Viral GPCR US28 can signal in response to chemokine agonists of nearly unlimited structural degeneracy
Figures
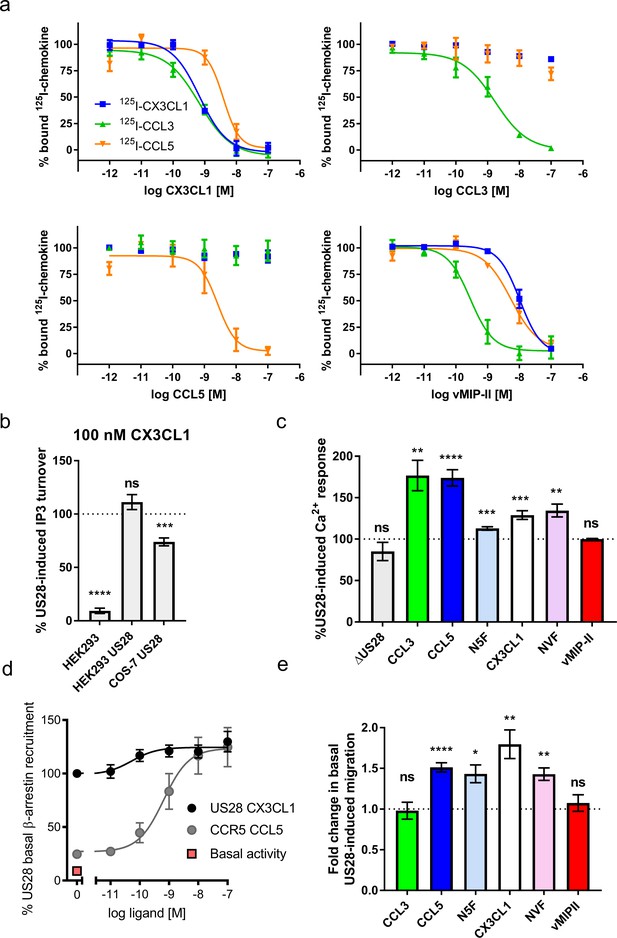
Natural and chimeric chemokines are agonists for US28.
(a) Radioligand binding competition experiments with labeled CX3CL1, CCL3, and CCL5. (b) CX3CL1-induced IP3 turnover is US28 and cell type specific. Dotted line indicates US28 basal activity; all statistics are relative to this. (c) US28-induced calcium responses of 100 nM natural and chimeric chemokines. Dotted line indicates US28 basal activity; all statistics are relative to this. (d) US28 basal β-arrestin recruitment leaves narrow dynamic range in which to observe ligand effects. (e) Migration effects of 100 nM natural and chimeric chemkines at US28. Dotted line indicates US28 basal activity; all statistics are relative to this. All data are given as mean ± s.e.m. of at least three independent biological replicates. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001 with respect to basal activity using one sample t-test, two-tailed.
-
Figure 1—source data 1
Full statistics for wild type and chimeric chemokines.
Exact p-values for chemokines with respect to basal activity using one sample t-test, two-tailed.
- https://doi.org/10.7554/eLife.35850.006
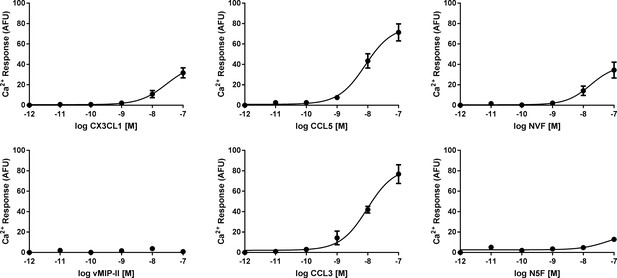
Calcium signaling dose-response plots for wild type and chimeric chemokines.
All data are given as mean ± s.e.m. of at least three independent biological replicates.
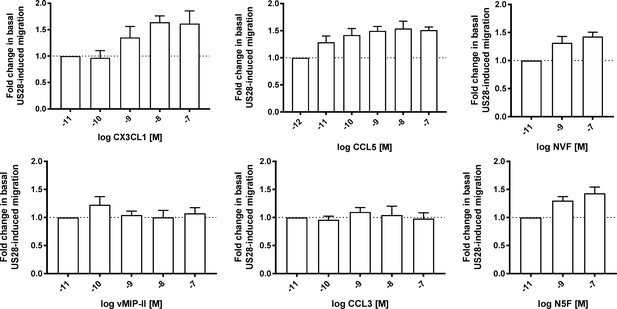
Cell migration dose-response plots for wild type and chimeric chemokines.
All data are given as mean ± s.e.m. of at least three independent biological replicates.
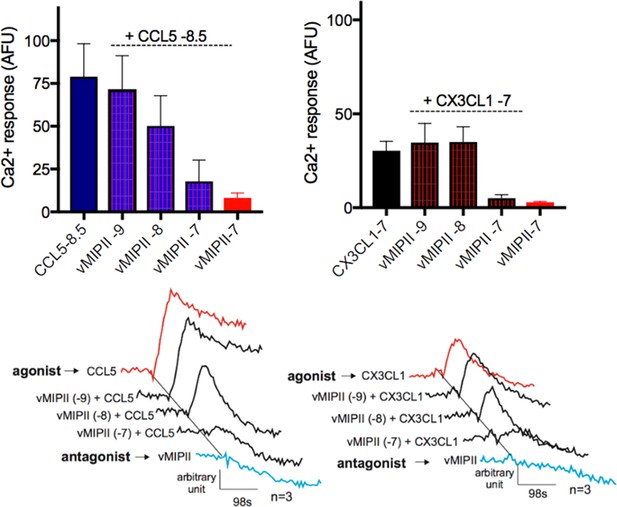
vMIP-II antagonizes chemokine signaling via US28.
top, CCL5 and CX3CL1 Ca2+ responses at a given log concentration decrease as vMIP-II concentration increases. bottom, Representative traces. All data are given as mean ± s.e.m. of at least three independent biological replicates.
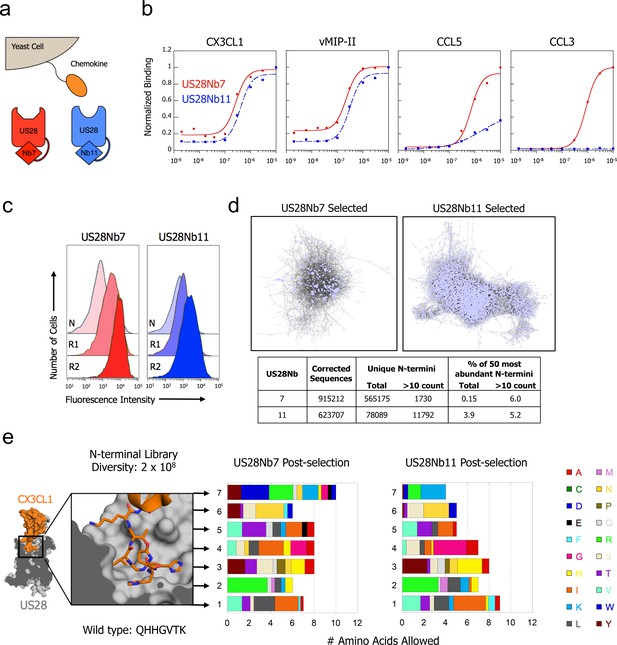
CX3CL1 library reveals chemokine sequence promiscuity and biased agonism.
(a) Illustration of yeast-displayed chemokine and nanobody-stabilized receptors. (b) Effect of intracellular nanobody 7 or 11 on US28 binding to yeast-displayed chemokines. All data points are normalized to binding at 1 µM US28Nb7 (c) Increase in binding at 1 nM US28 after selection with either US28Nb7 or US28Nb11. (d) Clustering of CX3CL1 N-terminal sequences revealed by deep-sequencing after selection. Each point is a unique N-terminus and sequences sharing 6 of 7 amino acids are connected. (e) Degree of amino acid convergence at each position of the CX3CL1 N-terminus. Amino acids with >3% abundance after selection are considered allowed and the size of a shaded region corresponds to that amino acid’s frequency (PDB: 4xt1).
-
Figure 2—source data 1
Unique CX3CL1 N-termini identified by deep sequencing after selection with US28Nb7 or US28Nb11.
Sequence identity and number of reads given for each unique sequence appearing more than 10 times.
- https://doi.org/10.7554/eLife.35850.012
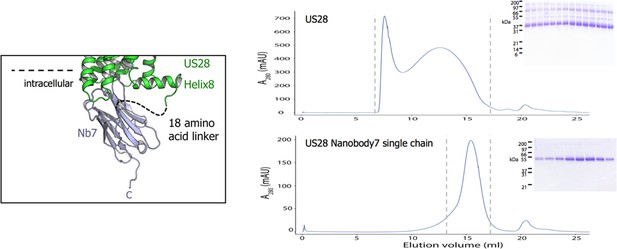
US28-nanobody single chain constructs enable purification of apo-receptor.
left, Structure of nanobody7 bound to US28 with a dashed line showing a linker creating a fusion construct. right, Size exclusion chromatograms of US28 and US28Nb7. SDS-PAGE gels depict the elution fractions between vertical hashes.
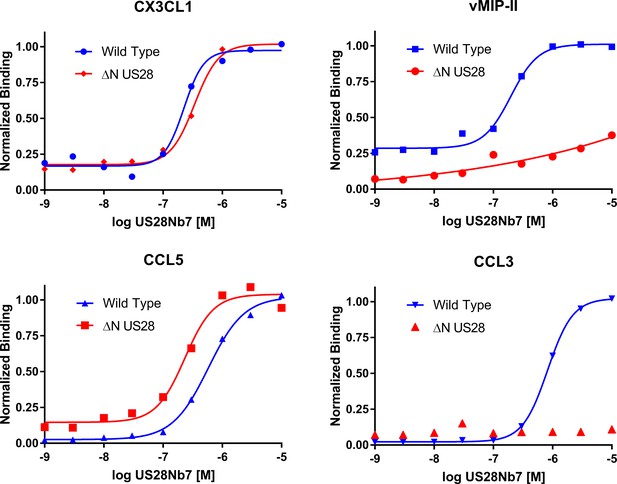
Effects of US28Nb7 N-terminal deletion on yeast-displayed wild-type chemokine binding.
ΔN US28 is US28Nb7 which has been engineered to delete the receptor N-terminus up to Cys22 (See methods for details). All data points are normalized to binding at 1 µM US28Nb7.
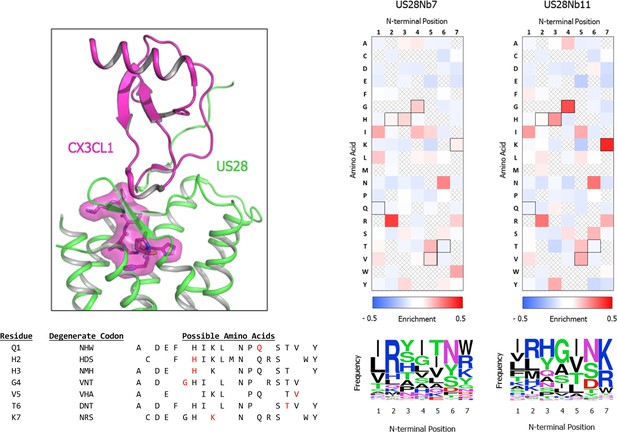
CX3CL1 ‘Site 2’ library design.
top left, CX3CL1 amino acids included in library design are highlighted (PDB: 4xt1). bottom left, Codons used and amino acids included at each position of the library (wild-type amino acid identity in red). right, Enrichment of amino acid frequency by chemokine N-terminal position after selection with either US28Nb7 or US28Nb11 as compared to the naive, unselected CX3CL1 library as determined by deep sequencing of the respective samples.
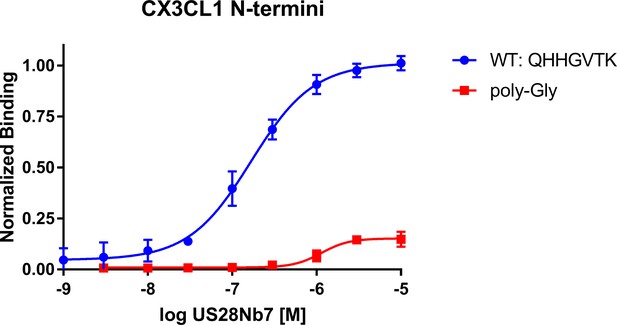
Effect of minimal chemokine Site 2 on US28Nb7 binding.
CX3CL1 in which the 7 N-terminal residues are all mutated to glycine is yeast-displayed and stained with US28Nb7. All data points are normalized to binding at 1 µM US28Nb7 with wild-type CX3CL1.
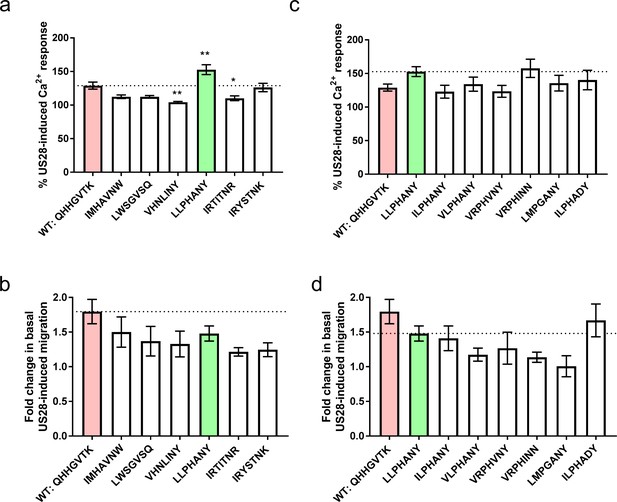
Signaling behavior or selected CX3CL1 library variants.
(a) Calcium and, (b) migration responses of diverse chemokines revealed by deep sequencing at 100 nM ligand. Dotted lines indicate wild-type CX3CL1 activity (red); all statistics are relative to this. CX3CL1.35, selected for further study, is indicated in green. (c) Calcium and, (d) migration responses of CX3CL1.35-related sequences from deep sequencing at 100 nM ligand. Dotted lines indicate CX3CL1.35 (green) activity; all statistics are relative to this. All data are given as mean ± s.e.m. of at least three independent biological replicates. * p<0.05, ** p<0.01, *** p<0.001 by one-way ANOVA (Dunnett’s test).
-
Figure 3—source data 1
Full statistics for selected CX3CL1 library variants.
Exact p-values for chemokines with respect to the listed reference using one-way ANOVA (Dunnett’s test).
- https://doi.org/10.7554/eLife.35850.017
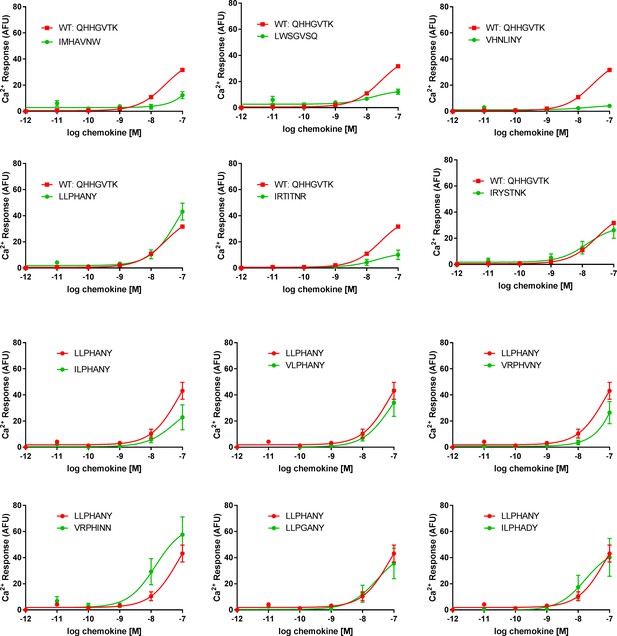
Calcium signaling dose-response plots for selected CX3CL1 library variants.
All data are given as mean ± s.e.m. of at least three independent biological replicates.
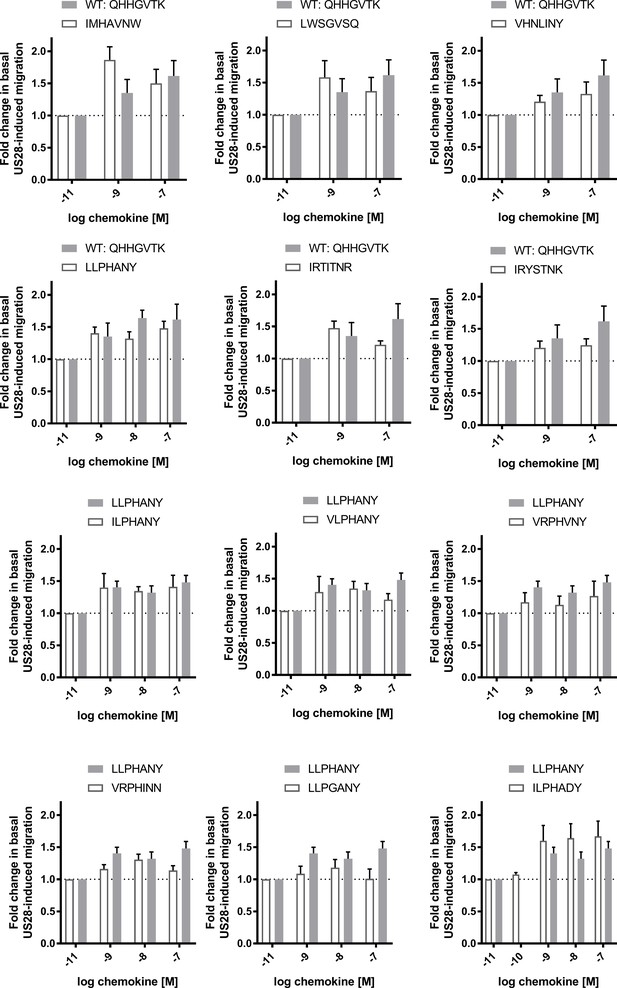
Cell migration dose-response plots for selected CX3CL1 library variants.
All data are given as mean ± s.e.m. of at least three independent biological replicates.
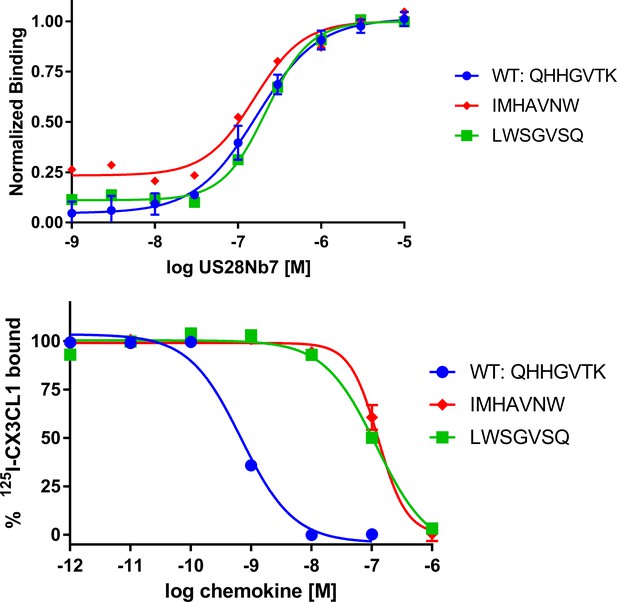
Characterization of representative CX3CL1 library variants for receptor binding.
top, Dose-response plots of yeast displayed CX3CL1 library variants stained with US28Nb7. bottom, Radioligand competition plots of cold, recombinant CX3CL1 library variants with I125-wild-type CX3CL1.
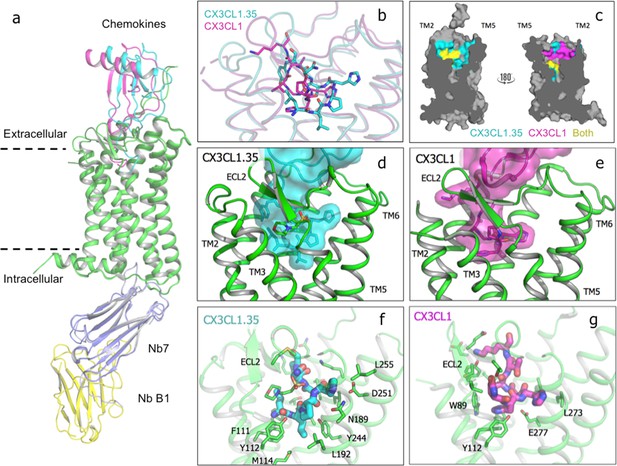
Crystal structure of US28 bound to engineered CX3CL1 and comparison with the wild-type chemokine.
(a) CX3CL1.35-US28 and CX3CL1-US28 structural alignment based on US28 transmembrane helices (PDB:4xt1). (b) Wild type and engineered chemokine N-termini trace different paths in US28’s binding pocket. (c) Cutaway image of US28 with chemokine contacts highlighted for CX3CL1.35 (teal), CX3CL1 (purple), or both (yellow). (d) CX3CL1.35 fills the entire binding pocket. (e) CX3CL1 hugs the TM2 side of US28. (f) US28 side chains contacted by CX3CL1.35 in the receptor binding pocket. (g) US28 side chains contacted by CX3CL1 in the receptor-binding pocket.
-
Figure 4—source data 1
CX3CL1.35 interactions with US28Nb7.
- https://doi.org/10.7554/eLife.35850.024
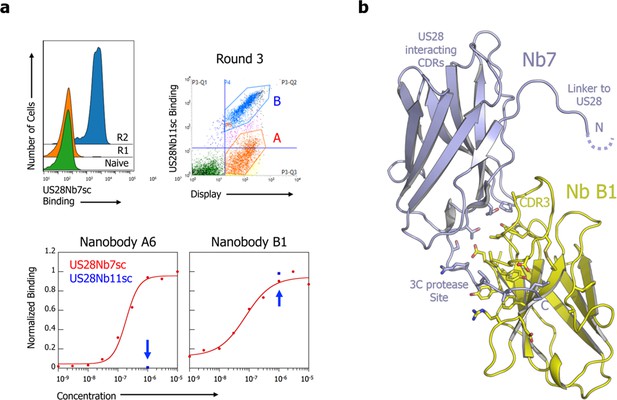
Nanobody B1 selections and binding interface.
(a) top, Selections increased US28Nb7 binding of the yeast-displayed nanobody library and yielded promiscuous binders (population B in round 3), including B1, and those specific to US28Nb7 (population A in round 3) like A6. bottom, On-yeast titration of the selected nanobodies against US28Nb7 (red). Single concentration staining of the nanobodies against US28Nb11 (blue) was performed to confirm specificity. (b) Details of the CX3CL1.35-US28Nb7-Nanobody B1 structure showing the interface between nanobodies 7 and B1. Nanobody B1 recognizes the conserved nanobody tail with the scar of a 3C protease site.
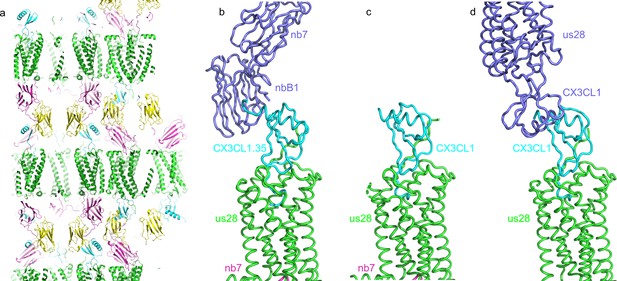
Crystal packing of CX3CL1.35-US28Nb7-nanobody B1 complex structure.
(a) Crystal lattice packing of the CX3CL1.35-US28Nb7-nanobody B1 complex. (b) The crystal lattice is stabilized by contact between CX3CL1.35 (cyan) and a symmetry-related nanobody B1 (slate blue). (c) In CX3CL1-US28-Nb7, CX3CL1 doesn’t make any crystal contacts. (d) In CX3CL1-US28, the C-terminus of CX3CL1 contacts a symmetry-related copy of itself (slate blue).
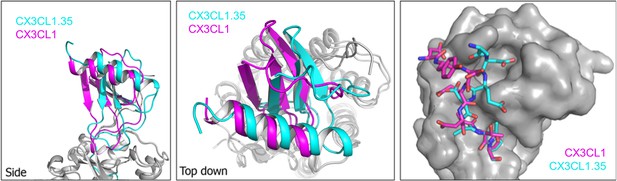
US28 structural comparisons.
left, Side view of structural alignment based on US28 transmembrane helices showing CX3CL1 (purple)- and CX3CL1.35 (teal)-bound US28 structures. middle, Top-down view. right, Alignment of the identical globular cores (residues 8–62) of CX3CL1 and CX3CL1.35 with the N-terminal peptide of US28 (purple: CX3CL1-bound, teal: CX3CL1.35-bound).
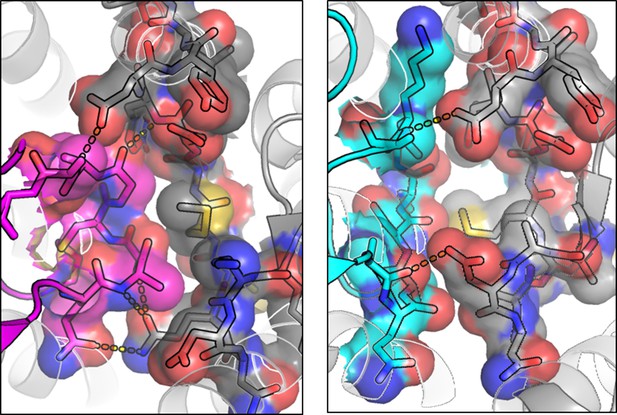
CX3CL1.35 ‘Site 1.5’ ECL2 contacts.
left, Top-down view of CX3CL1 (purple) interactions with US28 ECL2 (gray). right, Top-down view of CX3CL1.35 (teal) interactions with US28 ECL2 (gray).
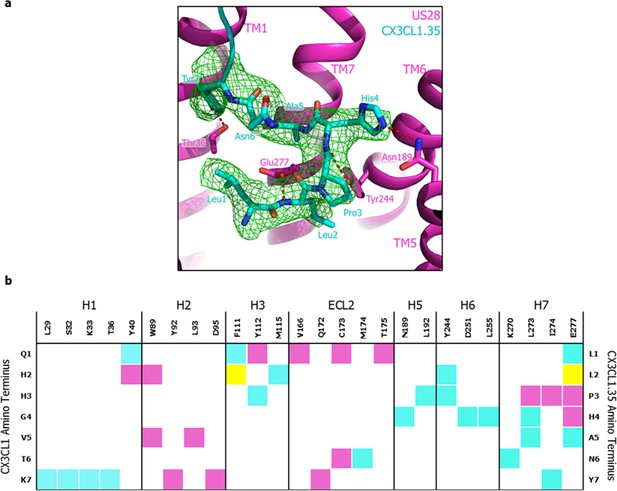
CX3CL1.35 ‘Site 2’ and receptor contacts.
(a) mFo-DFc simulated annealing omit map (green) is contoured at 3.0 sigma around the N-terminal residues of CX3CL1.35. (b) ‘Site 2’ contacts between US28 and wild-type CX3CL1 (purple), CX3CL1.35 (teal), or both chemokines (yellow).
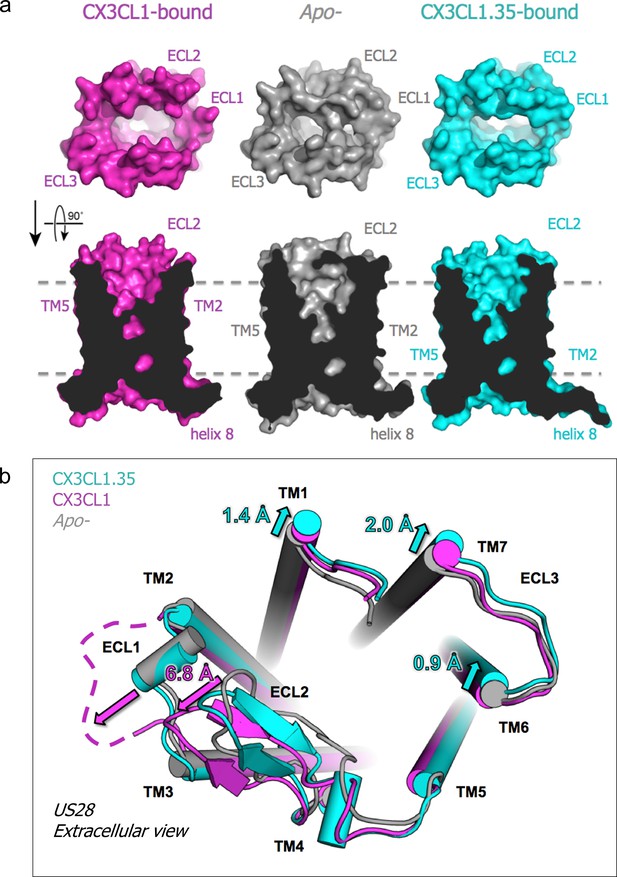
Chemokine-dependent conformational changes in US28.
(a) Top-down and side views of apo-, CX3CL1-, and CX3CL1.35-bound US28 showing expanded access to the receptor binding pocket when chemokine is present. (b) Structural alignment based on US28 transmembrane helices showing unique US28 conformational changes caused by each chemokine. (PDB: 4xt1). Crystallographic data and refinement statistics are summarized in Figure 5—source data 1.
-
Figure 5—source data 1
Summary of crystallization and crystallographic statistics.
- https://doi.org/10.7554/eLife.35850.027
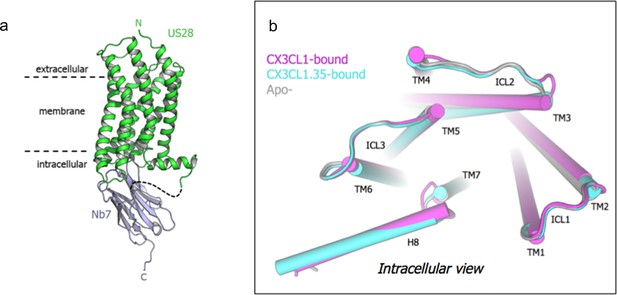
Apo- structure and comparison of intracellular faces.
(a) Overall structure of apo-US28Nb7. (b) Intracellular view of US28 in apo-, CX3CL1-bound, and CX3CL1.35-bound structures aligned on US28 transmembrane helices.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (human cytomegalovirus) | US28 (unique short region) | PMID 7961796 | strain TOWNE | |
| Strain, strain background (Saccharomyces cerevisiae) | EBY100 | Gift from Prof. Dane Wittrup (PMID 17406305) | Yeast cells | |
| Cell line (Spodoptera frugiperda) | SF9 | ATCC | CTL-1711 | Insect cells used for baculovirus production |
| Cell line (Trichoplusia ni) | Hi5 | Invitrogen | BTI-TN-5B1-4 | Insect cells used for baculovirus expression of NbB1 |
| Cell line (Homo sapiens) | HEK293 | ATCC | CRL-1573 | Mammalian cells used for Ca2+ signaling assay |
| Cell line (Homo sapiens) | HEK293-US28 wt | PMID 23303826 | Mammalian cells used for Ca2+ signaling assay and IP3 assays | |
| Cell line (Homo sapiens) | HEK293S GnTI- | Gift from Prof. H. Gobind Khorana (PMID 12370423) | Mammalian cells used for baculovirus expression of US28 variants and chemokines | |
| Cell line (Homo sapiens) | Flp-In TREx 293 | Invitrogen | R78007 | Hamster cells used for β-arrestin assay |
| Cell line (Chinese hamster ovary) | CHO-K1 EA-arrestin | DiscoverixRx | 93–0164 | |
| Transfected construct (β-arrestin recruitment) | US28 wt/ProLink/b-galactose | This report | Vector provided by DiscoverixRx | |
| Antibody | Anti-protein C (mouse IgG1) | ATCC | HB-9892 | Antibody used for staining yeast bound to protein C tagged target proteins. Purified from HPC-4 MOUSE HYBRIDOMA for Alexa647 labeling. |
| Antibody | Anti-FLAG M1 (mouse IgG2a) | Gift from Prof. Brian Kobilka (PMID 17962520) | Antibody used for US28 purification. Purified from M1 HYBRIDOMA to prepare anti-FLAG M1 affinity resin | |
| Antibody | Myc-Tag (9B11) Mouse mAb (Alexa Fluor 488 Conjugate) | Cell Signaling Technology | 2279 | Antibody used for staining yeast properly displaying proteins of interest with Myc-tag |
| Recombinant DNA reagent | BestBac Linearized Baculovirus DNA 2.0, Exp ression Systems, 91–002 | Expression Systems | 554739 | |
| Peptide, recombinant protein | CCL3 | Peprotech | 300–08 | |
| Peptide, recombinant protein | CCL5 | Peprotech | 300–06 | |
| Peptide, recombinant protein | CX3CL1 | Peprotech | 300–31 | |
| Commercial assay or kit | PathHunter β-arrestin assay | DiscoverixRx | ||
| Commercial assay or kit | MiSeq v2 2 × 150 | Illumina | MS-102–2002 | |
| Commercial assay or kit | MiSeq v2 2 × 250 | Illumina | MS-102–2003 | |
| Chemical compound, drug | Alexa Fluor 647 NHS Ester (Succinimidyl Ester) | Thermo Fisher Scientific | A37573 | Labeling reagent for anti-protein C antibody |
| Chemical compound, drug | monoolein | Sigma | M7765 | For LCP |
| Chemical compound, drug | cholesterol hemisuccinate tris salt | Anatrace | CH210 | For membrane protein purification and yeast staining buffer |
| Chemical compound, drug | cholesterol | Sigma | C8667 | For LCP |
| Chemical compound, drug | n-dodecyl-β-D-maltoside | Anatrace | D310 | For membrane protein SEC buffer |
| Chemical compound, drug | n-dodecyl-β-D-maltoside | Anatrace | D310S | For membrane protein solubilization buffer, affinity column buffer, and yeast staining buffer |
| Software, algorithm | Prism7 | GraphPad | ||
| Software, algorithm | XDS | PMID 20124692 | Data integration, scaling, space-group assignment and merging | |
| Software, algorithm | Phaser | PMID 19461840 | Molecular replacement | |
| Software, algorithm | Phenix suite | PMID 20124702 | Structure refinement | |
| Software, algorithm | Coot | PMID 20383002 | Structural model building | |
| Software, algorithm | PyMol | Schrödinger | Structural visualization/ figure preparation | |
| Software, algorithm | Pandaseq | PMID 22333067 | ||
| Software, algorithm | Geneious | Biomatters | ||
| Software, algorithm | Matlab | Mathworks | ||
| Software, algorithm | Cytoscape | PMID 14597658 | Cluster analysis visualization | |
| Software, algorithm | Cytobank | Cytobank, Inc. | Flow cytomettry visualization | |
| Software, algorithm | KaleidaGraph | Synergy Software | ||
| Software, algorithm | Clustering algorithm | PMID 24855945 | ||
| Other | CNBr-Activated Sepharose 4 Fast Flow | GE Healthcare | 17098101 | |
| Other | LS columns | Miltenyi | 130-042-401 | |
| Other | Anti-Cy5/Anti-Alexa Fluor 647 MicroBeads | Miltenyi | 130-091-395 | |
| Other | MidiMACS Magnetic Separator | Miltenyi | 130-042-302 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.35850.028





