent degradation of WT cells or cells with the endogenous knock-in of the mAID to the TBP locus. (C) Immunofluorescence using α-TBP of cells with endogenous mAID-TBP and stably expressing H2B-GFP without IAA (top) or after 6 hr of IAA treatment (bottom). (D) ATAC-seq analysis of reads under 100 bp. Global average signal (top) surrounding the TSSs of all genes were generated for asynchronous samples (left) that were untreated (– IAA; dark blue) and TBP-degraded (+IAA; light blue), and for mitotic samples (right) that were untreated (– IAA; red) and TBP-degraded (+IAA; orange). Bottom, the corresponding log2 ratio of (– IAA/+IAA) plotted in 4 kb surrounding the TSS. (E) ATAC-seq analysis of 180–250 bp reads (mono-nucleosome-sized). The scheme is the same as in (D), with the exception that the plots are centered 1 kb surrounding the TSS. (F) Western blot analysis of wild-type (WT) ES cells or Halo-Pol II knock-in (C64) using α-Flag to detect the Halo-Flag knock-in and α-N-20, an antibody against the N-terminal of Pol II large subunit, to detect all Pol II levels. The unphosphorylated and phosphorylated bands for endogenous and Halo-Pol II are marked. (G) Live imaging of Halo-Pol II showing nuclear localization as marked with H2B-GFP.
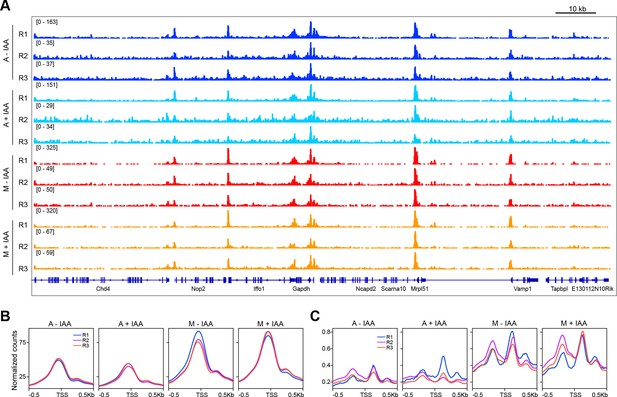
ATAC-seq replicates.
(A) Genome browser snapshot of a genomic region in Chr 6, showing each replicate for each sample. A, asynchronous. M, mitosis. R1, replicate 1. R2, replicate 2. R3, replicate 3. (B–C) Average plots for reads under 100 bp (B) and for 180–250 bp reads (C) in a 1 kb region centered at the TSS for each replicate of each sample.
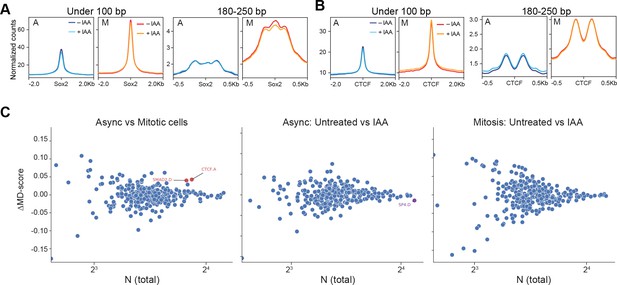
ATAC-seq at enhancer and CTCF sites.
(A) After combining all replicates, average signal around Sox2 sites were plotted for reads under 100 bp and 180–250 bp. (B) Same as in A, but for CTCF sites. (C) MA-plots. For all transcription factor motif models in mm10 (each dot), the change in MD-score in Async vs Mitosis (left), untreated vs IAA in Async cells (middle), and untreated vs IAA in Mitotic cells (right), are plotted relative to the number of motifs within 1.5 kb of any ATAC-seq peak center (x-axis). MD-scores are defined as the enrichment of a TF sequence motif within a small radios (150 bp) of ATAC-seq peaks relative to a larger local window (1500 bp) (Tripodi et al., 2018). Significantly different MD-scores are highlighted in red and purple (p-value<1×10−6). These analyses show that the majority of TF motifs show no change in enrichment in each condition tested. In Asynchronous vs Mitotic cells, only two factors show higher enrichment in Asynchronous vs Mitosis: CTCF and SMAD2.D. In Asynchronous cells, only SP4.D shows higher enrichment in TBP degraded vs untreated cells.
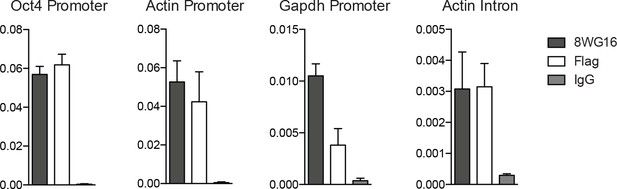
Halo-Pol II binds to promoters of active genes.
We performed ChIP using α-Pol II (8WG16 antibody), α-Flag (Flag is fused to Halo-Rpb1), and IgG control. We measured ChIP-levels at the Oct4, Actin beta, and Gapdh promoters as well as at an Actin beta intron. α-Flag (Halo-Rpb1) binds to similar regions as α-Pol II.
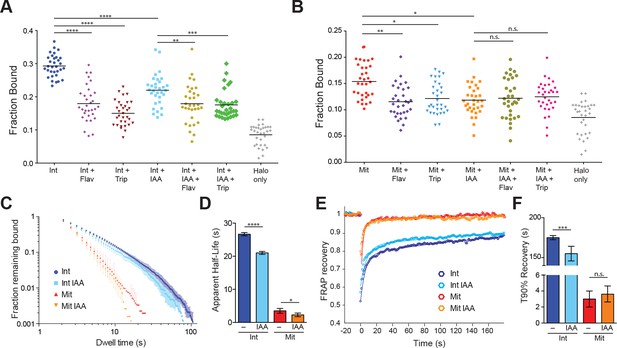
TBP-dependent dynamics of Pol II in live cells.
(A) Cells were labeled with PA-JF646 and individual molecules were tracked over 25,000 frames. Scatter plot showing the fraction bound (extracted from model fitting, see Materials and methods), for each individual interphase (Int) cell, and interphase cells treated with various drugs. Flav, Flavopiridol. Trip, Triptolide, IAA, Indole Acetic Acid (TBP-degradation). Halo only corresponds to cells stably expressing the HaloTag, showing non-specific levels of ‘bound’ molecules. n = 32 cells over four biological replicates. Black line represents mean fraction bound. (B) Scatter plot as in (A), but for mitotic cells. Halo-only sample in (A) is replotted for direct comparison. n = 32 over four biological replicates. (C) Dwell time histogram of the fraction of endogenously-tagged Halo-Pol II molecules remaining bound for interphase (blues) and mitotic (reds) cells either with or without IAA treatment, as indicated in legend. (D) Quantification of the apparent half-life (see Materials and methods) in seconds of Halo-Pol II in interphase (blues) and mitotic (reds) cells. n = 30 cells. (E) Quantification of fluorescence recovery at the bleach spot for Halo-Pol II in interphase (blues) and mitosis (reds), either with or without IAA treatment. n = 30 cells. (F) From (E), the average time to reach 90% recovery for Halo-Pol II in interphase (blues) or mitosis (reds), either with or without IAA treatment. Data are represented as mean ± SEM. *p-value<0.05, **p-value<0.01. ***p-value<0.001, ****p-value<0.00001, n.s., not significant.
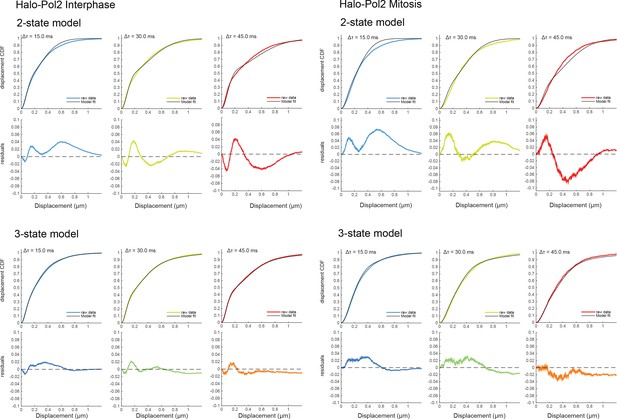
Model-fitting for Halo-Pol II in interphase (left) and mitotic (right) cells.
The displacement CDFs for time frames, from Δτ = 15, 30, and 45 ms, are plotted for Halo-Pol II as colored lines. The displacement CDF is then fitted with a 2-state (top) or a 3-state (bottom) model. The resulting fit is plotted as a black line, and the corresponding residuals from the fit is plotted below for each consecutive frame. The 2-state model resulted in large residuals after fitting, necessitating a 3-state model.
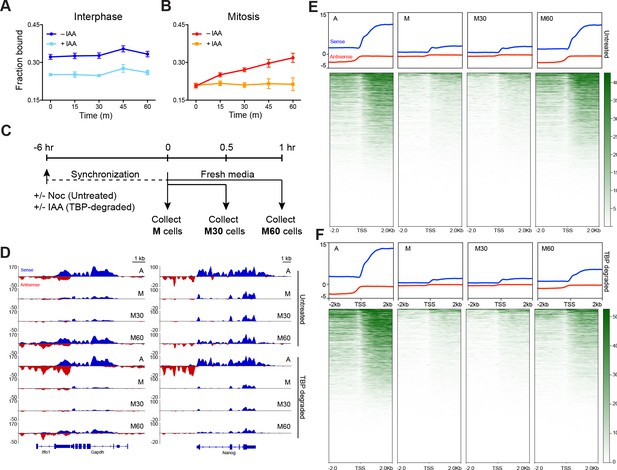
TBP recruits Pol II during mitosis.
(A) The fraction of bound Halo-Pol II molecules as a function of time (t = 0, release from DMSO/IAA treatment) under no IAA (WT conditions) or after 6 hr of IAA treatment (teal) for interphase cells. (B).Same as (A), but for mitotic cells. (C) Schematic of time course regimen for extracting chromatin-associated nascent RNA. Asynchronous cells are treated with DMSO or IAA for 6 hr prior to chromatin-associated nascent RNA extraction (not depicted). For other samples, cells are synchronized with Nocodazole for 6 hr, and treated with DMSO (untreated) or IAA (TBP-degraded) during Nocodazole synchronization. For mitotic samples (M), cells are immediately collected. For time course after mitosis, synchronized M cells (untreated or TBP-degraded) are replaced into fresh media to release from mitotic arrest and TBP degradation and are collected either after 30 min (M30) or 60 min (M60) following fresh media resuspension. (D) Extracted chromatin-associated nascent RNA were sequenced in strand-specific manner, and the sense (blue) and anti-sense (red) reads are plotted for Gapdh (left) and Nanog (right) loci for all corresponding samples. (E) Genome-wide average plots for all TSS and surrounding regions for sense (blue) and anti-sense (red) reads for each indicated untreated sample, and the corresponding heatmaps for all reads (bottom). (F) Same as E but for TBP-degraded samples.
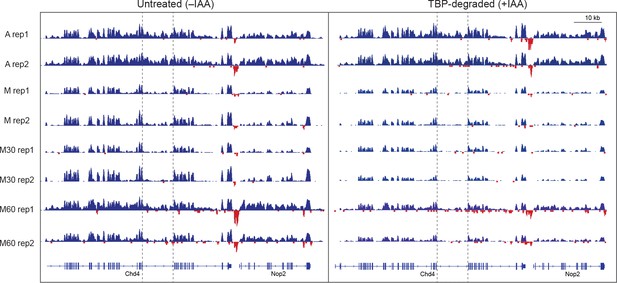
Nascent chr-RNA-seq is highly reproducible.
Genomic browser snapshots of sense (blue) and anti-sense (red) reads of each sample and the corresponding replicates for untreated cells and TBP-degraded cells. An intronic region (boxed) is highlighted to indicate levels of newly transcribed RNA.
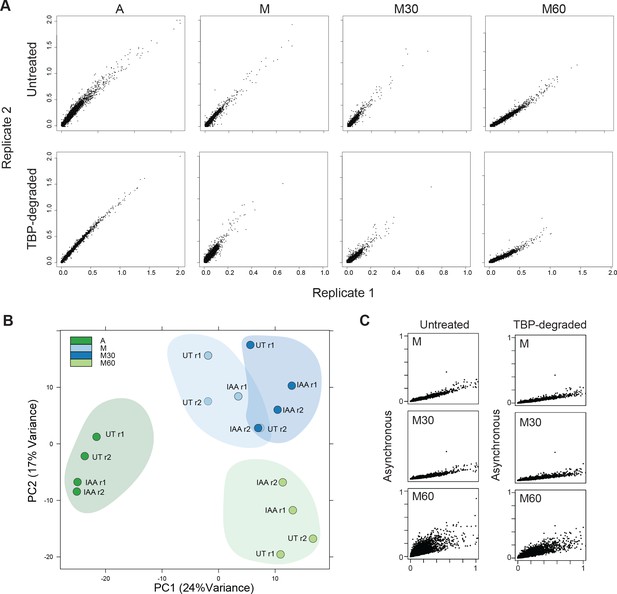
Nascent chr-RNA-seq is highly reproducible genome-wide.
(A) For each gene, we quantified the intronic transcripts per million (TPM) reads and plotted a scatter plot with replicate one on the x-axis and replicate two on the y-axis for all samples. (B) Using the intronic TPM, we performed principal component analysis (PCA). This analysis shows that the replicates from each time point cluster together. (C) After combining reads from replicates, we calculated the TPB for each gene as before and plotted a scatter plot with the Asynchronous sample on the y-axis, and the indicated sample on the x-axis. This analysis shows that the M and M30 samples show decreased TPM relative to asynchronous (A) samples. The M60 of untreated samples are getting closer to the diagonal, suggesting that gene expression has largely re-established the asynchronous profile. For TBP-degraded samples (right), the scatter plot shows levels largely under the diagonal, suggesting a global delay in gene reactivation.
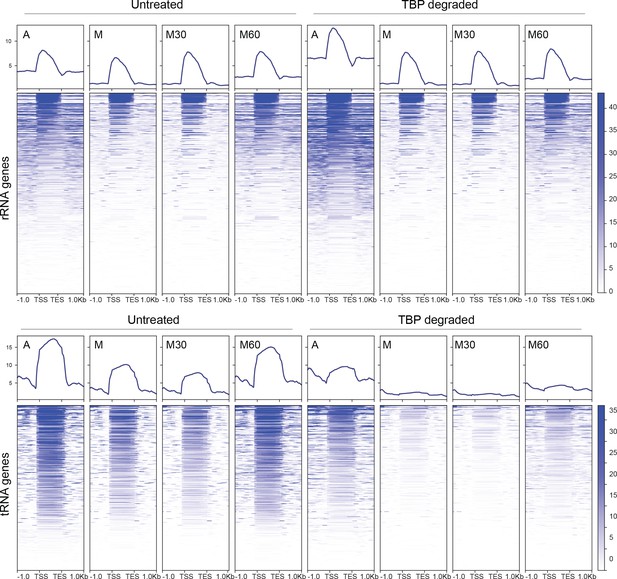
Nascent chr-RNA-seq profiles for rRNA (top) and tRNA (bottom) genes for each sample.
https://doi.org/10.7554/eLife.35621.020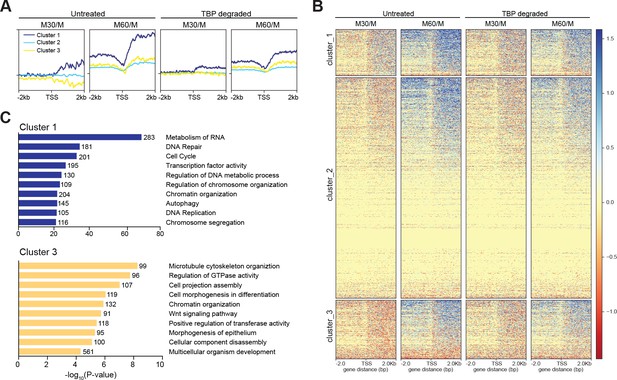
TBP promotes efficient reaction of transcription following mitosis.
(A) The log2 ratio of M30 and M60 reads relative to M reads were calculated for untreated and TBP-degraded samples (M30/M and M60/M, respectively). The M30/M samples were clustered using k-means clustering with k = 3, and the rest of the samples are ordered by this clustering, and average plots surrounding the TSS are shown for each cluster. (B) Heat map analysis of each cluster for each sample in (A). (C) GO term analysis for genes present in cluster 1 (top) and cluster 3 (bottom). Numbers correspond to the number of genes within the custer that is labeled with the specific GO term.
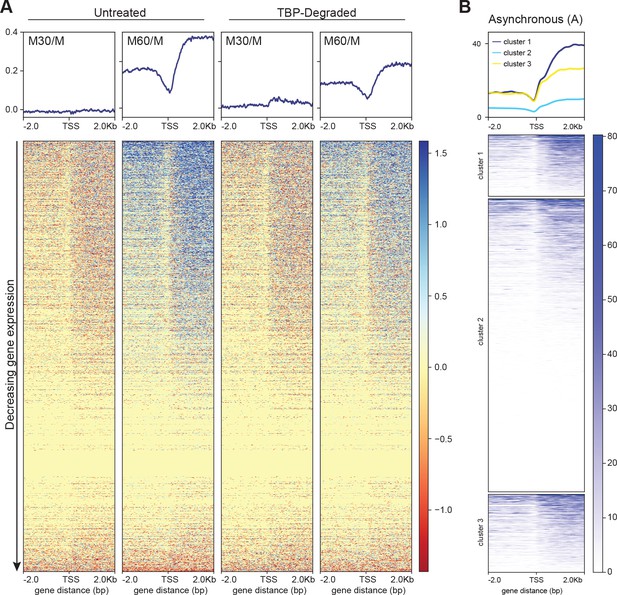
TBP degradation affects global reactivation following mitosis.
(A) The log2 ratio of M30 and M60 reads relative to M reads were calculated for untreated and TBP-degraded samples (M30/M and M60/M, respectively). Global average of the log2 ratios were obtained and plotted for all genes centered at the TSS (top), and the corresponding heatmap for all genes are shown (bottom). (B) The normalized reads for the untreated asynchronous (A) sample was clustered using the same grouping as in Figure 6F.
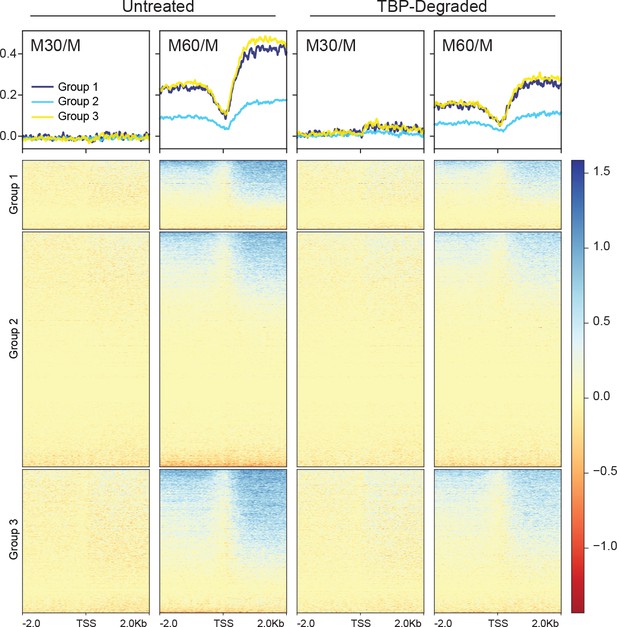
The log2 ratio of M30 and M60 reads relative to M reads in untreated and TBP-degraded samples were grouped according to k = 3 clustering as in Figure 3D.
https://doi.org/10.7554/eLife.35621.023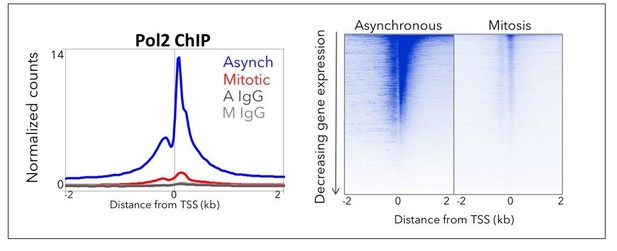
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (mouse cell line) | JM8.N4 mouse ES cells | KOMP repository | RRID: CVCL_J962 | Parental cell line used for all genetic manipulations |
| Antibody | TBP | Abcam | Abcam #ab51841; RRID:AB_945758 | 1:250 for Western; 10 μg for ChIP |
| Antibody | N-20 RNA Polymerase II | Santa Cruz | Santa Cruz #sc-899; RRID:AB_632359 | 1:250 for Western |
| Antibody | 8WG16 Pol II | Santa Cruz | RRID:AB_785522 | 1:250 for Western |
| Antibody | Flag | Sigma Aldrich | Sigma-Aldrich Cat# F3165, RRID:AB_259529 | 1:5000 for Western |
| Cell line (mouse) | Halo-TBP KI C41 | This paper | Halo-TBP KI C41 | Endogneous knock-in of HaloTag to N-ternimal of TBP in JM8.N4 cells |
| Cell line (mouse) | mAID-TBP KI C94 | This paper | mAID-TBP KI C94 | Endogenous knock-in of the minimal auxin inducible degron to N-terminal of TBP in JM8.N4 cells |
| Cell line (mouse) | Halo-Pol II C64 | This paper | Halo-Pol II C64 | Endogenous knock-in of the HaloTag to C-terminal of Rbp1, largest subunit of Pol II, in mAID-TBP C94 cells |
| Commercial assay or kit | Nextera DNA Library Preparation Kit | Illumina | FC-121–1030 | ATAC-seq reagent for Tn5 transposition |
| Software, algorithm | Spot-On | doi: 10.7554/eLife. 25776 | Spot-on software used to analyze spaSPT data |
Additional files
-
Supplementary file 1
Primers and guide RNAs for Cas9-mediated knock-ins.
Primers were ordered as custom DNA oligos from IDT in 25 nmole amounts using standard desalting methods.
- https://doi.org/10.7554/eLife.35621.024
-
Supplementary file 2
Statistics on spaSPT imaging analysis.
For all Halo-TBP and Halo-Pol II spaSPT imaging experiments, statistics on total number of trajectories, number of trajectories greater than 3, total number of localizations, localization per frame, total number of jumps, and jumps used for modeling are reported.
- https://doi.org/10.7554/eLife.35621.025
-
Supplementary file 3
Scaling factors for chr-RNA-seq replicates.
For each chr-RNA-seq replicate, the total number of sequenced ERCC spike-in control was determined. The scaling factor for each replicate was determined by 1000 divided by the number of sequenced ERCC spike-in control.
- https://doi.org/10.7554/eLife.35621.026
-
Transparent reporting form
- https://doi.org/10.7554/eLife.35621.027





