A population of adult satellite-like cells in Drosophila is maintained through a switch in RNA-isoforms
Figures
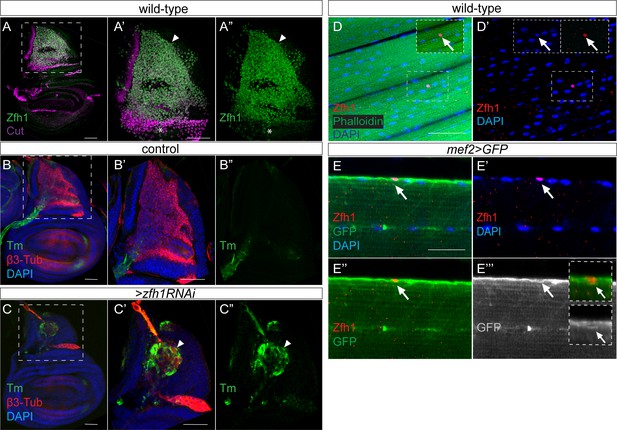
Zfh1 expression and function in MPs and in adult pMPs.
(A–A”) Zfh1 (Green) and Cut (Purple) expression in MPs associated with third instar wing discs, (A’–A’’) higher magnification (3X) of boxed region in A. Zfh1 is present in all MPs, but those with highest Cut expression have lower levels of Zfh1 (asterisk). Scale bars: 50 μM, (n > 30 wing discs from three biological replicates). (B–C) Down regulation of zfh1 induces premature differentiation of the MPs (arrowhead in C’-C’’). β3-Tubulin (β3-Tub, Red) and Tropomyosin (Tm, Green) expression in control (B, 1151-Gal4 > wRNAi) and Zfh1 depleted (C, 1151-Gal4 > zfh1 RNAi) third instar wing discs, (B’–C’’) higher magnification (3X) of boxed regions in B and C. (n > 20 wing discs; from three biological replicates). (D–D’) Zfh1 expression (red) indicates the existence of persistent muscle progenitors (pMPs; arrows) associated with the muscle fibres (Phalloidin (Green), DNA/Nuclei (Blue); n > 10 heminota; from three biological replicates). The immune cell marker P1 was included in the immunostaining and is absent from the pMPs (see Figure 1—figure supplement 2). Scale bars: 50 μM. (E–E’’’) Zfh1 (Red) expressing pMPs (e.g. arrows in E’’’) are closely embedded in the muscle lamina of the adult indirect flight muscles and express Mef2 (myogenic cells; Mef2-Gal4 >Src::GFP, green). Nuclei (Blue), Scale bars: 25 μM, (n > 10 heminota; from two biological replicates).
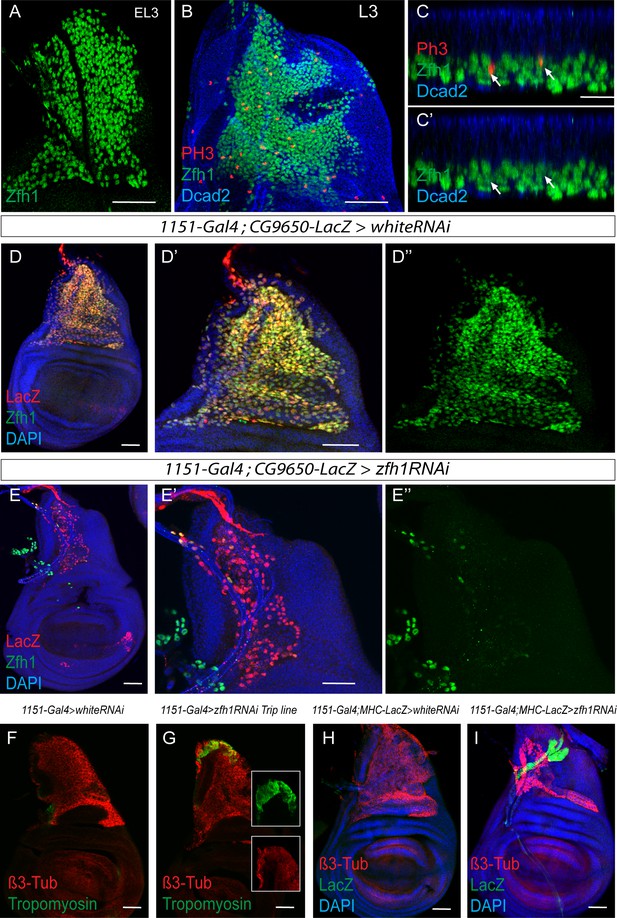
Zfh1 expression and down regulation in MPs.
(A) Zfh1 (Green) expression in MPs associated with early third instar wing discs showing that at this stage Zfh1 is uniformly expressed in all the MPs. Scale bar: 50 μM. (B) Third instar wing discs stained for Zfh1 (Green), pH3 (Red) and Cadherin (DCad2; Blue) revealing that some MPs are undertaking mitotic divisions. Scale bar: 50 μM. (n = 12 wing discs from two biological replicates). (C-C’) x-z optical section of image B showing active mitotic division of Zfh1 MPs in the most proximal layer to the disc epithelium (Arrows). Scale bar: 25 μM. (D-E) Zfh1 (Green), β-Gal (CG9650-LacZ, Red, marks MPs) and DAPI (Blue) expression in control (D, 1151-Gal4; CG9650-LacZ > white RNAi) and zfh1 down regulation (1151-Gal4 > zfh1 RNAi) with KK 103205 RNAi line. Scale bars: 50 μM. The level of Zfh1 protein is strongly reduced after zfh1 RNAi expression in the MPs (D’’ and E’’). (F-G) β3-Tubulin (β-Tub, Red) and Tropomyosin (Green) expression in control (F, 1151-Gal4 > white RNAi) and zfh1 down regulation (1151-Gal4 > zfh1 RNAi) with the Val10 TRiP RNAi line. Premature differentiation of MPs is observed using the TRiP Val10 zfh1 RNAi line comparing to the control (arrow; n > 10 wing discs for each genotype from two biological replicates). (H-I) β3-Tubulin (β-Tub, Red) and MHC-lacZ (Green) expression in control (H, 1151-Gal4; MHC-lacZ >white RNAi) and zfh1 down regulation (I, 1151-Gal4; MHC-lacZ >zfh1 RNAi) with the KK 103205 RNAi line. Premature differentiation of the MPs is observed by the expression of MHC-LacZ reporter line after zfh1 down regulation (n = 20 wing discs from two biological replicates).
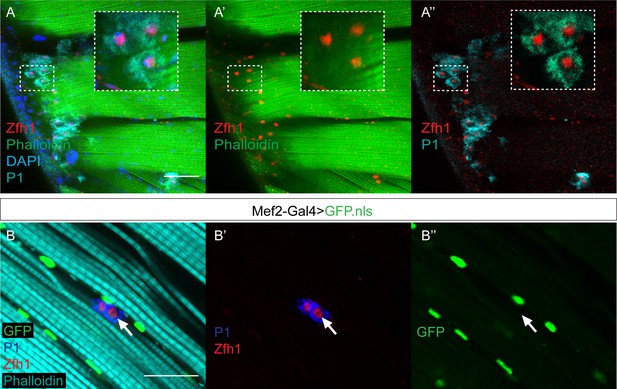
A population of plasmatocytes associated with the adult flight muscles expresses Zfh1 but not Mef2.
(A-A’’) Wild type indirect flight muscles stained for P1 (Cyan), Zfh1 (Red), Phalloidin (Green) and DAPI (Blue). P1 expression indicates the presence of phagocytic immune cells, which are also positive for Zfh1. Insets: boxed regions magnified 4X. Scale bar: 50 μM. (n = 27 heminota from two biological replicates). (B-B’’) Indirect flight muscles from flies expressing Mef2-Gal4 > UAS GFPnls stained for GFP (Green), P1 (Blue), Zfh1 (Red) and Phalloidin (Cyan). The Zfh1 immune cells detected in the IFMs lack Mef2 expression (Arrows B-B’’). Scale bar: 25 μM. (n = 11 heminota from two biological replicates).
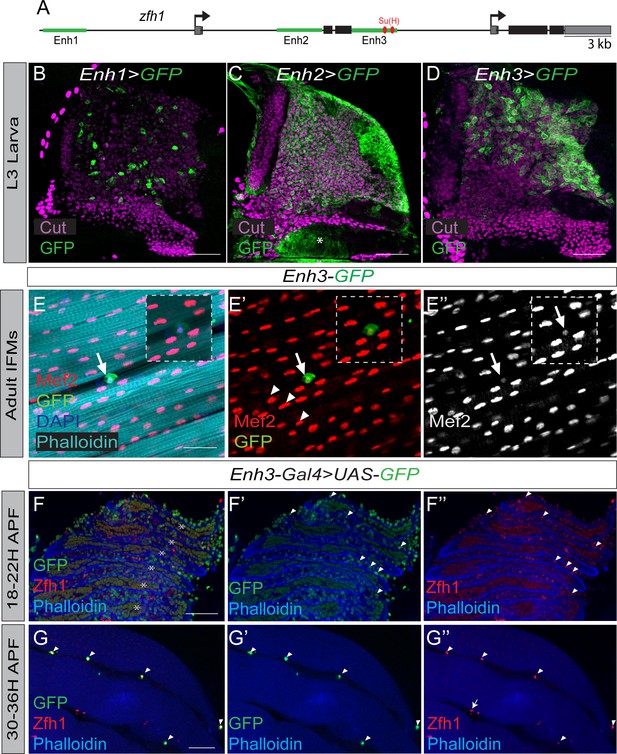
Regulation of zfh1 in MPs and adult pMPs.
(A) Schematic view of zfh1 genomic region, zfh1 regulatory enhancers are represented by green rectangles and arrows indicate transcription start-sites. Coding exons and untranslated regions are represented in black and grey boxes, respectively. (B-D) Three different zfh1 enhancers are active in the MPs (labelled with Cut, purple). Enh1 (VT050105, B) drives GFP (Green) in a subset of scattered MPs; Enh2 (VT050115, C) drives GFP throughout the MPs, and in some non-MP cells (asterisk); Enh3 (GMR35H09, D) is highly expressed in a subset of MPs located in the posterior region of the notum. Scale bars: 50 μM. (n = 30 wing discs). (E-E’’) Enh3-GFP (Green) expression is maintained in adult pMPs (characterised by low levels of Mef2, red; arrows E’-E’’) but not in differentiated muscle nuclei (high Mef2, red; arrowheads G’-G’’). Phalloidin marks muscles (Cyan) and DAPI labels all nuclei (Blue). Insets: boxed regions magnified 12.5 X. Scale bars 25 μM. (n = 10 heminota; from two biological replicates). (F-G) Muscle (IFM) preparation isolated from Enh3-Gal4 > UAS GFP pupae at 18–22 hr APF (F) or at 30–36 hr APF (G). Enh3-GFP (Green) and Zfh1 (Red) are detected in MPs. (F) At 18–22 hr Enh3-GFP activity is higher (arrowheads in E’) in some undifferentiated MPs located between muscle templates (muscles are labeled with Phalloidin, Blue, asterisks). (G) At 30–36 hr APF, Enh3-GFP (Green) activity and Zfh1 (Red) are detected in pMPs (arrowheads) and not in differentiated muscle nuclei. A few Zfh1 +ve cells do not express Enh3-GFP (Arrows G’’). Note: anti-P1, an immune cell marker, was included in the staining to exclude plasmatocytes from the analysis (see Figure 1—figure supplement 2).

Identification of zfh1 enhancers active in MPs and pMPs.
(A) Chromosome 3R: 30,765,926–30,789,202 showing the zfh1 genomic region with coding exons and untranslated regions represented in black and grey boxes, respectively. Arrows represent the transcription start-sites. Su(H) and Twist ChIP enriched regions in DmD8 cells are represented in Blue and Purple, respectively (Data from Bernard et al., 2010) (ChIP Twi) and Krejcí et al. (2009) (ChIP Su(H)). Black lines represent all GMR and VT lines tested in our study. Green lines indicate the enhancers that are active in MPs in vivo. (B-D) Zfh1 (white) expression in MPs is significantly reduced in discs from Enh3 deletion ∆Enh3 (C) compared to controls (B). In each condition light and dark shading indicates data points from two independent replicates (*p<0.05, Student t-test; n = 14 wing discs for each genotype). Scale bars: 50 μM. (E) Effect of Enh3 deletion (ΔEnh3) on zfh1 mRNA level in MPs as determined by quantitative RT-PCR. zfh1 mRNA level is significantly decreased by Enh3 deletion. **p=0.0042 was obtained using unpaired Student’s t-test. Error bars represent s.e.m of three independent experiments.
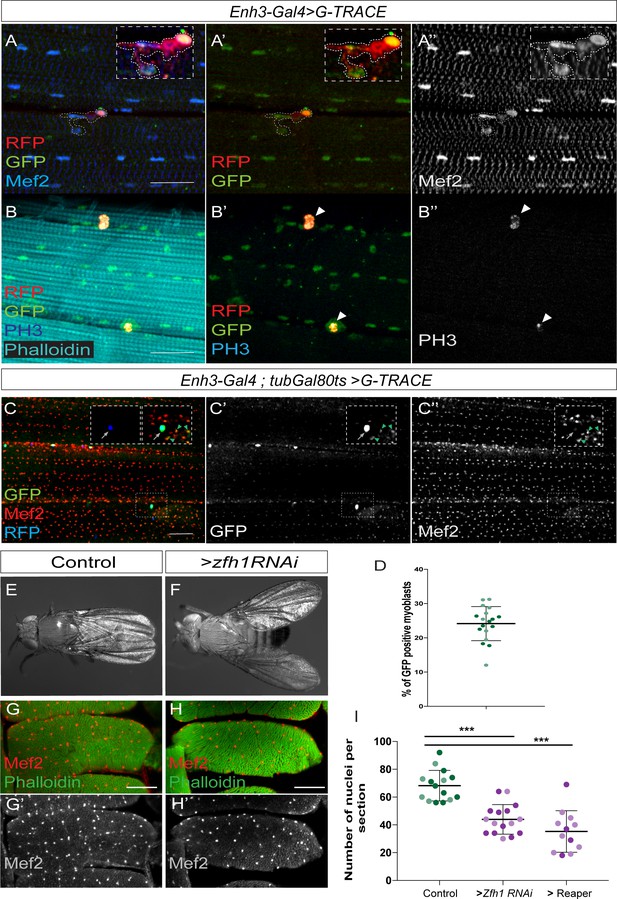
Adult pMPs contribute to muscle homeostasis.
(A-A’’) Lineage tracing shows that adult pMPs contribute to muscles. Cross-section of indirect flight muscles from adult flies where Enh3-Gal4 drives expression of the G-Trace cassette; GFP (Green) indicates myoblasts that have expressed Enh3-Gal4, RFP (Red) indicates myoblasts where Gal4 is still active, Mef2 labels muscle nuclei (Blue). Note that the RFP (Red, detected with anti-RFP in A’) persists in the recently born myoblasts (Mef2), which are closely localized to the pMPs. Insets: boxed regions magnified 20 X (n = 15 heminota; from three biological replicates). (B-B’’) pMPs are mitoticaly active, indicated by anti-phosphH3 (White in B’’). (PH3 detected in 47% of pMPs, n = 80; from three biological replicates). (C-C’’) Cross-section of indirect flight muscles from adult flies (Enh3-Gal4; tubGal80ts > G Trace) where Enh3-Gal4 directed G-Trace activity was induced for 10 days after animal hatching. GFP (Green, Arrowheads, C’) indicates descendants of pMPs (Blue, Arrows); Mef2 labels muscle nuclei (Red, White). (D) Proportion of newly born myoblasts (marked by GFP; e.g. C’) relative to total number of myoblasts (marked by Mef2; e.g. C’’) in muscle preparations. (n = 18 heminota; light and dark shading indicates data points collected from two independent replicates replicates). Scale bar: 60 μM. (E-F) Prolonged zfh1 depletion in pMPs (10 days after adult hatching) leads to a ‘held out’ wings posture; dorsal view of (E) control (Enh3-Gal4; tubGal80ts > UAS wRNAi;) and (F) zfh1 depletion (Enh3-Gal4; tubGal80ts > UAS-zfh1RNAi (KK 103205)) adult flies. (G-H) Transverse sections of DLM4 muscle stained with Phalloidin (Green) and Mef2 (Red, White) from the indicated genotypes. Fewer Mef2 +ve nuclei are present in muscles when zfh1 is depleted. Scale bars: 50 μM. (I) Similar reductions in muscle nuclei occur following zfh1 depletion (Enh3-Gal4; tubGal80ts > UAS-zfh1RNAi) or following genetic ablation of pMPs, via expression of the pro-apoptotic gene reaper (rpr; Enh3-Gal4;tubGal80ts > UAS rpr). The number of nuclei per section in the indicated conditions was significantly different, light and dark shading indicates data points collected from two independent replicates (>zfh1 RNAi ***p=0.0013, n = 16;>Rpr ***p<0.0001, n = 12).
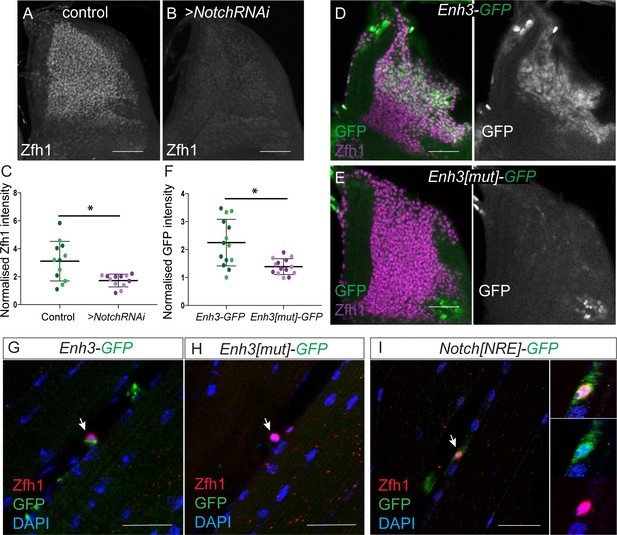
Notch directs Zfh1 expression in MPs and pMPs.
(A-C) Zfh1 level (White) is significantly reduced when Notch is down regulated. Expression of Zfh1 in MPs (A) is severely reduced in the presence of Notch RNAi (B, 1151-Gal4 > UAS NotchRNAi), Scale Bars: 50 μM. (C) Quantification of Zfh1 expression levels (*p<0.05, n = 12 wing discs in each condition, light and dark shading indicates data obtained from two independent replicates). (D-F) Enh3 (D, Enh3-GFP, Green) expression in MPs (Purple, Zfh1) is abolished when Su(H) motifs are mutated (E, Enh3[mut]-GFP). Scale bars: 50 μM. (F) Quantification of expression from Enh3 and Enh3[mut] (*p=0.022, n = 14 wing discs in each condition, light and dark shading indicates data obtained from two independent replicates). (G-H) Enh3 (G, Enh3-GFP, Green) expression in adult pMPs (red, Zfh1) is abolished when Su(H) motifs are mutated (H, Enh3[mut]-GFP, Green), DAPI (Blue) reveals all nuclei. (I) Notch[NRE]-GFP (Green) is co-expressed with Zfh1 (Red) in the pMPs associated with the indirect flight muscles; DAPI (Blue) detects all nuclei. (n = 12 heminota; from two independent replicates). In G-I anti-P1 was included to label immune cells and exclude them from the analysis. Scale bars: 25 μM.
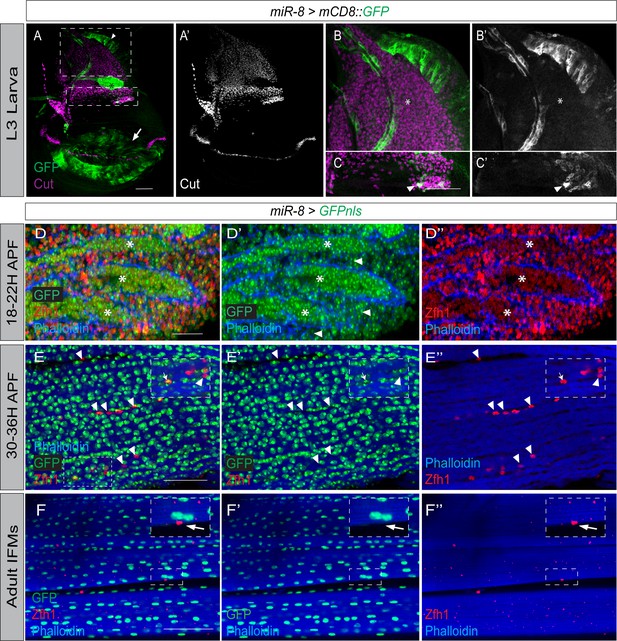
Expression dynamics of miR-8 and Zfh1 in MPs and pMPs during indirect flight muscle development.
(A-C) miR-8 (Green, miR-8-Gal4 > UAS-mCD8::GFP) is not highly expressed in MPs (Cut, Purple) but is prevalent in the wing disc pouch (Arrow), notum (Arrowhead) and air sac (Asterisk). Higher magnification shows that low level of miR-8 expression can be detected in the subset of MPs where Zfh1 is normally low (Arrowhead in C) but not in other MPs. Scale bars: 50 μM. (n > 20 wing discs; from three biological replicates). (D-D’’) IFM preparation isolated from miR-8-Gal4 > UAS-nlsGFP pupae at 18–22 hr APF. At this stage miR-8 (Green; D and D’) is co-expressed with Zfh1 (Red; D and D’’) in MPs but miR-8 is more highly expressed in differentiated myoblasts (Asterisks in D’), found within the muscles (labeled with Phalloidin, Blue), compared to the undifferentiated MPs, found between the muscles (Arrowheads in D’). In contrast, Zfh1 (D, D’) is detected at lower levels in the differentiated myoblasts compared to the undifferentiated MPs (Asterisks in D’). Scale bar: 25 μM. (E-E’’) IFM preparation isolated from miR-8-Gal4 > UAS-nlsGFP pupae at 30–36 hr APF. At this stage, mir-8 is highly detected in the differentiated muscle nuclei. The majority of the Zfh1 +ve pMPs (Red) do not express mir-8 (Green) (Arrowheads). Few Zfh1 +ve pMPs express mir-8 (Arrows). Scale bar: 50 μM. (F-F’’) In adult IFMs, mir-8 (Green) expression is absent from Zfh1 +ve (red) pMPs (Arrows, D-D’) but is present at uniformly high levels in IFMs, (Phalloidin, Blue). Scale bars: 50 μM. (n = 20 heminota; from three biological replicates).
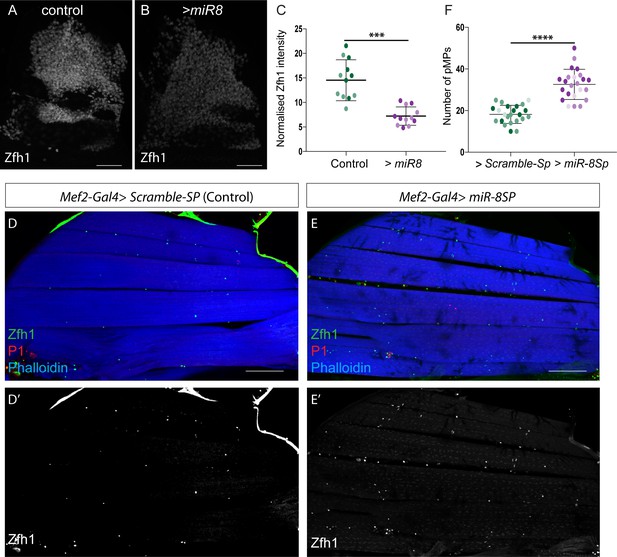
The conserved microRNA miR-8/miR-200 antagonizes zfh1 to promote muscle differentiation.
(A-C) Effect of miR-8 overexpression (1151-Gal4 > UAS-miR-8) on Zfh1 (White) protein level in MPs. Scale Bars: 50 μM. (C) Zfh1 expression is significantly reduced by miR-8 over-expression. (***p=0.0009, n = 12 wing discs in each condition, light and dark shading indicates data points from two independent replicates). (D-E) Sagittal sections of adult IFMs stained for Phalloidin (Blue), Zfh1 (Green) and P1 (Red). Down regulating miR-8 during muscle differentiation (Mef2-Gal4 > UAS-miR-8-Sp) increases the final number of adult pMPs. (F) The number of pMPs in adult IFMs in the indicated conditions was significantly different. (****p<0.0001, n = 18 adults for each genotype; light, dark and intermediate shading indicates data points from three independent replicates). Scale bars: 100 μM.
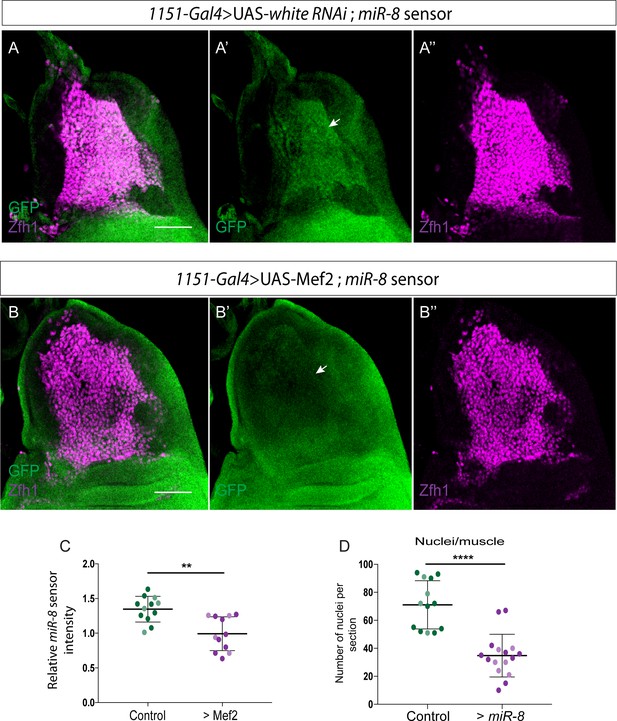
miR-8 responds to high level of Mef2.
miR-8 activity was detected using miR-8-EGFP sensor (Kennell et al., 2012). (A-B) Effect of Mef2 overexpression (1151-Gal4 > UAS-Mef2) on miR-8 sensor (Green) expression level in MPs. miR-8 sensor expression in MPs is significantly reduced by Mef2 overexpression (Arrows in A’ and B’). Zfh1 expression (Red) was used to visualize the MPs. (C) Quantification of miR-8 Sensor intensity in the indicated conditions. (**p<0.01, n = 12 wing discs for each indicated genotype). In each condition light and dark shading indicates data points from two independent replicates. Scale bars = 50 μM. (D) Prolonged miR-8 overexpression in the adult pMPs (Enh3-Gal4; Gal80ts > UAS-miR-8) significantly affects the muscle homeostasis. (****p<0.0001, n = 16 for each genotype). In each condition light, dark and intermediate shading indicates data points from three independent replicates.
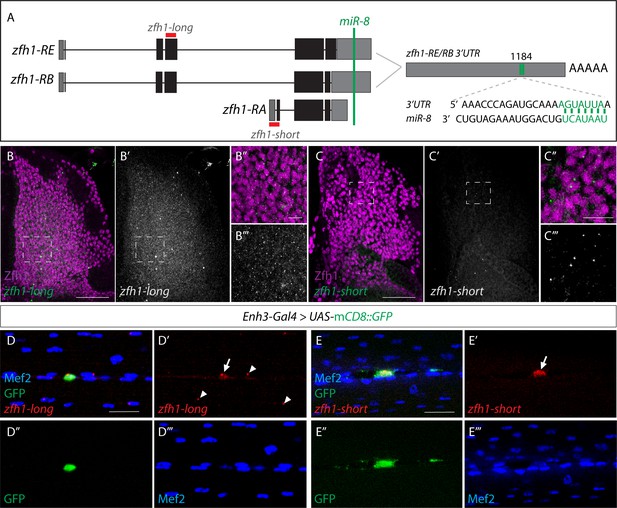
Transcriptional dynamics of zfh1 isoforms in MPs and pMPs.
(A) Schematic representation of zfh1 isoforms. zfh1-short (zfh1-RA) is initiated from a different transcription start site and has shorter 3’UTR that lacks the target site for miR-8 (Green; Antonello et al., 2015) present in zfh1-long isoforms (zfh1-RB, zfh1-RE); the position of the miR-8 seed sites in zfh1-long 3’ UTR are depicted. Non-coding exons and coding exons are depicted by grey and black boxes respectively, red lines indicate the probes used for FISH experiments in B-E. (B-C) zfh1-long is present uniformly in MPs. (n > 10 wing discs from two replicates; B, Green and B’, White) whereas zfh1-short is only detected in a few MPs (n > 15 wing discs from three replicates; C, Green and C’, White), detected by in situ hybridisation in wild type third instar wing discs stained for Zfh1 (Purple). Scale bars: 10 μM. (B’ B’’, C’ C’’) Higher magnifications of boxed regions (Scale bars: 50 μM). (D-E) In adult IFMs zfh1-long is detected in the pMPs (n = 11 pMPs from two replicates, arrow in D’) and in some differentiated nuclei located in their vicinity (arrowheads in D’) whereas zfh1-short is only present in pMPs (n = 15 pMPs from two replicates, arrow in E’). Enh3 expression (Green, Enh3-Gal4 > UAS-mCD8GFP) labels adult pMPs and Mef2 (Blue) labels all muscle nuclei. Scale bars: 20 μM.

Zfh1-short isoform is capable of blocking muscle differentiation.
(A) Domain structure of Drosophila Zfh1 protein isoforms E, B and A. Purple bars indicate the predicted zinc fingers, Green bar indicates the predicted homeodomain. All three protein-isoforms contain the core zinc finger and homeodomains needed for Zfh1 DNA-binding activity. Isoforms E and B contain two additional N-terminal zinc fingers (Postigo et al., 1999). (B-H) Zfh1-short protects the MPs from differentiation. B. Overexpression of Mef2 in the MPs (1151-Gal4 > Mef2; white-RNAi) induces premature muscle differentiation revealed by the ectopic expression of the MHC-lacZ reporter (Purple). This phenotype is associated with reduced MPs (β3-Tub, Green). (C) Co-expression of Mef2 along with zfh1-short (1151-Gal4 > Mef2; zfh1-short) rescues the premature differentiation phenotype. (D-E) Quantification of the differentiation area (D) and the progenitor area (I) in the indicated genotypes. (D, p=0.0025 and E, p=0.0002. n = 12 wing discs; from two biological replicates).
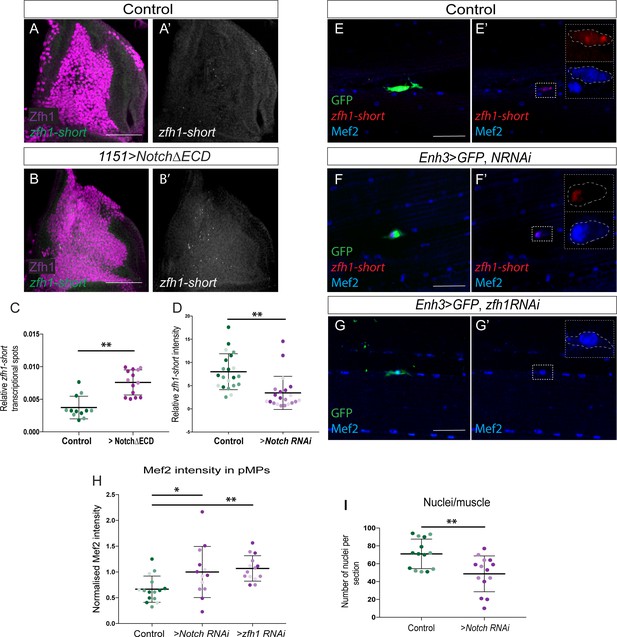
zfh1-short regulation by Notch is important to maintain muscle homeostasis.
(A-C) Expression of an activated Notch (1151-Gal4 > UAS-NΔECD) in MPs induces ectopic zfh1-short transcription. In situ hybridisation detecting zfh1-short (Green) in MPs (Zfh1, Purple) with wild type (A-A’) or elevated Notch activity (B-B’). Scale bars: 50 μM. (C) Quantification showing significant increase in zfh1-short transcriptional dots upon Notch up regulation, relative to total number of MPs (**p<0.01, Student t-test; n = 14 wing discs for each genotype, light and dark shading indicates data points from two independent replicates). (D-H) Notch depletion leads to a severe decrease in zfh1-short (Red) (E’-F’ and D) in pMPs (Green; Enh3-Gal4; UAS-mCD8GFP > UAS Notch-RNAi; tubGal80ts) and to an increase in Mef2 levels (Blue) (E-F’ and H). (G-G’ and H) zfh1 depletion in the pMPs (Enh3-Gal4; UAS-mCD8GFP > UASzfh1 RNAi; tubGal80ts) leads to an increase in Mef2 levels (Blue). Scale bars: 25 μM. Quantifications of zfh1-short (D) and Mef2 (H) in the indicated conditions show that the levels are significantly different (D, n = 21 pMPs for each genotype (**p<0.01); H, n = 15 pMPs for Control RNAi and n = 13 pMPs for Notch RNAi (*p<0.05) and n = 14 pMPs for zfh1 RNAi (**p<0.01)). In each condition light, dark and intermediate shading indicates data points from three independent replicates. (I). Prolonged Notch depletion in the pMPs (Enh3-Gal4; Gal80ts > UAS Notch RNAi) affects the muscle homeostasis. (**p<0.01, n = 14 for each genotype, light and dark shading indicates data points from two independent replicates).
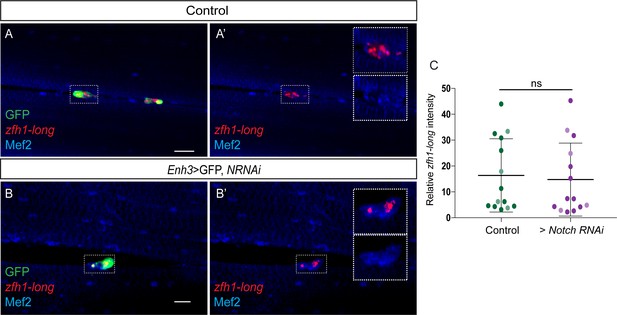
Notch activity is not necessary for zfh1-long (Red) transcription (A’-B’ and C) in pMPs (Green; Enh3-Gal4; UAS-mCD8GFP > white RNAi).
Enh3-Gal4 was used to drive expression of control RNAi (A-A’), Notch RNAi (B,B’) specifically in pMPs (Enh3-Gal4; UAS-mCD8GFP > UAS Notch RNAi; tubGal80ts). Mef2 (Blue) marks all muscle and pMPs nuclei. Scale bars: 25 μM. (C) Quantification of zfh1-long intensity in the indicated conditions. zfh1-long transcriptional intensity measured from control RNAi and Notch RNAi were not statistically different (ns) (p=0.8190, n = 15 (control RNAi) and n = 14 (Notch RNAi)). zfh1-long transcriptional intensity quantifications were generated from two independent replicates, indicated by light and dark shading.
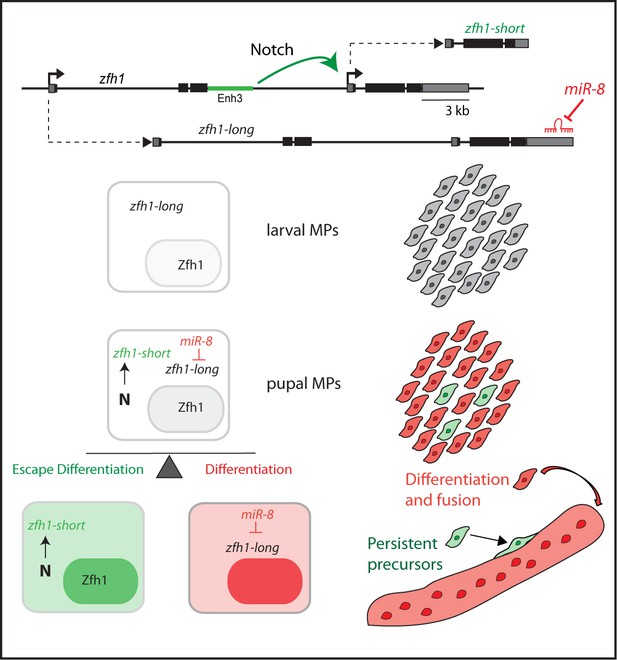
Model summarizing the role of alternate zfh1 isoforms in the maintenance of adult pMPs.
zfh1-long (Grey) is expressed in all MPs at larval stage. Silencing of zfh1-long by miR-8 (Red) facilitates the MPs differentiation. zfh1-short (Green) transcription is driven and maintained in pMPs by a Notch responsive element (Enh3, Green rectangle), which may also contribute to zfh1-long regulation. Because zfh1-short is insensitive to miR-8, Zfh1 protein is maintained in pMPs, enabling them to escape differentiation and persist as MPs in the adult.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (D. melanogaster) | zfh1 | NA | FLYB:FBgn0004606 | |
| Gene (D. melanogaster) | Notch | NA | FLYB:FBgn0004647 | |
| Gene (D. melanogaster) | miR-8 | NA | FLYB:FBgn0262432 | |
| Genetic reagent (D. melanogaster) | Enh3-Gal4 | Janelia Research Campus | BDSC: 49924, FLYB: FBtp0059625 | FlyBase symbol: P{GMR35H09-GAL4}attP2 |
| Genetic reagent (D. melanogaster) | zfh1 RNAi (kk 103205) | Vienna Drosophila RNAi Center | VDRC: 103205 | |
| Genetic reagent (D. melanogaster) | miR-8-Gal4 | Kyoto Stock Center | DGRC: 104917 | Genotype: y[*] w[*]; P{w[+mW.hs]=GawB}NP5247/CyO, P{w[-]=UAS lacZ.UW14}UW14 |
| Genetic reagent (D. melanogaster) | UAS-miR-8-Sp | Bloomington Stock Center | BDSC: 61374, FLYB: FBst0061374 | Genotype: P{UAS-mCherry.mir-8.sponge.V2}attP40/CyO; P{UAS-mCherry.mir-8.sponge.V2}attP2 |
| Genetic reagent (D. melanogaster) | Enh3-GFP | This paper | ||
| Genetic reagent (D. melanogaster) | ∆Enh3 | This paper | ||
| Genetic reagent (D. melanogaster) | UAS-zfh1-short | This paper | UAS-Zfh1-short construct provided by BDGP, Clone # UF5607 | |
| Genetic reagent (D. melanogaster) | Notch[NRE]-GFP | Sarah Bray (Cambridge, UK) | ||
| Antibody | anti-Zfh1 | Ruth Lehmann (New York, USA) | ||
| Antibody | anti-Mef2 | Eileen Furlong (Heidelberg, Germany) | ||
| Recombinant DNA reagent | pCFD4 | Addgene | Addgene # 49411 | |
| Recombinant DNA reagent | pDsRed-attP | Addgene | Addgene # 51019 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.35954.017





