Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome
Figures
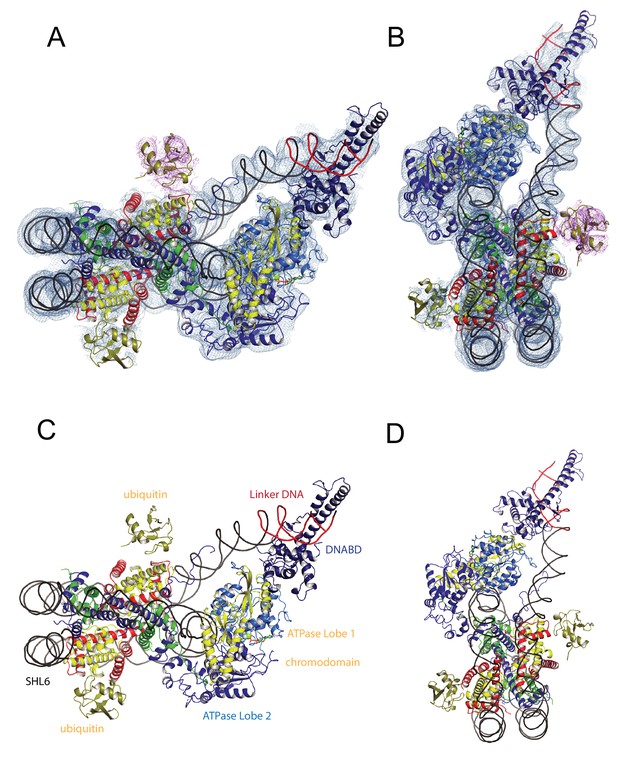
A Chd1-Nucleosome complex.
(A, B) Overall fit of nucleosome bound Chd1 to density map. Chd1 chromodomains – yellow, DNABD – dark blue, ATPase lobe one cyan, ATPase lobe two blue, Ubiquitin dark yellow, H2B yellow, H2A red, H3 green, H4 blue. (C, D) Two views of the structural model.
-
Figure 1—source data 1
Raw data for Figure 1—figure supplement 1C.
- https://doi.org/10.7554/eLife.35720.009
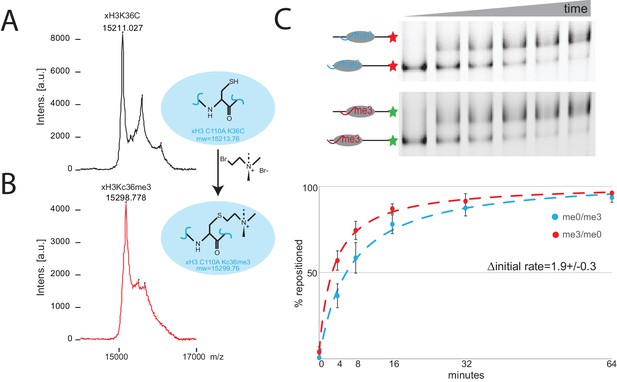
Preparation of nucleosomes with an alkylation mimic of histone H3 K36 methylation.
Xenopus laevis histone H3K36C was alkylated to generate a mimic of trimethyl lysine (Simon et al., 2007). MALDI mass spectrometry shows that the mass of free histone H3 (A) increases by 86 Da following alkylation (B) consistent with efficient conversion. (C) Chd1 directed nucleosome repositioning was assessed using a two colour nucleosome repositioning assay in which differently modified octamers are assembled onto DNA fragments labelled with different fluorescent dyes assays. Comparison of repositioning rates from four repeats indicates that the initial rate of repositioning is 1.9 fold greater for the trimethyl mimic.
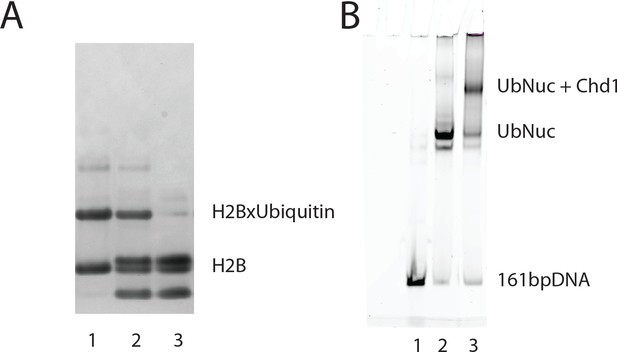
Generation of nucleosomes with ubiquitin cross-linked to histone H2B K120.
Ubiquitin was cross-linked to H2BK120C as described previously (Long et al., 2014; Morgan et al., 2016). Ubiquitin modified H2B was purified by ion exchange chromatography and refolded into H2A:H2Bub dimer (lane 1), and H2Bub octamer (lane 2), which is compared to native octamer (lane 3) (A). Nucleosomes were assembled on DNA including an asymmetric 14 base pair linker using ubiquitin modified H2B and alkylated H3K36. (B) The assembly of nucleosomes was assessed by native PAGE (lane 2). Incubation with Chd1 resulted in a major species consistent with binding of a single Chd1 to a nucleosome (lane 3).
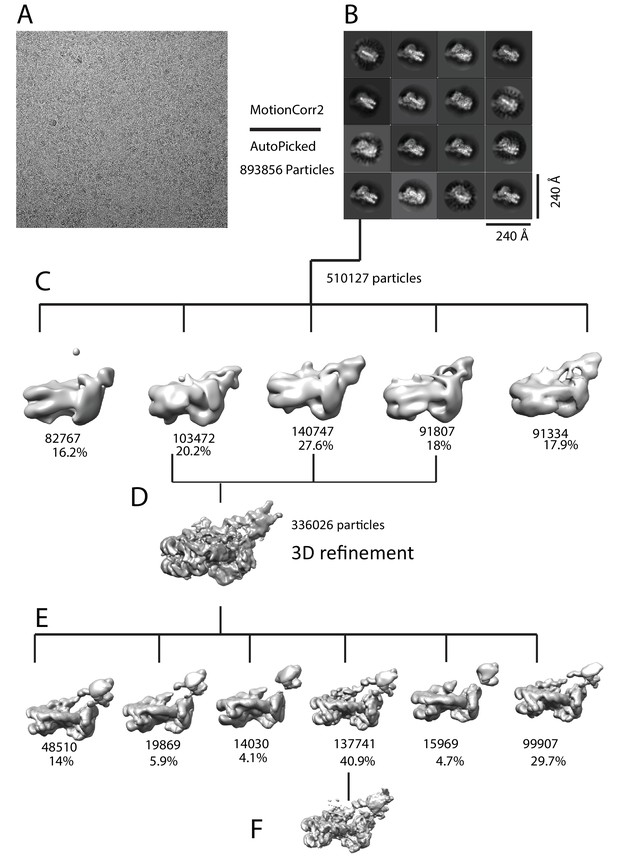
Image processing for Chd1-nuclesome complex.
(A) An example motion corrected micrograph. (B) 2D classification resulted in identification of 16 classes with Chd1 bound to nucleosomes. (C) Initial 3D classification resulted in five classes three of which were combined (D), reclassified (E) and refined (F).
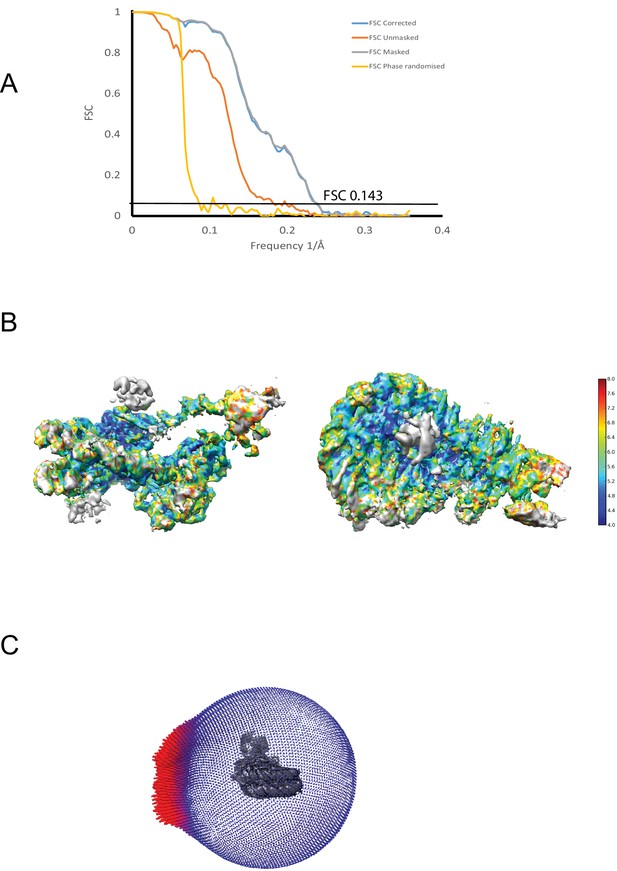
Properties of the Chd1-nuclesome complex.
(A) The fourier shell correlation between two half maps is plotted for corrected, unmasked, masked and phase randomised data as indicated. The average resolution was estimated from the corrected FSC at 0.143 as 4.5A. (B) Plot showing local distribution of resolution calculated from the unfiltered half maps. (C) Angular distribution of particles used for final refinement.
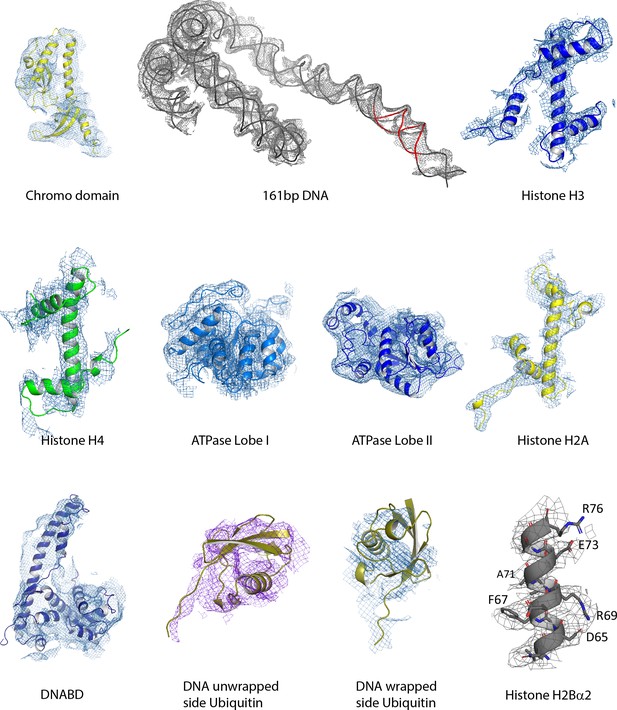
Fit of individual components.
The indicated components fitted individually to relevant regions of the smoothed density map. In the case of H2Bα2 the fit is to the unsmoothed map.
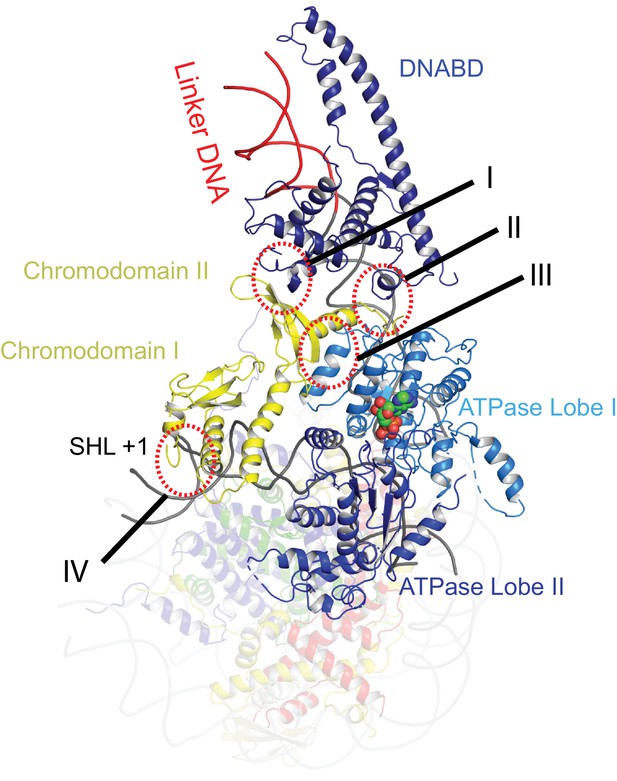
Contacts constraining Chd1 chromodomains.
Overview of the major contacts constraining the positioning of the Chd1 chromodomains. Colours of domains as for Figure 1. Key contacts are highlighted. (I) Chromodomain II SLIDE, (II) Chromodomain linker helix to SANT, (III) Chromodomain II to ATPase lobe 1, (IV) Chromodomain I to nucleosomal DNA at SHL + 1.
-
Figure 2—source data 1
Raw data for Figure 2—figure supplement 5A.
- https://doi.org/10.7554/eLife.35720.016
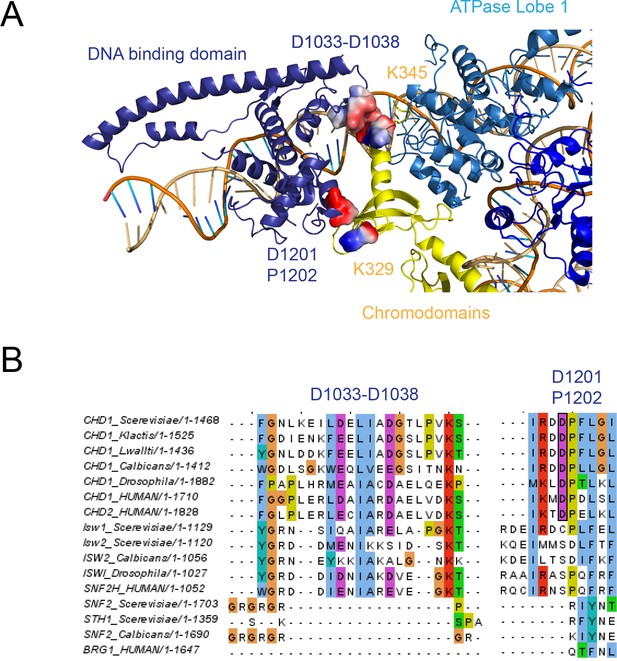
The chromodomain contacts with SANT and SLIDE domains.
(A) Residues involved in the interactions are coloured by surface charge. (B) Sequence alignments showing conservation of residues making these contacts in Chd1 proteins.
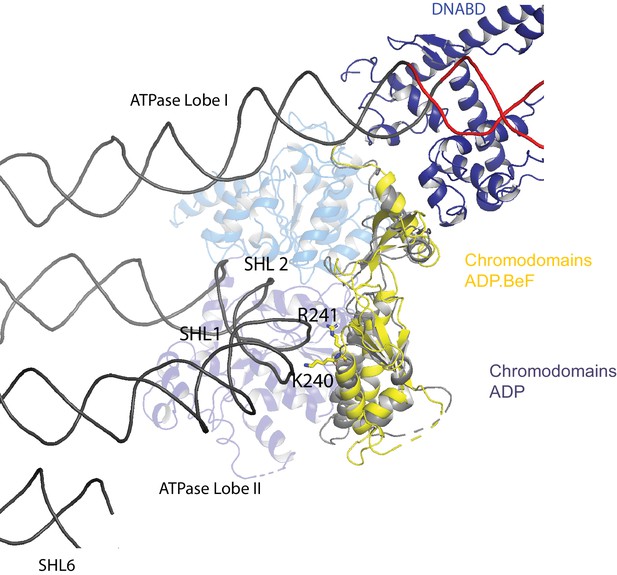
Chromodomain interactions with nucleosomal DNA at SHL + 1.
Interaction of Chd1 chromodomains with nucleosomal DNA at SHL + 1. R241 and K240 come in close contact with DNA.
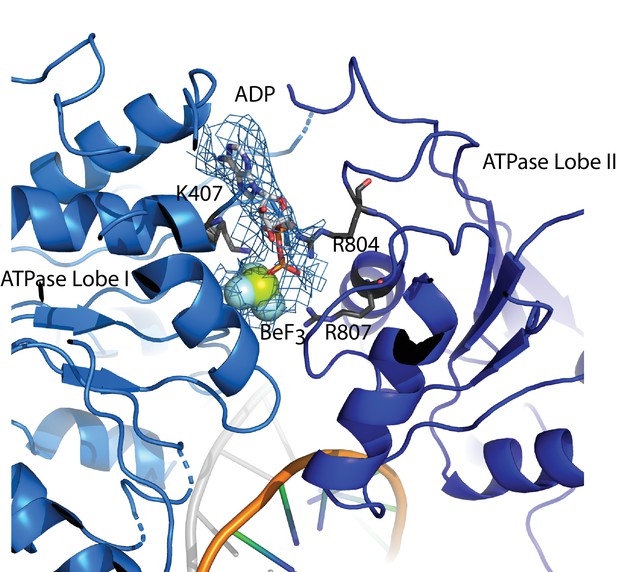
The ATP binding site.
ADP Berylium Fluoride fitted to density in a pocket formed by conserved residues contributed by both ATPase domains which are positioned in a closed state. Residues observed to contribute to catalysis in related proteins are well positioned to function similarly in Chd1.
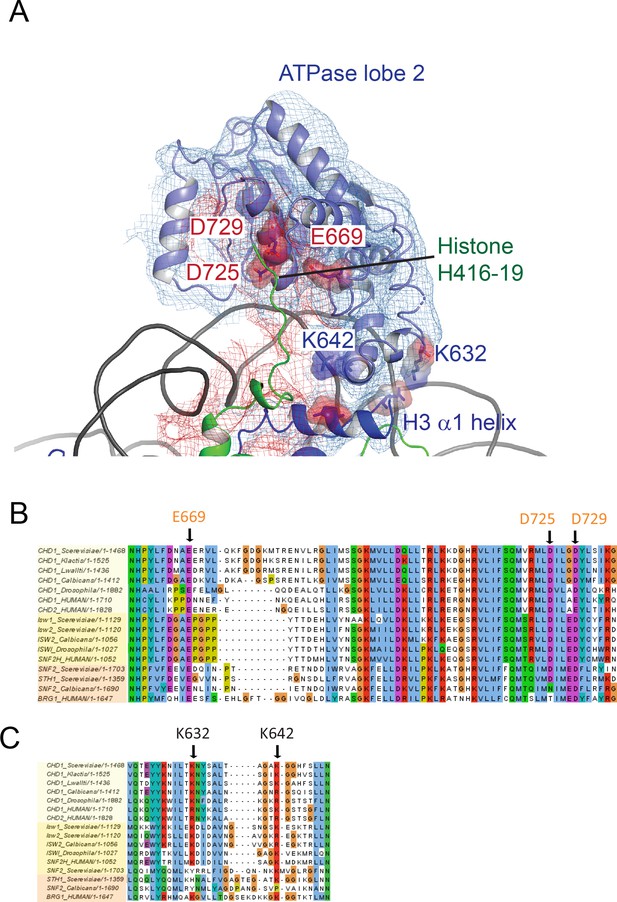
ATPase lobe two interaction with histone H4 tail.
(A) Fit of ATPase lobe 2, histone H3 and Histone H4 tail to density map. Residues coming into proximity with the H4 tail, D275, D279 and E669 are indicated by surface charge. The contact between ATPase lobe two and the histone H3 alpha one helix is also shown. (B) and (C) Sequence alignments show that the sequences participating in these interactions are conserved in Chd1 proteins and to some extent in Iswi and Snf2 proteins as well.
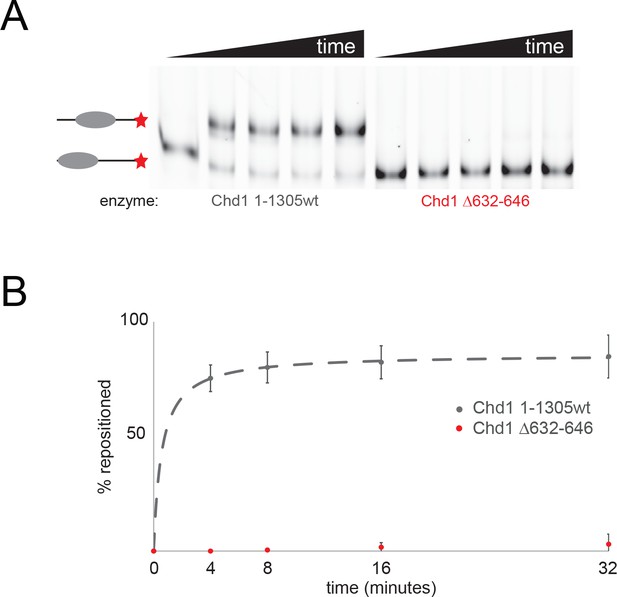
Mutation of ATPase residues contacting histone H3 abolishes Chd1 activity.
(A) The ability of Chd1-1305 and Chd1 1–3505 Δ 632–646 to reposition nucleosomes initially located asymmetrically on 0W47 cy5 labelled DNA fragments was assessed by native gel electrophoresis. (B) Quantitation of nucleosome repositioning following triplicate repeats indicates that the deletion abolishes nucleosome sliding detectable using this assay.
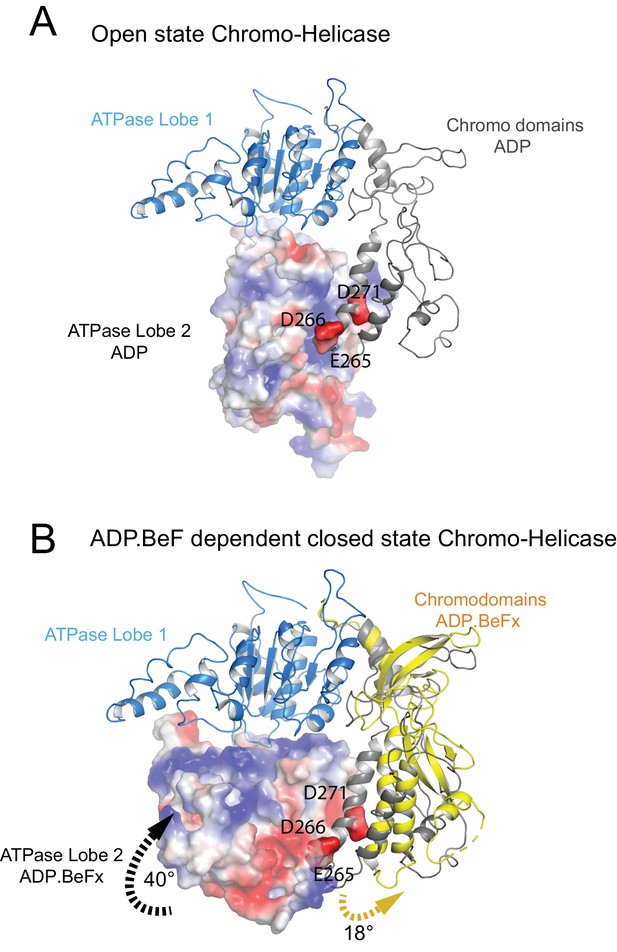
Closure of the ATPase lobes changes the chromodomain interaction surface.
(A) The long acidic helix within chromodomain I interacts with a basic surface on ATPase lobe two in the open state (3MWY). (B) In the closed state, the basic surface on lobe two is rotated towards DNA and replaced with an acidic region. The long acidic helix within chromodomain I is repositioned away from this acidic surface.
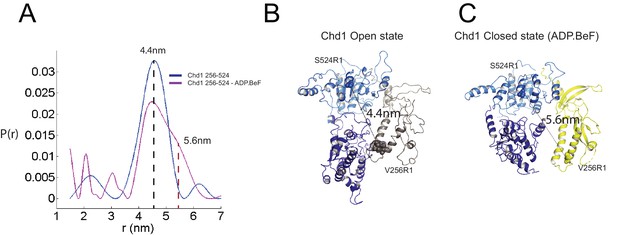
Nucleotide dependent reconfiguration of Chd1 chromodomains.
Pulsed electron paramagnetic measurements were used to measure the distance between nitroxyl reporter groups attached to Chd1 at ATPase lobe 1 (S524) and chromodomain I (V256). (A) The probability distribution P(r) at different separations was measured in the presence (purple) and absence (blue) of ADP.BeF. The distance corresponding to the major distance is shown for both measurements. The distance corresponding to the additional distribution appears as a shoulder in the presence of ADP.BeFx and is also indicated. Modelled distances between these labelling sites in the open state (3MWY), and the closed state observed in the Chd1 bound nucleosome are indicated in (B) and (C), respectively.
-
Figure 4—source data 1
PELDORDistanceDistribution.csv.
- https://doi.org/10.7554/eLife.35720.019
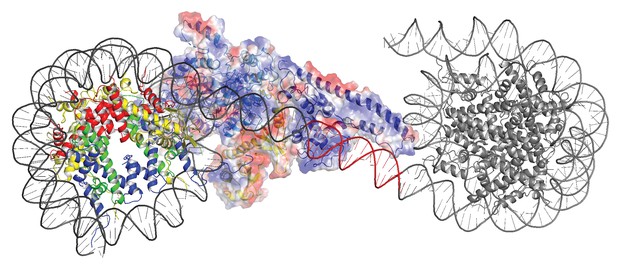
Modelling the interaction of Chd1 between adjacent nucleosomes.
The structure of the Chd1 bound nucleosome was used to model binding to the linker between an adjacent nucleosome (grey) by extending the liner DNA to 19 bp. With shorter linkers steric clashes with the adjacent nucleosome become progressively more severe.
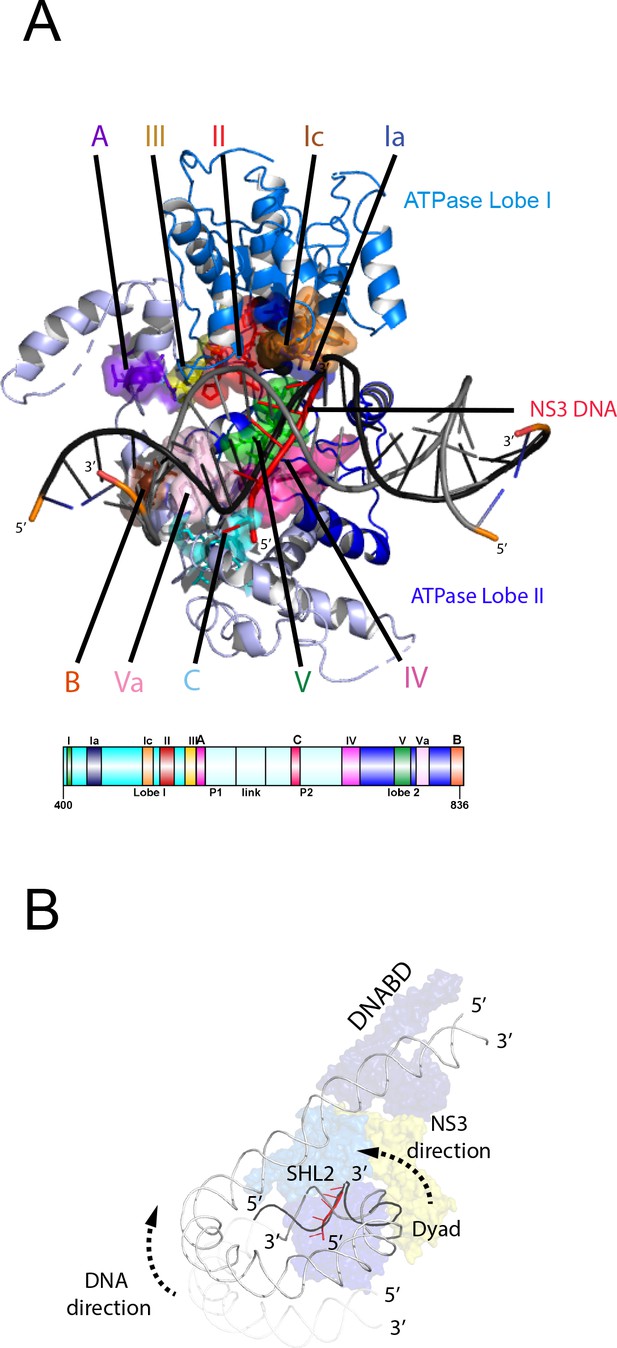
Comparison of NS3 and Chd1 interactions with DNA.
(A) Contacts between motifs conserved with DNA at SHL2. Motifs and colouring are indicated on the structure. The ssDNA from NS3 aligned to Chd1 is shown docked into Chd1 (red). In Chd1 motifs II and III contact the opposite (3’−5’) strand. Contacts made by remodelling enzyme specific extension are labelled A, B and C. The locations of each sequence are indicated in the schematic guide. (B) Schematic indicating directionality in context of a nucleosome. The directionality of NS3 translocation inferred from docking the ssDNA is 3’−5’ away from the nucleosome dyad. Assuming movement of Chd1 around the nucleosome is constrained (for example via contact with linker DNA, the H4 tail and histone H3) translocation of Chd1 with this directionality is anticipated to drive DNA in the opposite direction towards the long linker as indicated.
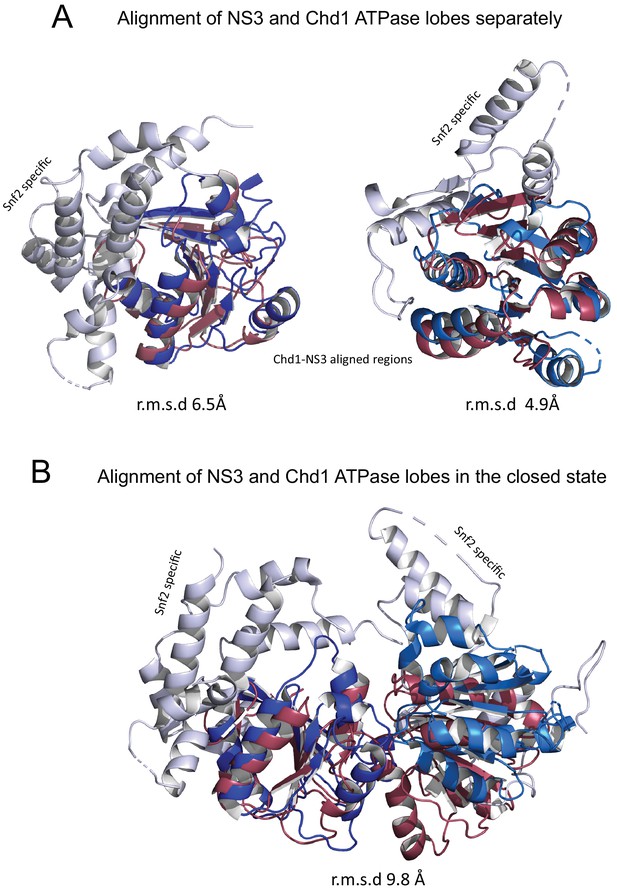
Comparison of NS3 and Chd1 ATPase domains.
ATPase domains of Chd1 and NS3 docked individually and together. ATPase lobes 1 and 2 are shown in light and dark blue. Regions specific for Snf2 proteins are shown in white. NS3 ATPase domains in raspberry.
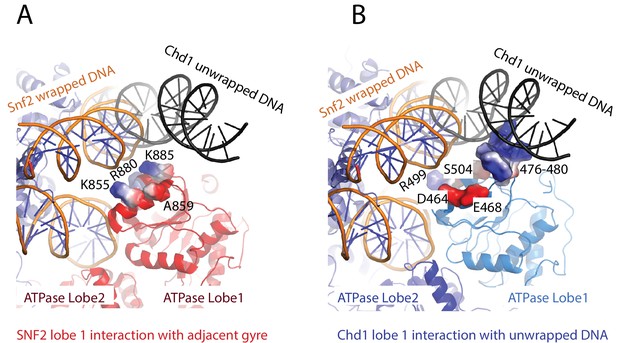
Snf2 and Chd1 interactions with the adjacent DNA gyre.
(A) Contacts between Snf2 and DNA adjacent to SHL2 at SHL6 K855, R880 and K885 (PDB 5 × 0Y). (B) These contacts are not conserved in Chd1 and instead an acidic surface is presented by D464 and E468. The region 476 to 480 contacts the unravelled DNA. In both panels normally wrapped nucleosomal DNA is indicated in orange, and the gyre unwrapped by Chd1 in black.
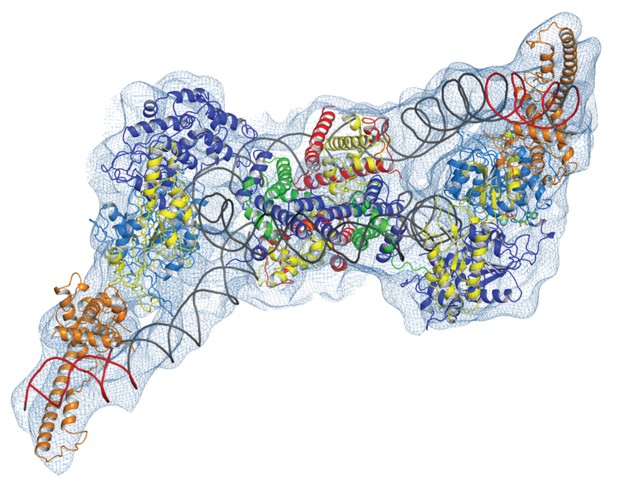
Complex of two Chd1 bound to one nucleosome.
The volume obtained for the 2Chd1:1Nucleosome complex is shown contoured to 0.0027. Two Chd1 molecules in the conformation observed in Figure 1 and a nucleosome on which DNA is unwrapped from both sides to the same degree as observed on the Chd1 bound side of the 1:1 complex are shown ridged body fitted into the volume. The correlation coefficient for fitting is 0.95. The colouring of domains is as for Figure 1.
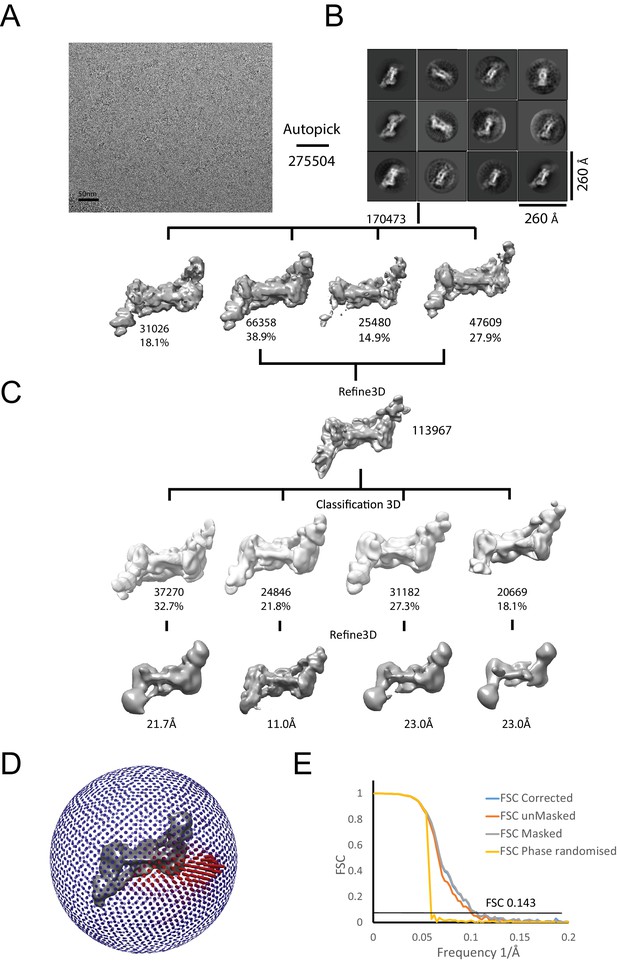
Generation of density map for two Chd1 molecules bound to a single nucleosome.
Complexes consisting of two Chd1 molecules bound to a single nucleosome with 14 base pair linkers on each side were prepared on EM grids. (A) Example micrograph used to identify 275504 particles from which 12 2D classes were identified as shown in (B). (C) 3D classes were processed to obtain a final volume based on 24857 particles as indicated. Particles display an orientation bias (D) The fourier shell correlation between two half maps is plotted for corrected, unmasked, masked and phase raondomised data as indicated. The average resolution was estimated from the corrected FSC at 0.143 as 10 Å (E).
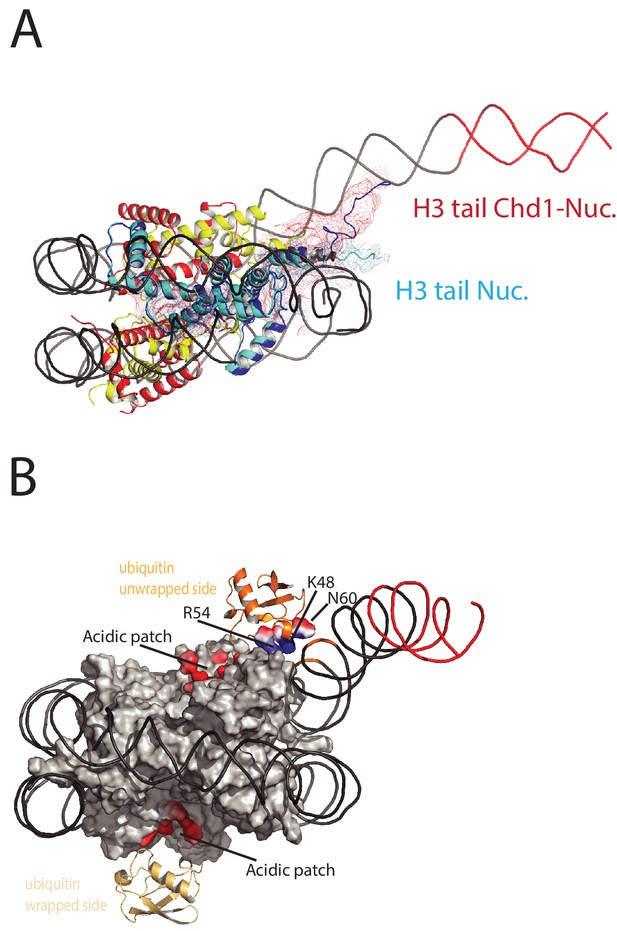
Histone epitopes are repositioned on Chd1 bound nucleosomes.
(A) The histone H3 tail on the unwrapped side of a Chd1 bound nucleosome is shown in dark blue fitted to electron density shown in red. The density extends to residue 26 on this side of the nucleosome and follows a path towards the unwrapped DNA. On the wrapped side of the nucleosome, the H3 tail is less well defined and follows a path similar to that observed on nucleosomes bound by 53BP1 (shown in light blue). H3 tail. (B) Ubiquitin on the wrapped side of the nucleosome is located over the acidic patch. On the unwrapped side of the nucleosome it is repositioned away from the acidic patch and interacts with the unravelled DNA.
-
Figure 8—source data 1
Raw data for Figure 8—figure supplement 1A.
- https://doi.org/10.7554/eLife.35720.029
-
Figure 8—source data 2
Raw data for Figure 8—figure supplement 1B.
- https://doi.org/10.7554/eLife.35720.030
-
Figure 8—source data 3
Raw data for Figure 8—figure supplement 1B(2).
- https://doi.org/10.7554/eLife.35720.031
-
Figure 8—source data 4
Raw data for Figure 8—figure supplement 1B(3).
- https://doi.org/10.7554/eLife.35720.032
-
Figure 8—source data 5
Raw data for Figure 8—figure supplement 2.
- https://doi.org/10.7554/eLife.35720.033
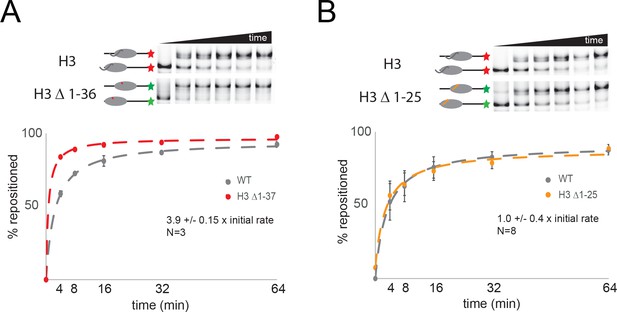
The histone H3 tail inhibits Chd1 activity.
(A) Nucleosomes with intact H3 assembled onto 0W47 DNA fragments labelled with cy5 were mixed with nucleosomes with histone H3 truncated to K36 assembled on cy3 labelled DNA. Following repositioning by Chd1 for the times indicated repositioning of each nucleosome was assessed by scanning the gels with appropriate excitation and emission filters as shown in the top panels. Quantitation of the proportion of nucleosomes repositioned shows that H3 Δ1–37 nucleosomes are repositioned 4-fold faster. (B) The approach described in A was used to show that truncation of H3 to 25 does not affect the rate of repositioning by Chd1.
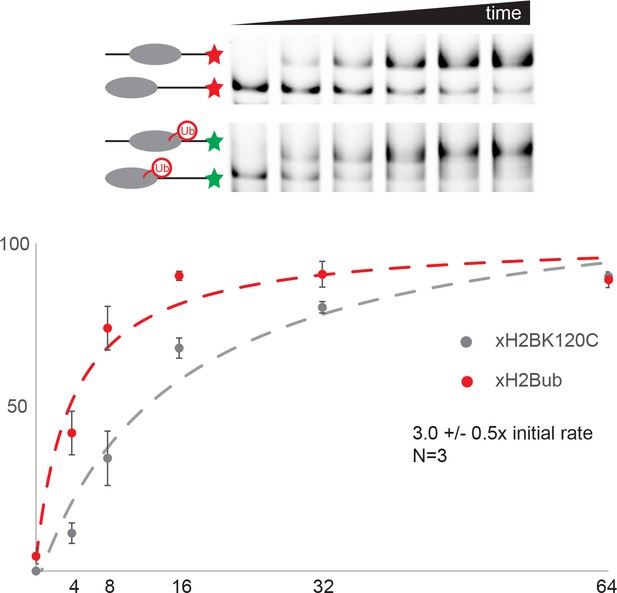
Coupling ubiquitin to Histone H3 K123 directly stimulates Chd1 activity.
The dual fluorescence labelling assay described for Figure 8—figure supplement 1 was applied to study the effect of cross-linking ubiquitin to histone H3 K123. Ubiquitinylated nucleosomes were observed to be repositioned three fold faster than non ubiquitinylated nucleosomes.
Tables
Data collection and processing summary
https://doi.org/10.7554/eLife.35720.034| Data collection and processing | UbNucleosome-Chd1 complex | Nucleosome-2Chd1 complex |
|---|---|---|
| Microscope | Titan Krios | Titan Krios |
| Detector | Gatan K2 Summit | Falcon 3 |
| Mode | Electron Counting | Integrating |
| Magnification | 105000X | 59000X |
| Voltage (kV) | 300 | 300 |
| Number of frames | 32 | 40 |
| Electron exposure (e-/Å2) | 40 | 62.5 |
| Exposure time (Sec) | 10 | 1.5 |
| Dose per fraction | 1.25 | 1.56 |
| Pixel Size | 1.4 | 1.42 |
| Defocus Range | 1.5–3.5 | 1.7–3.5 |
| Symmetry Imposed | C1 | C1 |
| Initial Particle images (no.) | 1050000 | 275504 |
| Final Particle images (no.) | 137,000 | 24846 |
| Final Map resolution (Å) | 4.5 | 10 |
| Map Sharpening B factor (Å2) | −200 | −800 |
Additional files
-
Supplementary file 1
Primers used in generating the deletion ∆632–646 on Chd1. DNA sequence of Chd1 ∆632–646. Protein sequence of Chd1 ∆632–646. Primers used to generate H3D1-25. Protein sequence of H3D1-25. DNA sequence of H3D1-25. Protein sequence of H3D1-37. DNA sequence of H3D1-37.
- https://doi.org/10.7554/eLife.35720.035
-
Transparent reporting form
- https://doi.org/10.7554/eLife.35720.036





