A synthetic peptide that prevents cAMP regulation in mammalian hyperpolarization-activated cyclic nucleotide-gated (HCN) channels
Figures
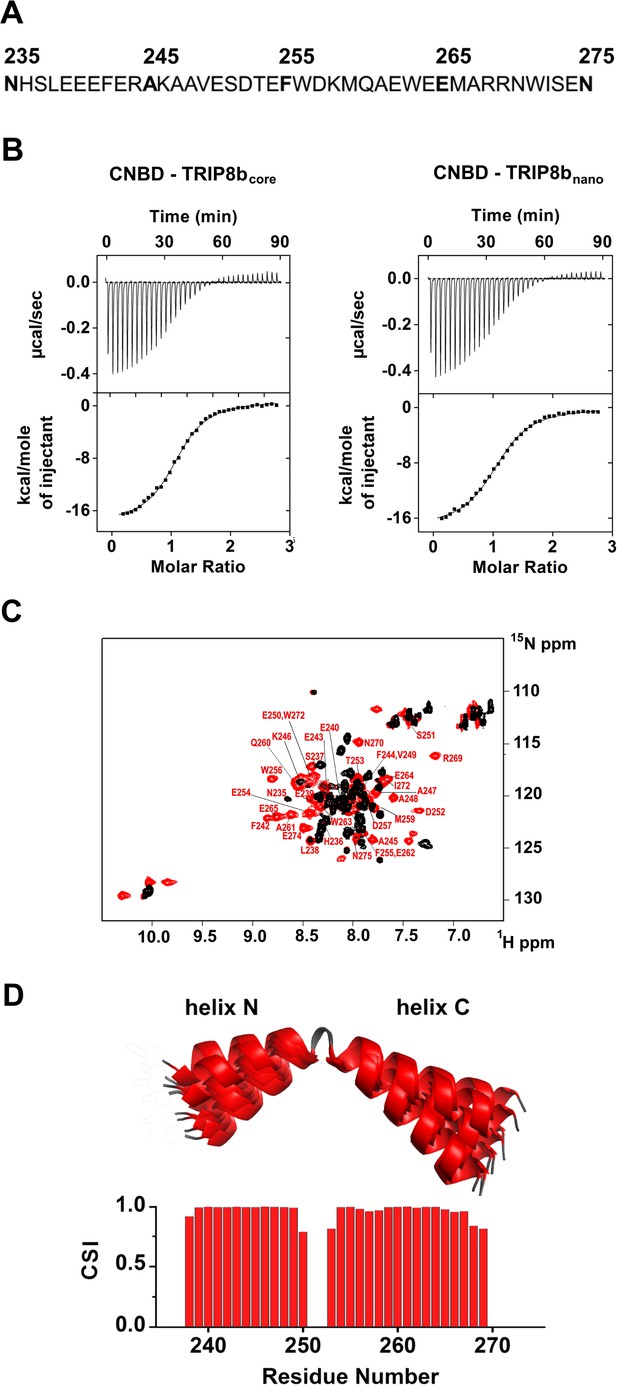
Functional and structural characterization of TRIP8bnano.
(A) Primary sequence of TRIP8bnano. Amino acid numbering refers to full length mouse TRIP8b (1a4). (B) Binding of TRIP8bcore and TRIP8bnano to purified His6-MBP-CNBD measured by Isothermal titration calorimetry (ITC). Upper panel, heat changes (μcal/sec) during successive injections of 8 μL of the corresponding TRIP8b peptide (200 μM) into the chamber containing His6-MBP-CNBD (20 μM). Lower panel, binding curve obtained from data displayed in the upper panel. The peaks were integrated, normalized to TRIP8b peptide concentration, and plotted against the molar ratio (TRIP8b peptide/His6-MBP-CNBD). Solid line represents a nonlinear least-squares fit to a single-site binding model, yielding, in the present examples, a KD = 1.2 ± 0.1 μM for TRIP8bcore and KD = 1.4 ± 0.1 μM for TRIP8bnano. (C) Evidence for TRIP8bnano folding upon CNBD binding based on the superimposition of the [1H, 15N] heteronuclear single quantum coherence (HSQC) NMR spectrum of CNBD-free TRIP8bnano (black) and CNBD-bound TRIP8bnano (red). The latter experiment was performed at the molar ratio ([CNBD]/[TRIP8bnano])=3. The backbone amide (HN) signals of the residues of CNBD-bound TRIP8bnano are labelled in red. (D) (Top) Ribbon representation of the 10 lowest energy conformers of TRIP8bnano bound to CNBD used for in silico modelling of CNBD-TRIP8bnano complex. The unfolded regions at the N- and C-termini of the construct (residues 235–237 and 270–275) are omitted for clarity. (Bottom) Chemical Shift Index (CSI, calculated using TALOS+) plotted as a function of the residue number of TRIP8bnano bound to CNBD. Positive values represent helical propensity.
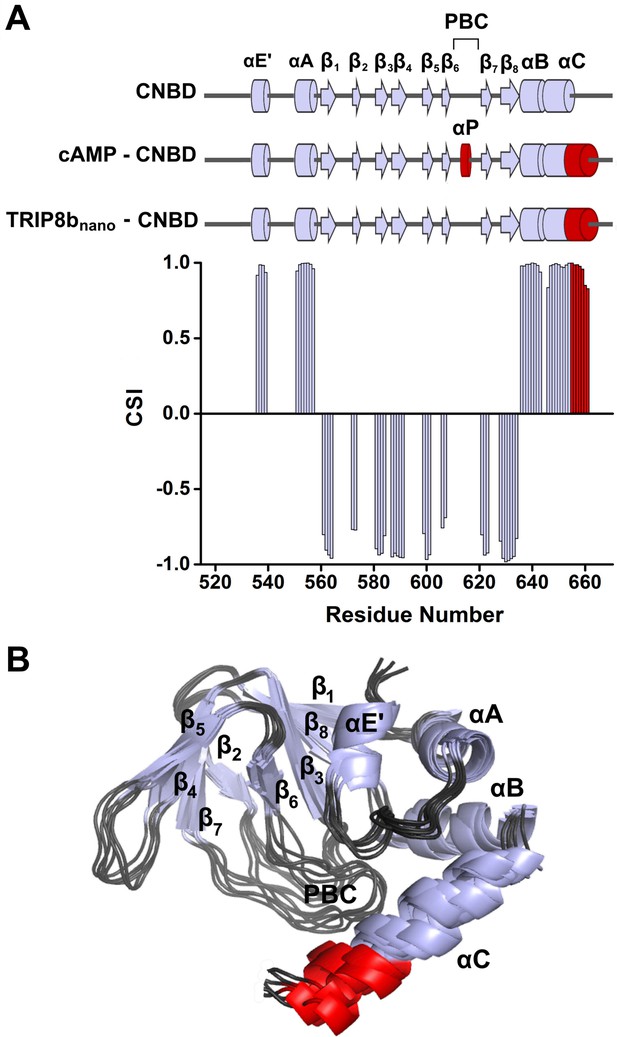
NMR structure of CNBD bound to TRIP8bnano.
(A) (Top) comparison of secondary structure elements of cAMP-free CNBD (Saponaro et al., 2014), cAMP-bound CNBD (Zagotta et al., 2003) and cAMP-free CNBD bound to TRIP8bnano (this study). Secondary structure elements are indicated by arrows (β-strands) and cylinders (α-helices) and labeled. The loop between β6 and β7 constitutes the Phosphate Binding Cassette (PBC). The elements that fold upon binding of cAMP and TRIP8bnano are shown in red. (Bottom) Chemical Shift Index (CSI, calculated using TALOS+) plotted as a function of the residue number of CNBD bound to TRIP8bnano. Positive values represent helical propensity, while negative values represent strands. (B) Ribbon representation of the 10 lowest energy conformers of CNBD bound to TRIP8bnano used for in silico modeling of CNBD - TRIP8bnano complex. Secondary structure elements are coloured in light gray and labeled. Loop regions are colored in dark gray. The distal region of the C-helix (residues 657–662), which is unfolded in the free form of the CNBD (Saponaro et al., 2014) and folds upon TRIP8bnano binding, is coloured in red. The unfolded regions at the N- and C-termini of the construct (residues 521–532 and 663–672 respectively) are omitted for clarity.
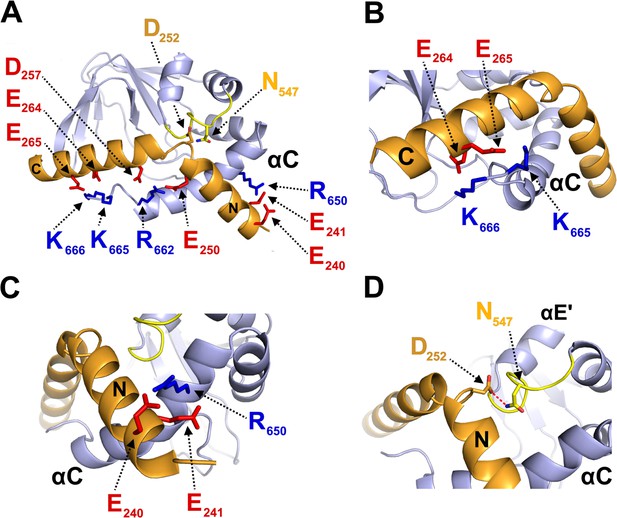
Structural model of CNBD – TRIP8bnano complex.
(A) Ribbon representation of the complex where CNBD is in gray and TRIP8bnano is in orange. Helix N (N) and helix C (C) of TRIP8bnano are labeled. C-helix of CNBD (αC) is labeled, while N-bundle loop is colored in yellow. Positively charged residues of C-helix CNBD (blue) and negatively charged residues of TRIP8bnano (red) involved in salt bridges are shown as sticks and labeled. N547 of the N-bundle loop (yellow) and D252 of TRIP8bnano (orange) are shown as sticks and labeled. (B) Close view of K665 and K666 of CNBD that interact, respectively, with E265 and E264 of TRIP8bnano. C-helix (αC) of CNBD, and Helix C (C) of TRIP8bnano are labeled. (C) Close view of R650 of CNBD that is positioned between E240 and E241 of TRIP8bnano. C-helix (αC) of CNBD, and Helix N (N) of TRIP8bnano are labeled. (D) Close view of N547 of N-bundle loop that forms a hydrogen bond (red dashed line) with D252 of TRIP8bnano. Helix E’ (αE') and C-helix (αC) of CNBD, and Helix N (N) of TRIP8bnano are labeled.
-
Figure 3—source data 1
Acquisition parameters for NMR experiments performed on cAMP-free human HCN2 CNBD in complex with TRIP8bnano and vice-versa.
- https://doi.org/10.7554/eLife.35753.011
-
Figure 3—source data 2
Docking calculation.
- https://doi.org/10.7554/eLife.35753.012
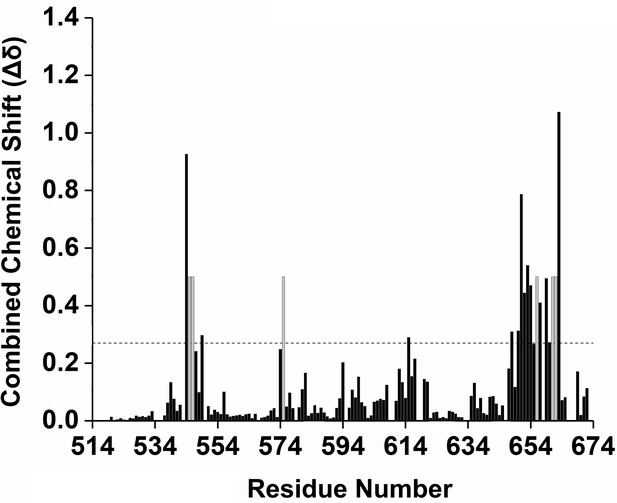
CNBD residues involved in TRIP8bnano binding.
Combined chemical shift variations of NMR signals between CNBD unbound and TRIP8bnano-bound state. Combined chemical shift variations are calculated from the experimental 1H and 15N chemical shift changes (Δδ(1H) and Δδ (15N), respectively) between the corresponding peaks in the two forms, through the following equation (Garrett et al., 1997): . Residues experiencing intermediate exchange regime (whose NMR signal becomes broad beyond detection upon addition of TRIP8bnano) are shown in grey. The horizontal dotted line indicates the average value plus one standard deviation. Residues above the line were set as ‘active’ in the docking calculation described in the text (see Materials and methods).
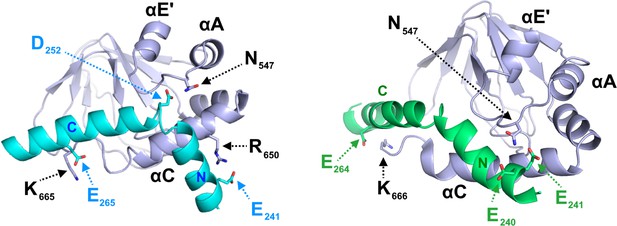
Representative families of clusters obtained from the first docking calculation.
The clusters obtained can be grouped in two classes, shown here, on the basis of the position of helix N of TRIP8bnano (N) relative to the CNBD. In the left representative structure, helix N is oriented in a way that allows E241 to interact with R650 of C-helix. In the right representative structure, E240 - E241 interact with N547 of CNBD. In all clusters K665 or K666 of CNBD establish a contact with either E264 or E265 of TRIP8bnano. The structural elements of CNBD are labeled: αE’, αA and αC. Helices N and C of TRIP8bnano are labeled.
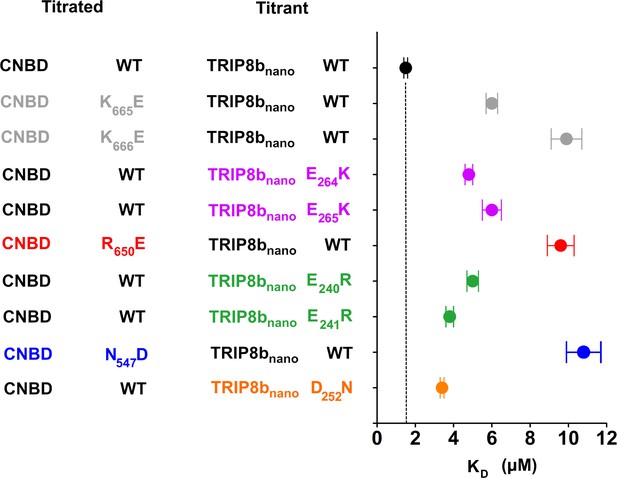
Biochemical validation of CNBD – TRIP8bnano complex.
Dissociation constant (KD) of the interaction between the indicated CNBD and TRIP8bnano peptides were measured by means of Isothermal Titration Calorimetry (ITC). CNBD WT - TRIP8bnano WT (black filled circle)=1.4 ± 0.1 µM; CNBD K665E - TRIP8bnano WT (grey filled circle)=6 ± 0.3 µM; CNBD K666E - TRIP8bnano WT (grey filled circle)=9.9 ± 0.8 µM; CNBD WT - TRIP8bnano E64K (purple filled circle)=4.8 ± 0.2 µM; CNBD WT - TRIP8bnano E65K (purple filled circle)=6 ± 0.5 µM; CNBD R650E - TRIP8bnano WT (red filled circle)=9.6 ± 0.7 µM; CNBD WT - TRIP8bnano E40R (green filled circle)=5 ± 0.3 µM; CNBD WT - TRIP8bnano E41R (green filled circle)=3.8 ± 0.2 µM; CNBD N547D - TRIP8bnano WT (blue filled circle)=11 ± 0.9 µM; CNBD WT - TRIP8bnano D52N (orange filled circle)=3.4 ± 0.1 µM. Data are presented as mean ± SEM. Number of experiments (N) ≥ 3. KD values of all tested combinations are statistically different from the KD of CNBD WT for TRIP8bnano WT (*p≤0.05; **p<0.01). Statistical analysis performed with ANOVA, followed by post-hoc Tukey test. Dashed black vertical line indicates the KD of WT peptides.
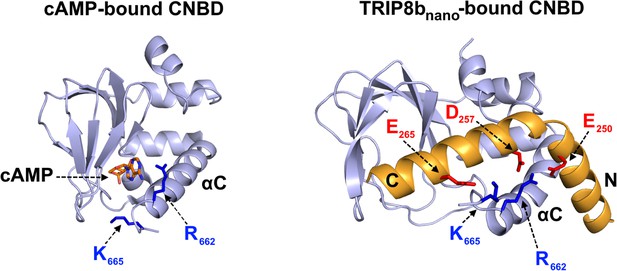
Different orientation of R662 and K665 in the cAMP-bound and TRIP8bnano-bound conformation of the CNBD.
Residues R662 and K665 of CNBD C-helix (αC) which interact with cAMP (left, [Lolicato et al., 2011]) and TRIP8bnano (right, this study) are represented as blue sticks and labeled. TRIP8bnano residues E250 and D257 interacting with R662 and residue E265 interacting with K665 are shown in red sticks and labeled.
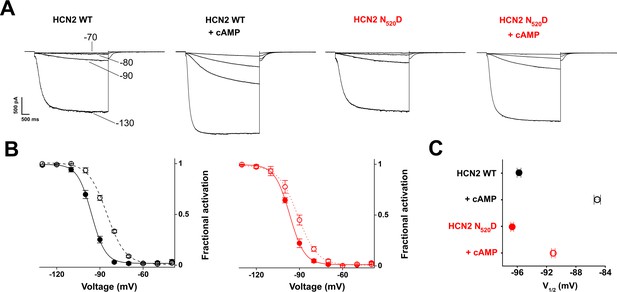
Mutation N520D affects cAMP affinity in full-length HCN2 channel.
Note that in mouse HCN2 N520D corresponds to N547D in human HCN2 (used in our structural studies). (A) Representative whole-cell HCN2 currents recorded, at the indicated voltages (−70, –80, -90, –130 mV) in HEK 293 T cells transfected with WT and N520D mutant, in the absence and presence of 5 µM cAMP in the pipette. (B) Mean activation curves obtained from the tail currents in (A) and other recordings in control solution (filled circle) and in the presence of cAMP (open circles), with HCN2 wt (black) and N547D mutant. Data were fitted to a Boltzmann equation (solid and dotted lines) (see Materials and methods). (C) Half activation potential (V1/2) values derived from Boltzmann equation fitting. HCN2 WT (black filled circle) V1/2 = -95.9 ± 0.3 mV; HCN2 WT + 5 µM cAMP (black open circle) V1/2 = 85.2 ± 0.4 mV; HCN2 N520D (red filled circle) V1/2 = -96.8 ± 0.2 mV; HCN2 N520D + 5 µM cAMP (red open circle) V1/2 = -91.2 ± 0.2 mV. Data are presented as mean ± SEM. Number of experiments (N) ≥ 6. Statistical analysis performed with two-way ANOVA, followed by post-hoc Tukey test (p<0.001) showed that wt and mutant channel are identical and that the cAMP-induced shifts are different.
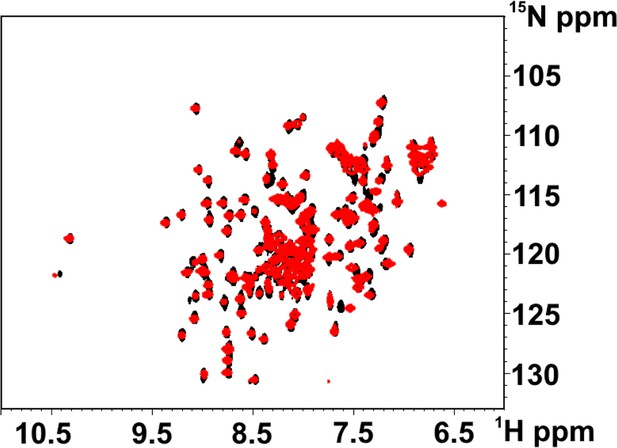
Structural characterization of N547D CNBD protein.
Superimposition of the [1H, 15N] HSQC spectra of CNBD wt (black) and CNBD N547D mutant (red).
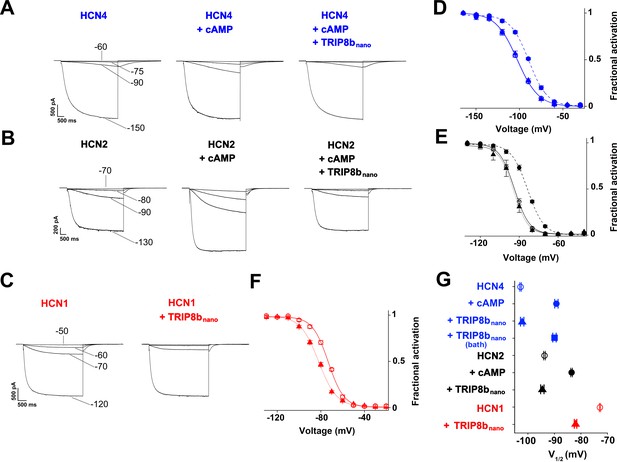
TRIP8bnano abolishes cAMP effect on HCN channel gating.
(A-C) Representative whole-cell HCN4, HCN2 and HCN1 currents recorded, at the indicated voltages, with control solution or with cAMP (1 μM for HCN4 and 5 μM for HCN2) or with cAMP + 10 µM TRIP8bnano in the patch pipette (for HCN1, 10 µM TRIP8bnano only was added). (D-F) Mean activation curves measured from HCN4, HCN2 and HCN1 in control solution (open circles), cAMP (filled circles), cAMP +TRIP8bnano, or TRIP8bnano only in the case of HCN1 (filled triangles). Solid and dashed lines indicate Boltzmann fitting to the data (see Materials and methods). (G) Half activation potential (V1/2) values of HCN4 (blue), HCN2 (black) HCN1 (red) in control solution (open circle), cAMP (filled circle) and cAMP +TRIP8bnano, or TRIP8bnano only in the case of HCN1 (filled triangle). HCN4, control = −102.8 ± 0.3 mV; HCN4 +1 µM cAMP = −89.2 ± 0.6 mV; HCN4 +1 µM cAMP +10 µM TRIP8bnano = −102.1 ± 0.6 mV, HCN4 +1 µM cAMP in the patch pipette +10 µM TRIP8bnano in the bath solution = −91.7 ± 0.3 mV; HCN2, control = −93.7 ± 0.3 mV; HCN2 +5 µM cAMP = −83.5 ± 0.3 mV; HCN2 +5 µM cAMP +10 µM TRIP8bnano = −94.5 ± 0.6 mV; HCN1, control = −72.8 ± 0.2 mV; HCN1 +10 µM TRIP8bnano = −82 ± 0.5 mV. Data are presented as mean ± SEM. Number of cells (N) ≥ 11. There is no significant difference between the controls and the addition of TRIP8bnano with (HCN4, HCN2) or without (HCN1) cAMP in the pipette. No significant difference was observed following the addition of TRIP8bnano in the bath. Statistical analysis performed with two-way ANOVA, followed by post-hoc Tukey test (p<0.001).
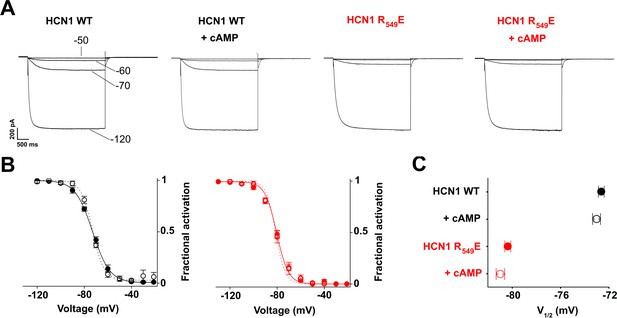
Comparison of half activation potentials (V1/2) of HCN1 WT and HCN1 R549E mutant (Chen et al., 2001).
(A) Currents were measured at the indicated voltages (- 50,–60, −70,–120 mV), from HEK 293 T cells with control solution or with 15 μM cAMP in the patch pipette. (B) Mean activation curves obtained from tail currents were fitted to a Boltzmann equation (see Materials and methods). (C) Half activation potential (V1/2) determined from Boltzmann fitting. HCN1 WT (black filled circle)=−72.7 ± 0.2 mV; HCN1 WT + cAMP (black open circle)=−73.1 ± 0.3 mV; HCN1 R549E (red filled circle)=−80.4 ± 0.2 mV; HCN1 R549E + cAMP (red open circle) = −81 ± 0.3 mV. Note that in human HCN1, used here, R549E corresponds to mouse HCN1 R538E used in Chen et al. (2001). Data are presented as mean ± SEM. Number of experiments (N) ≥ 8. Wt values are significantly different from mutant. Statistical analysis performed with two-way ANOVA, followed by post-hoc Tukey test (p<0.001).
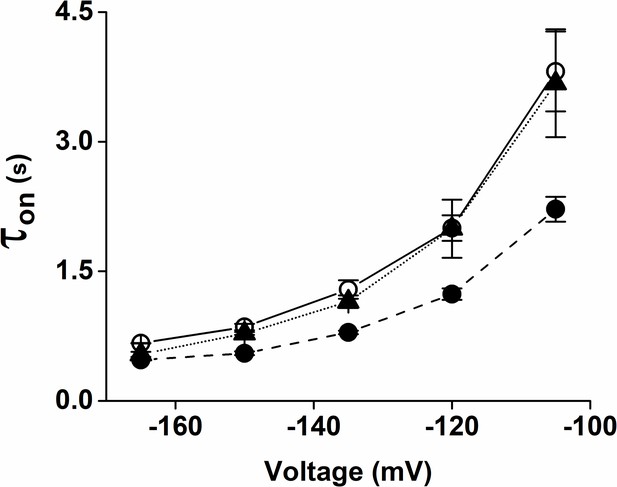
Voltage-dependency of activation time constant (τon) of HCN4 channels in control solution (open circles), 1 µM cAMP (filled circles), 1 µM cAMP + 10 µM TRIP8bnano (filled triangles).
Data are presented as mean ± SEM. Number of cells (N) = 5.
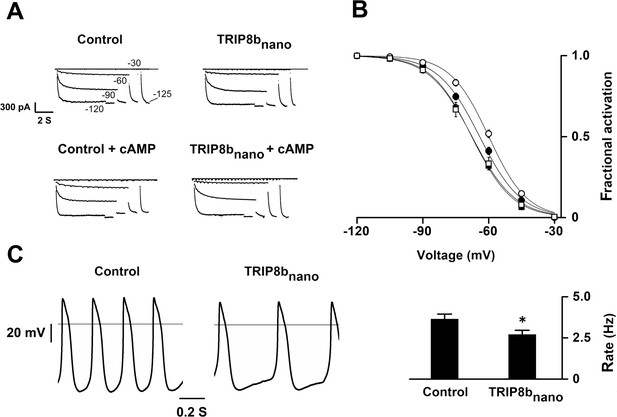
Effects of TRIP8bnano on voltage-dependent activation of If and spontaneous rate in rabbit sinoatrial node (SAN) myocytes.
(A) Representative whole-cell If currents recorded, at the indicated voltages, in the control solution and in the presence of 10 μM TRIP8bnano, without (top) and with 1 μM cAMP in the pipette (bottom). (B) Mean If activation curves were measured using a two-step protocol (see Materials and methods) in control (filled circles) or in the presence of: 1 µM cAMP (open circles); 10 µM TRIP8bnano (filled squares); 1 µM cAMP +10 µM TRIP8bnano (open squares). Ligands were added in the patch pipette. Half activation potential (V1/2) of If activation curves measured in control = −64.1 ± 0.4 mV or in the presence of: 1 µM cAMP = −59.9 ± 0.4 mV; 10 µM TRIP8bnano = −67.7 ± 0.4 mV; 1 µM cAMP +10 µM TRIP8bnano = −67.6 ± 0.7 mV. Data are presented as mean ± SEM. Number of cells (N) was ≥15. V1/2 values are significantly different between each other’s whit the exception of V1/2 obtained in the presence TRIP8bnano and cAMP +TRIP8 bnano. Statistical analysis performed with two-way ANOVA, followed by post-hoc Bonferroni test (*p<0.05) (C) (Left) Representative recordings of single SAN cell spontaneous activity in control and in the presence of 10 µM TRIP8bnano. (Right) Mean spontaneous rate (Hz) recorded in control solution = 3.65 ± 0.29 Hz and in the presence of 10 µM TRIP8bnano added to the pipette = 2.69 ± 0.27 Hz. Data are presented as mean ± SEM. Number of cells (N) was ≥7. Statistical analysis performed with t test (*p<0.05).
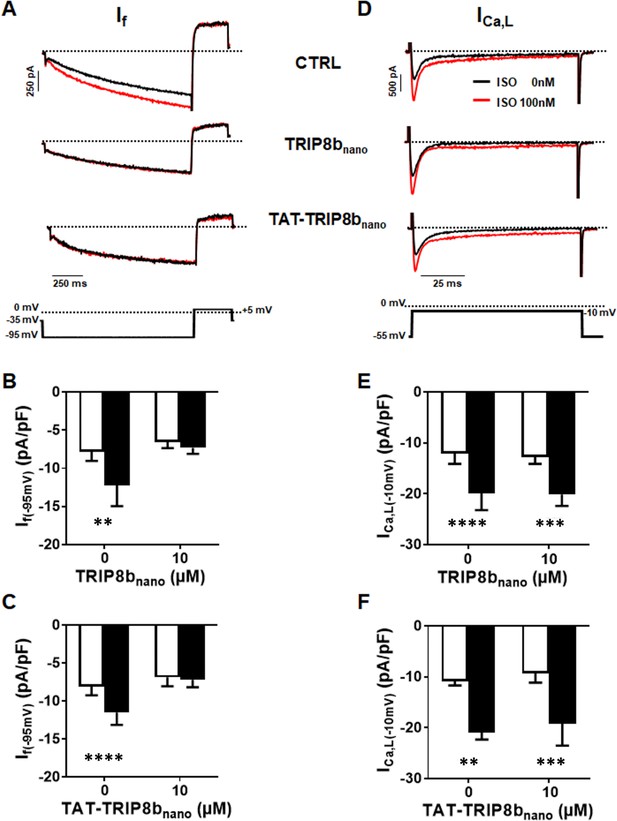
Effect of TRIP8bnano and TAT-TRIP8bnano on If and ICa,L in mouse sinoatrial node (SAN) myocytes.
(A) Representative examples of If recordings at −95 mV in control conditions (top), in 10 µM TRIP8bnano dialyzed cell (middle) and in cells perfused with 10 µM TAT-TRIP8bnano (bottom), before (black trace) and after (red trace) application of ISO 100 nM. The voltage-clamp protocol used for recordings is shown above current traces. (B) Mean normalized If current density recorded at −95 mV in absence and in presence of 10 µM TRIP8bnano in the patch pipette, before (open bars) and after (filled bars) 100 nM ISO perfusion. Data are presented as mean ± SEM. Number of cells (N) ≥ 6. Statistical analysis performed with two-way ANOVA test, followed by Sidak multiple comparisons test (**p<0.01). (C) Mean normalised If current density recorded at −95 mV in control solution or in the solution containing 10 µM TAT-TRIP8bnano, in absence (open bars) and in the presence (filled bars) of 100 nM ISO. Data are presented as mean ± SEM. Number of cells (N) ≥ 8. Statistical analysis performed with two-way ANOVA test, followed by Sidak multiple comparisons test (****p<0.0001). (D) Representative examples of ICa,L recordings at −10 mV in control conditions (top), in 10 µM TRIP8bnano dialyzed cell (middle) and in cells perfused with 10 µM TAT-TRIP8bnano (bottom), before (black trace) and after (red trace) application of ISO 100 nM. The voltage-clamp protocol used for recordings is shown above current traces. (E) Mean normalized ICa,L current density recorded at −10 mV in absence and in presence of 10 µM TRIP8bnano in the patch pipette, before (open bars) and after (filled bars) 100 nM ISO perfusion. Data are presented as mean ± SEM. Number of cells (N) ≥ 8. Statistical analysis performed with two-way ANOVA test, followed by Sidak multiple comparisons test (***p<0.001; ****p<0.0001). (F) Mean normalized ICa,L current density recorded at −10 mV in control solution or in the solution containing 10 µM TAT-TRIP8bnano, in absence (open bars) and in the presence (filled bars) of 100 nM ISO. Data are presented as mean ± SEM. Number of cells (N) ≥ 7. Statistical analysis performed with two-way ANOVA test, followed by Sidak multiple comparison test (**p<0.01; ***p<0.001).
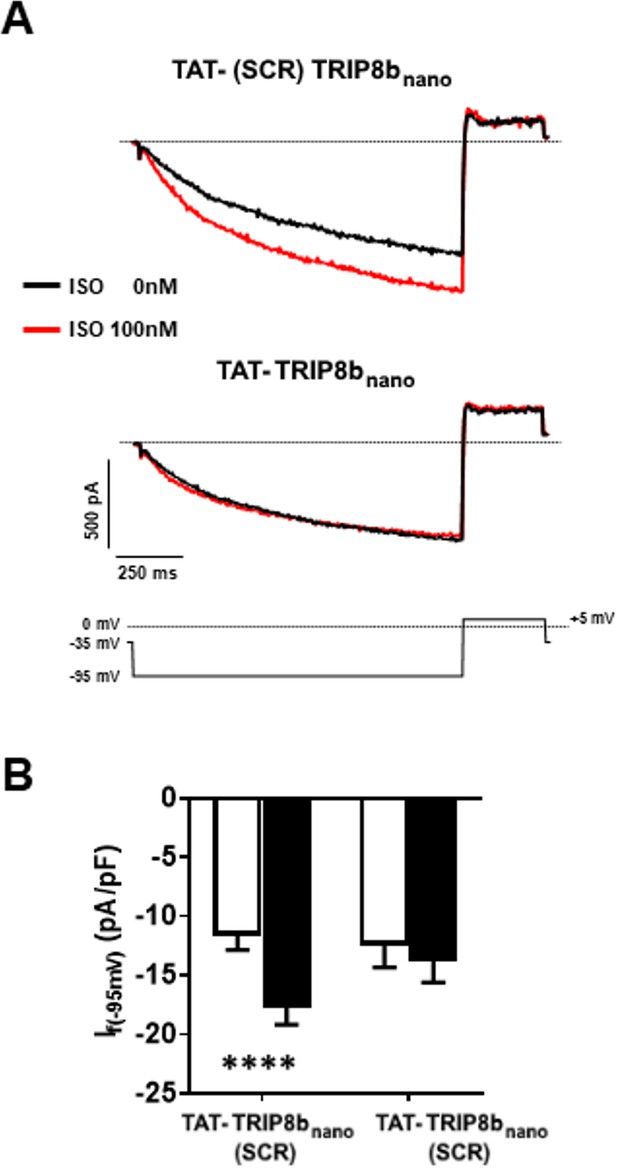
Effect of TAT- (SCRAMBLED) TRIP8bnano on If in mouse sinoatrial node (SAN) myocytes.
(A) Representative examples of If recordings (at −95 mV) in TAT-(SCRAMBLED)TRIP8bnano (top) or in TAT-TRIP8bnano (bottom) incubated cells. The voltage-clamp protocol used for recordings is shown above current traces. (B) Mean normalized If current density recorded at −95 mV in the presence of 10 µM TAT- (SCRAMBLED) TRIP8bnano or TAT-TRIP8bnano, before (open bars) and after (filled bars) 100 nM isoprenaline (ISO) perfusion. Data are presented as mean ± SEM. Number of cells (N) ≥ 11. Statistical analysis performed with two-way ANOVA test, followed by Sidak multiple comparisons test (****p<0.0001).
Tables
| Reagent type species | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (human) | HCN1 | Xention Ltd. (Cambridge, UK) | NM_021072.3 | |
| Gene (mouse) | HCN2 | PMID: 11331358 | NM_008226.2 | Laboratory of Steven A. Siegelbaum |
| Gene (rabbit) | HCN4 | PMID: 10212270 | NM_001082707.1 | |
| Gene (mouse) | TRIP8b | PMID: 19555649 | Laboratory of Steven A. Siegelbaum | |
| Strain, strain background (E. coli) | DH5α | Thermo Fisher Scientific | ||
| Strain, strain background (E. coli) | Stbl2 | Thermo Fisher Scientific | ||
| Strain, strain background (Mus musculus) | Male or female C57BL/6J mice | The Jackson Laboratory | RRID:MGI:5653012 | |
| Strain, strain background (Oryctolagus cuniculus) | New Zealand white female rabbits | Envigo | ID strain:HsdOkd:NZW | |
| Cell line (human) | HEK 293T | ATCC | RRID:CVCL_0063 | Tested negative for mycoplasma |
| Biological sample (Mus musculus) | Isolated adult Sinoatrial node (SAN) cardiomyocytes | PMID: 11557233 | ||
| Biological sample (Oryctolagus cuniculus) | Isolated adult Sinoatrial node (SAN) cardiomyocytes | PMID: 2432247 | ||
| Recombinant DNA reagent | pET-52b (plasmid) | EMD Millipore | ||
| Recombinant DNA reagent | modified pET-24b (plasmid) | Laboratory of Daniel L. Minor, Jr. | ||
| Recombinant DNA reagent | pcDNA 3.1 (plasmid) | Clontech Laboratories | ||
| Recombinant DNA reagent | pCI (plasmid) | Promega | ||
| Recombinant DNA reagent | TRIP8bnano (cDNA) | This paper | Made by PCR and cloning; see Constructs | |
| Recombinant DNA reagent | TRIP8bnano (E240R) (cDNA) | This paper | Made by site-directed mutagenesis of TRIP8bnano wt; see Constructs | |
| Recombinant DNA reagent | TRIP8bnano (E241R) (cDNA) | This paper | Made by site-directed mutagenesis of TRIP8bnano wt; see Constructs | |
| Recombinant DNA reagent | TRIP8bnano (E264K) (cDNA) | This paper | Made by site-directed mutagenesis of TRIP8bnano wt; see Constructs | |
| Recombinant DNA reagent | TRIP8bnano (E265K) (cDNA) | This paper | Made by site-directed mutagenesis of TRIP8bnano wt; see Constructs | |
| Recombinant DNA reagent | TRIP8bnano (D252N) (cDNA) | This paper | Made by site-directed mutagenesis of TRIP8bnano wt; see Constructs | |
| Recombinant DNA reagent | TRIP8bcore (cDNA) | PMID: 25197093 | ||
| Recombinant DNA reagent | human HCN2 CNBD (cDNA) | PMID: 25197093 | ||
| Recombinant DNA reagent | human HCN2 CNBD (N547D) (cDNA) | This paper | Made by site-directed mutagenesis of human HCN2 CNBD wt; see Constructs | |
| Recombinant DNA reagent | human HCN2 CNBD (K665E) (cDNA) | This paper | Made by site-directed mutagenesis of human HCN2 CNBD wt; see Constructs | |
| Recombinant DNA reagent | human HCN2 CNBD (K666E) (cDNA) | This paper | Made by site-directed mutagenesis of human HCN2 CNBD wt; see Constructs | |
| Recombinant DNA reagent | human HCN2 CNBD (R650E) (cDNA) | This paper | Made by site-directed mutagenesis of human HCN2 CNBD wt; see Constructs | |
| Recombinant DNA reagent | human HCN1 (cDNA) | Xention Ltd. (Cambridge, UK) | ||
| Recombinant DNA reagent | TRIP8b (1a4) (cDNA) | This paper | Made by PCR and cloning; see Constructs | |
| Recombinant DNA reagent | mouse HCN2 (cDNA) | Laboratory of Steven A. Siegelbaum | ||
| Recombinant DNA reagent | rabbit HCN4 (cDNA) | PMID: 10212270 | ||
| Recombinant DNA reagent | mouse HCN2 (N520D) (cDNA) | This paper | Made by site-directed mutagenesis of mouse HCN2 wt; see Constructs | |
| Sequence-based reagent | human HCN1 (R549E) (cDNA) | This paper | Made by site-directed mutagenesis of human HCN1 wt; see Constructs | |
| Peptide, recombinant protein | TAT-TRIP8bnano (YGRKKRRQRRRGG- NHSLEEEFERAKAAVESTEFWDKMQA EWEEMARRNWISEN) | CASLO ApS | ||
| Peptide, recombinant protein | TAT-(SCRAMBLED)TRIP8bnano (YGRKKRRQRRRGG-RNEAEAAE VAQKDMINERARTHEFEWESWE MWENLSESFK) | CASLO ApS | ||
| Commercial assay or kit | QuickChange Lightning Site- Directed Mutagenesis Kit | Agilent | ||
| Commercial assay or kit | Thermo Scientific TurboFect Transfection Reagent | Thermo Fisher Scientific | ||
| Chemical compound, drug | Adenosine 3', 5'-cyclic monophosphate (cAMP) | SIGMA | ||
| Software, algorithm | Clampfit 10.5/10.7 | Molecular Devices | RRID:SCR_011323 | |
| Software, algorithm | CYANA-2.1 | L. A. Systems, Inc. | ||
| Software, algorithm | AMBER 12.0 | http://pyenmr.cerm.unifi.it/ access/index/amps-nmr | ||
| Software, algorithm | HADDOCK2.2 | www.wenmr.eu |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.35753.019





