Matrin 3-dependent neurotoxicity is modified by nucleic acid binding and nucleocytoplasmic localization
Figures
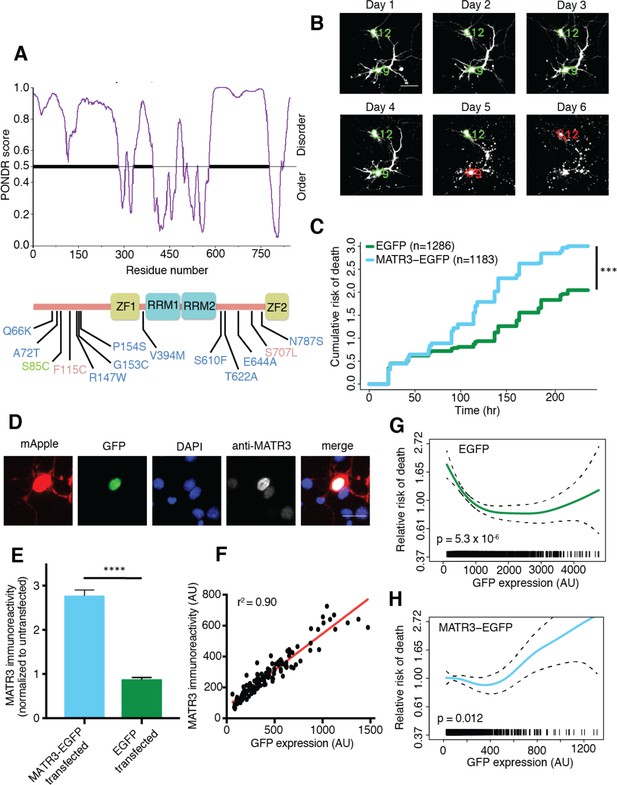
MATR3 overexpression results in dose-dependent neurodegeneration.
(A) Diagram of MATR3 showing nucleic acid-binding domains as well as the distribution of pathogenic mutations implicated in ALS (blue), ALS/FTD (red), and ALS/distal myopathy (green) within domains predicted to be disordered by PONDR VSL2 (Peng et al., 2006). (B) Longitudinal fluorescence microscopy (LFM) allows unique identification and tracking of thousands of primary neurons (green outlines) transfected with fluorescent proteins, as well as monitoring of cell death (red outlines), indicated by loss of fluorescence signal and changes in morphology. (C) MATR3-EGFP expressing neurons exhibited a higher risk of death compared to neurons expressing only EGFP, as quantified by the hazard ratio (HR) (HR = 1.48; EGFP n = 1286, MATR3-EGFP n = 1183; ***p<2×10−16, Cox proportional hazards). (D–E) Transfection of neurons with MATR3-EGFP resulted in a 2.8-fold increase in anti-MATR3 immunoreactivity over untransfected cells (MATR3-EGFP n = 133, untransfected n = 136, EGFP n = 113, ****p<0.0001, one-way ANOVA with Tukey’s post-hoc test). (F) On a single-cell basis, GFP fluorescence is directly proportional to anti-MATR3 reactivity (p<0.0001, r2 = 0.90; linear regression). (G) Penalized spline modeling confirmed a protective effect associated with higher EGFP expression that plateaus at ~1500 arbitrary units (AU); (p=5.3×10−6, penalized spline regression). (H) However, penalized spline analysis showed no relationship between expression and survival at low and medium expression but a significant increase in risk of death with high MATR3-EGFP levels (p=0.012; penalized spline regression). Scale bars in (B) and (D), 20 μm.
-
Figure 1—source data 1
Survival and GFP intensity data for Figures 1C,G, and H and GFP intensity and immunoreactivity data used for Figures 1E and F.
- https://doi.org/10.7554/eLife.35977.003
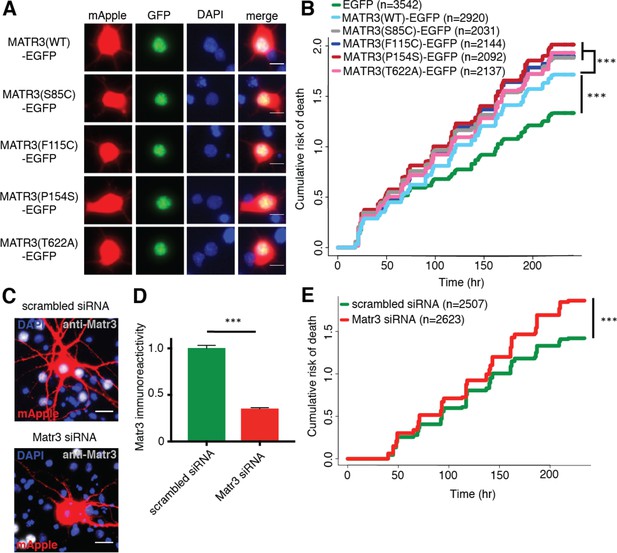
Neurons are susceptible to both gain-of-function and loss-of-function MATR3 toxicity.
(A) In primary rodent cortical neurons, the S85C, F115C, P154S, and T622A disease-associated MATR3 mutants have the same granular nuclear distribution as MATR3(WT)-EGFP. (B) All four disease mutations display a subtle but significant increase in toxicity compared to MATR3(WT)-EGFP (comparing to MATR3(WT)-EGFP, n = 2920: MATR3(S85C)-EGFP, HR = 1.16, n = 2031, ***p=3.79×10−6; MATR3(F115C)-EGFP, HR = 1.14, n = 2144, ***p=5.57×10−5; MATR3(P154S)-EGFP, HR = 1.24, n = 2092, ***p=1.77×10−11; MATR3(T622A)-EGFP, HR = 1.14, n = 2137, ***p=6.02×10−5; Cox proportional hazards). (C–D) siRNA targeting the endogenous rat Matr3 reduced MATR3 antibody reactivity by approximately 65% (scrambled siRNA, n = 576; anti-Matr3 siRNA, n = 508; p<0.0001, two-tailed t-test). (E) Neurons transfected with anti-Matr3 siRNA displayed a higher risk of death compared to those transfected with scrambled siRNA (HR = 1.20; scrambled siRNA, n = 2507; anti-Matr3, n = 2623; ***p=2.05×10−8, Cox proportional hazards). Scale bars in (A), 10 μm; scale bars in (C), 20 μm.
-
Figure 2—source data 1
Survival data for Figure 2B and E and immunoreactivity data for Figure 2D.
- https://doi.org/10.7554/eLife.35977.005
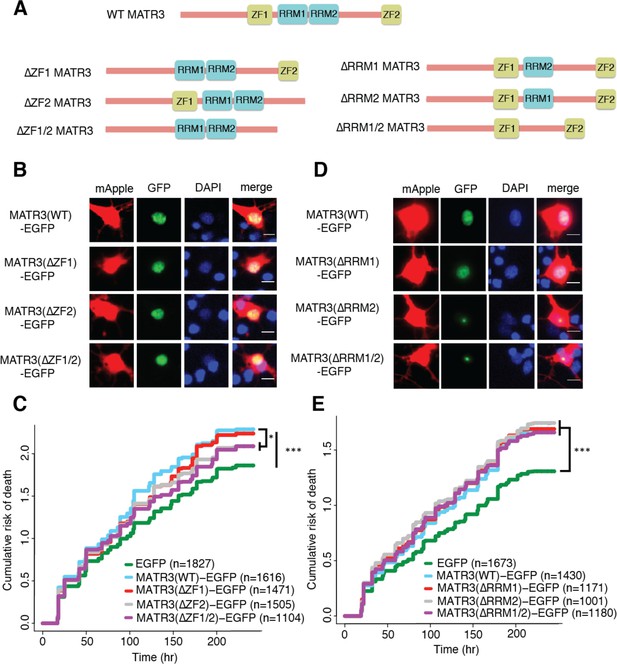
MATR3’s ZFs mediate overexpression toxicity, and its RRMs regulate subcellular distribution.
(A) Schematic of MATR3 domain deletion mutants. (B) Zinc finger (ZF) domain deletions do not change the localization of MATR3-EGFP compared to the full-length protein. (C) ZF2 deletion, either in isolation or combination with ZF1, results in modest rescue of overexpression toxicity (comparing to MATR3(WT)-EGFP, n = 1616: MATR3(∆ZF1)-EGFP, HR = 0.94, n = 1471, p=0.10; MATR3(∆ZF2)-EGFP, HR = 0.93, n = 1505, *p=0.040; MATR3(∆ZF1/2)-EGFP, HR = 0.90, n = 1104, **p=0.0093; Cox proportional hazards). (D) While MATR3(∆RRM1)-EGFP exhibits the same localization as MATR3(WT)-EGFP, deletion of RRM2 results in redistribution into intranuclear granules. (E) RRM deletion had little effect on MATR3-mediated toxicity (comparing to MATR3(WT)-EGFP n = 1430: MATR3(∆RRM1)-EGFP, HR = 1.05, n = 1171, p=0.25; MATR3(∆RRM2)-EGFP, HR = 1.09, n = 1001, p=0.066; MATR3(∆RRM1/2)-EGFP, HR = 1.04, n = 1180, p=0.42). Scale bars in (B) and (D), 10 μm.
-
Figure 3—source data 1
Survival data for Figures 3C and E.
- https://doi.org/10.7554/eLife.35977.008
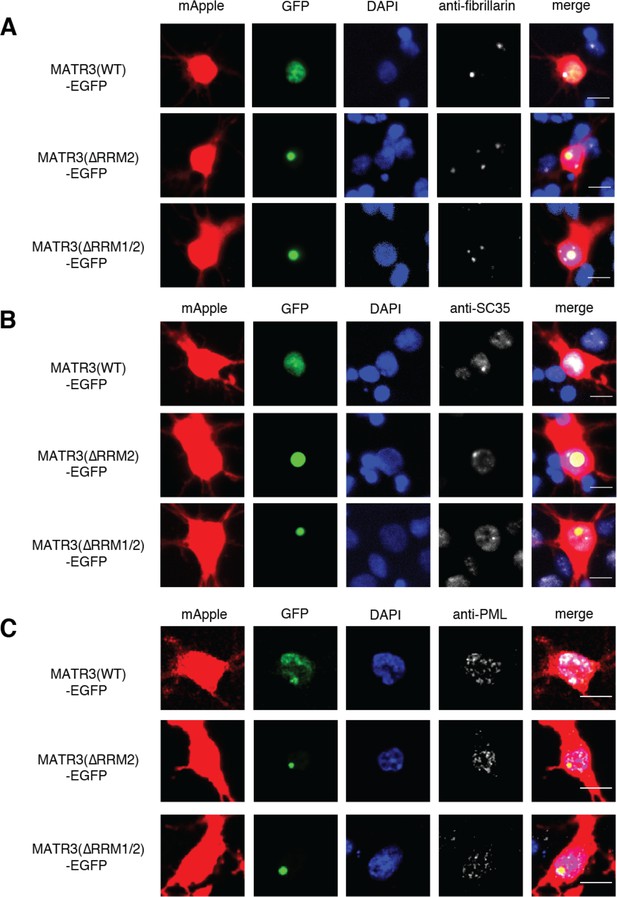
MATR3(∆RRM2)-EGFP and MATR3(∆RRM1/2)-EGFP do not join preexisting subnuclear organelles.
(A-C) Immunostaining for nucleoli, speckles, and PML bodies using antibodies against fibrillarin, SC35, and PML, respectively, show no colocalization between MATR3(∆RRM2)-EGFP and MATR3(∆RRM1/2)-EGFP granules and these nuclear structures. Scale bars in (A), (B), and (C), 10 μm.
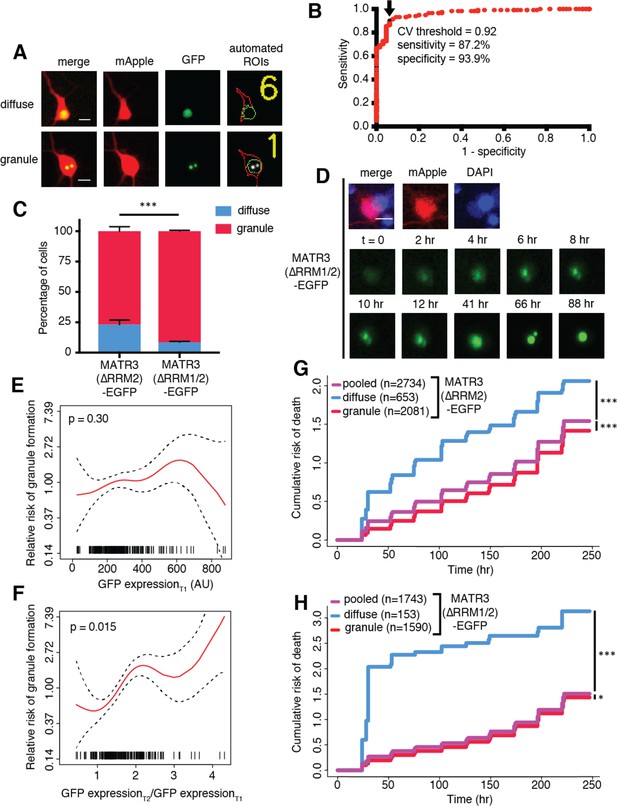
MATR3(∆RRM2)-EGFP and MATR3(∆RRM1/2)-EGFP are highly neurotoxic in their diffuse form.
(A) Automated analysis of MATR3-EGFP distribution in transfected primary cortical neurons. Regions of interest (ROIs) were drawn around the cell body (marked by mApple fluorescence, red) and diffuse MATR3-EGFP (indicated by GFP fluorescence, green), and used to calculate a coefficient of variation (CV) representing MATR3-EGFP distribution within each ROI. (B) Receiver operating characteristic (ROC) curve for MATR3-EGFP CV values. A CV threshold of 0.92 (arrow/black point) identified cells with intranuclear MATR3-EGFP granules with 87.2% sensitivity and 93.9% specificity. (C) Using this cutoff, we determined that 1 day after transfection, 76.1% (2081/2734) of MATR3(∆RRM2)-EGFP neurons displayed intranuclear granules compared to 91.2% (1590/1743) of MATR3(∆RRM1/2)-EGFP cells (***p<0.00001, Fisher’s exact test). (D) Intranuclear granules form in a time-dependent manner in neurons expressing MATR3(∆RRM2)-EGFP and MATR3(∆RRM1/2)-EGFP. (E–F) Penalized spline models depicting the relationship between MATR3(∆RRM2)-EGFP expression on day 1 (E) or change in GFP expression between day 1 and day 2 (F), and risk of developing an intranuclear granule by day 3. Expression level at day one was not significantly associated with risk of granule formation (E; p=0.30, penalized spline regression), but the relative increase in expression from day 1 to day 2 is (F; p=0.015, penalized spline regression). (G) For MATR3(∆RRM2)-EGFP, neurons exhibiting granules by day one displayed improved survival compared to the pooled combination of all cells. Conversely, neurons with diffusely distributed MATR3(∆RRM2)-EGFP fared far worse (comparing to the pooled condition: cells with granules n = 2081, HR = 0.86, ***p=1.02 × 10−5; cells with diffuse protein n = 653, HR = 1.75, ***p<2 × 10−16; Cox proportional hazards). (H) Neurons with MATR3(∆RRM1/2)-EGFP granules by day one similarly displayed a reduced risk of death in comparison to the pooled group, while diffuse MATR3(∆RRM1/2)-EGFP was highly toxic (comparing to the pooled condition: cells with granules, n = 1590, HR = 0.92, *p=0.03; cells with diffuse protein, n = 153, HR = 3.78, ***p=2 × 10−16; Cox proportional hazards). Scale bars in (A) and (B), 10 μm.
-
Figure 4—source data 1
Sensitivity/specifity data for Figure 4B, granule formation risk and GFP expression data for Figures 4E and F, and survival data for Figures 4G and H.
- https://doi.org/10.7554/eLife.35977.010
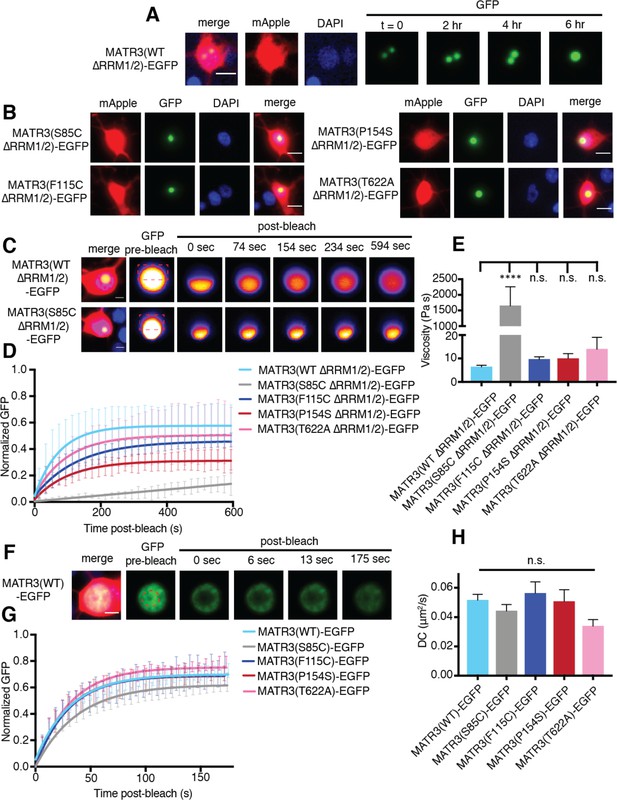
MATR3(∆RRM1/2)-EGFP droplets display liquid-like properties that are affected by the S85C mutation.
(A) MATR3(∆RRM2)-EGFP and MATR3(∆RRM1/2)-EGFP droplets show liquid-like properties such as mobility and fusion. (B) Pathogenic MATR3 mutations on the ∆RRM1/2 background result in similar phase-separated droplets. (C–D) Fluorescence recovery after photobleaching (FRAP) of MATR3(∆RRM1/2)-EGFP droplets shows internal rearrangement consistent with liquid-like behavior, but the recovery of MATR3(S85C ∆RRM1/2)-EGFP droplets was significantly delayed. (E) MATR3(S85C ∆RRM1/2)-EGFP droplets displayed significantly higher viscosity in comparison to MATR3(WT ∆RRM1/2)-EGFP (comparing to MATR3(WT ∆RRM1/2)-EGFP, n = 5: MATR3(S85C ∆RRM1/2)-EGFP, n = 5, ****p<0.0001; MATR3(F115C ∆RRM1/2)-EGFP, n = 5, p>0.9999; MATR3(P154S ∆RRM1/2)-EGFP, n = 5, p>0.9999; MATR3(T622A ∆RRM1/2)-EGFP, n = 4, p>0.9999; one-way ANOVA with Tukey’s post-hoc test). (F–G) FRAP experiments involving full-length MATR3-EGFP variants showed no differences in rates of return. (H) Similarly, there were no differences in diffusion coefficients (DC) among full-length MATR3 variants (MATR3(WT)-EGFP, n = 5; MATR3(S85C)-EGFP, n = 5; MATR3(F115C)-EGFP; n = 5, MATR3(P154S)-EGFP, n = 5; MATR3(T622A)-EGFP, n = 4; p=0.17, one-way ANOVA). Scale bars in (A) and (B), 10 μm; scale bars in (C) and (F), 5 μm. Curves in (D) and (G) show fitted curves ± SD.
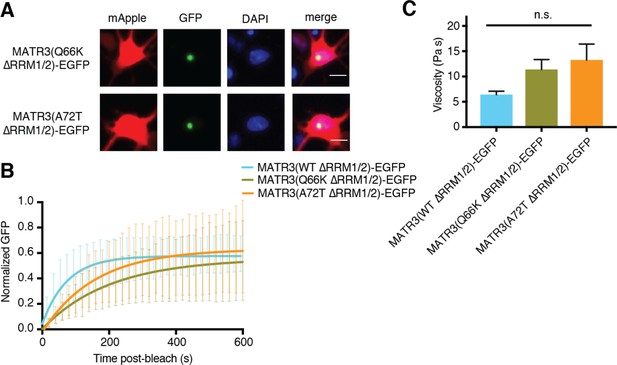
Pathogenic N-terminal domain mutations other than S85C on the ∆RRM1/2 background do not affect granule viscosity.
(A) When expressed in rodent primary cortical neurons, the Q66K and A72T disease-associated mutations on MATR3(∆RRM1/2)-EGFP form intranuclear granules. Scale bar, 10 μm. (B–C) However, these mutants do not alter droplet viscosity (MATR3(WT ∆RRM1/2)-EGFP n = 5, MATR3(Q66K ∆RRM1/2)-EGFP n = 6, MATR3(A72T ∆RRM1/2)-EGFP n = 6; p=0.11; one-way ANOVA). Curves in (B) show fitted curves ± SD.
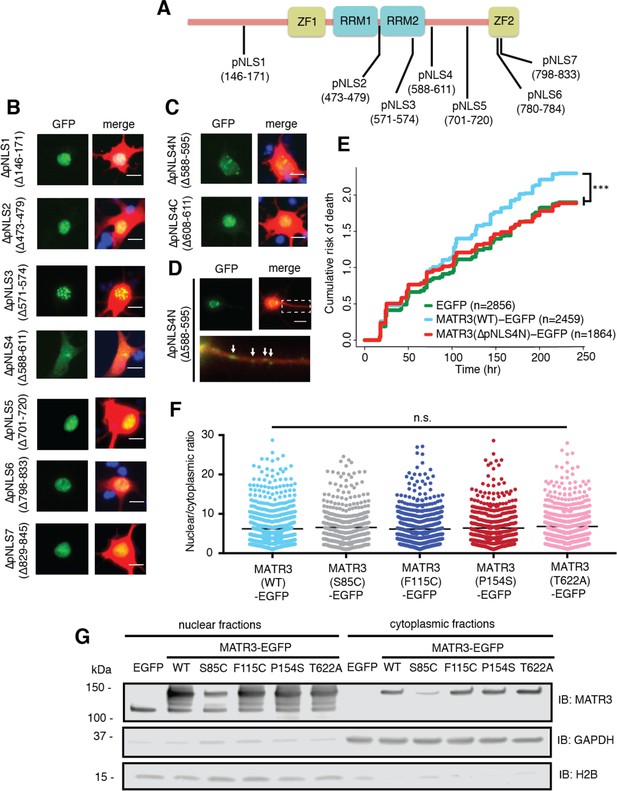
Reducing MATR3 nuclear localization mitigates overexpression toxicity.
(A) Schematic showing putative MATR3 nuclear localization signals (pNLS). (B–C) Deletion of the N-terminal arm of NLS4 (∆pNLS4N) led to nuclear MATR3 clearance in neurons. (D) MATR3(∆pNLS4N)-EGFP forms granular structures in the cytoplasm and neuronal processes (white arrows). (E) Disrupting nuclear localization of MATR3 prevents neurotoxicity from overexpression (compared to MATR3(WT)-EGFP, n = 2459: MATR3(∆pNLS4N)-EGFP, n = 1864, HR = 0.89, ***p=0.00041, Cox proportional hazards). (F–G) Pathogenic MATR3 mutants display no difference in subcellular protein localization as assessed by automated image nuclear/cytoplasmic analysis (F; MATR3(WT)-EGFP, n = 789; MATR3(S85C)-EGFP, n = 462; MATR3(F115C)-EGFP, n = 596; MATR3(P154S)-EGFP, n = 524; MATR3(T622A)-EGFP, n = 657; p=0.067, one-way ANOVA) or biochemical fractionation in transfected HEK293T cells (G). Western blot demonstrated reduced abundance of the S85C mutant in transfected HEK293T cells. Scale bars in (B) and (C), 10 μm; scale bar in (D), 50 μm.
-
Figure 6—source data 1
Survival data for Figure 6E and nucleocytoplasmic localization data for Figure 6F and Figure 6—figure supplement 1B.
- https://doi.org/10.7554/eLife.35977.015
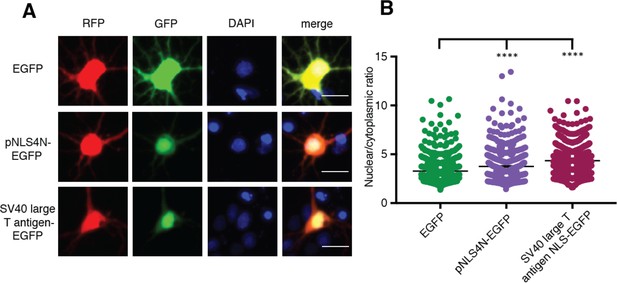
The N-terminal arm of MATR3’s bipartite NLS is capable of driving nuclear enrichment of a heterologous protein.
(A-B) Appending the eight amino acids of the pNLS4N sequence onto EGFP results in enhanced nuclear localization, comparable to the effect of the canonical SV40 large T cell antigen NLS (comparing to EGFP n = 877: pNLS4N-EGFP n = 601, p<0.0001; SV40 large T cell antigen NLS-EGFP n = 1095, ****p<0.0001; one-way ANOVA with Tukey’s post-hoc test). Scale bar, 20 μm.
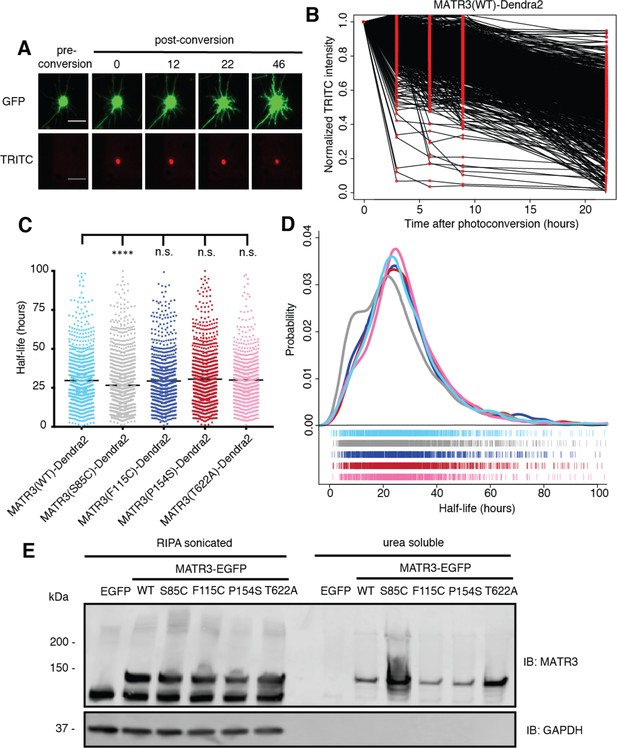
Pathogenic MATR3 mutations have little effect on MATR3 turnover, but a subset reduce solubility.
(A) Optical pulse labeling of Dendra2-tagged MATR3 variants. Each neuron is transfected with EGFP alone to outline the cell body, as well as MATR3-Dendra2, which fluoresces in the red channel (TRITC) upon photoconversion. Scale bar, 50 μm. (B) Normalized red fluorescence (TRITC) signal for individual neurons. The time-dependent decay of red fluorescence over time is used to calculate MATR3-Dendra2 half-life for each neuron. (C–D) MATR3(S85C)-Dendra2 displayed a subtle but significant reduction in half-life compared to MATR3(WT)-Dendra2 (comparing to MATR3(WT)-Dendra2, n = 1269: MATR3(S85C)-Dendra2, n = 1670, ****p<0.0001; MATR3(F115C)-Dendra2, n = 1122, p>0.9999; MATR3(P154S)-Dendra2, n = 1509, p=0.9309; MATR3(T622A)-Dendra2, n = 923, p=0.9989; one-way ANOVA with Tukey’s post-hoc test). (E) Sonication in RIPA resulted in equivalent amounts of all MATR3 variants by Western blotting. The S85C variant was markedly enriched in the RIPA-insoluble, urea-soluble fraction, while the T622A variant showed more modest enrichment.
-
Figure 7—source data 1
Half-life data for Figures 7C and D.
- https://doi.org/10.7554/eLife.35977.017
Tables
| Reagent type | Designation | Source | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | HEK293T | ATCC | CRL-3216; RRID:CVCL_0063 | |
| Antibody | Rabbit anti-MATR3 | Abcam | EPR10635(B); RRID:AB_2491618 | (1:1000) for ICC in Figure 2 and Western blot in Figure 6 and 7 |
| Antibody | Rabbit anti-MATR3 | Abcam | EPR10634(B) | (1:1000) for ICC in Figure 1 |
| Antibody | Mouse anti-fibrillarin | Abcam | 38F3; RRID:AB_304523 | (1:1000) |
| Antibody | Mouse anti-SC35 | Novus Biologicals | NB100-1774SS; RRID:AB_526734 | (1:2000) |
| Antibody | Mouse anti-PML | Santa Cruz Biotechnology | sc-377390 | (1:50) |
| Antibody | Goat anti-mouse 647 | Jackson Immuno Research | 115-605-003; RRID:AB_2338902 | (1:1000) |
| Antibody | Goat anti-rabbit 647 | Jackson Immuno Research | 111-605-003; RRID:AB_2338072 | (1:1000) |
| Antibody | Mouse anti-GAPDH | Millipore Sigma | MAB374; RRID:AB_2107445 | (1:1000) |
| Antibody | Rabbit anti-H2B | Novus Biologicals | NB100-56347; RRID:AB_838347 | (1:1000) |
| Antibody | AlexaFluor goat anti-mouse 594 | ThermoFisher | A-11005; RRID:AB_141372 | (1:10,000) |
| Antibody | AlexaFluor goat anti-rabbit 488 | ThermoFisher | A-11008; RRID:AB_143165 | (1:10,000) |
| Recombinant DNA reagent | MATR3 cDNA | Addgene | #32880 | |
| Recombinant DNA reagent | pGW1 MATR3(WT)- EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(S85C)- EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(F115C)- EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(P154S)- EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(T622A)- EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(∆ZF1)- EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(∆ZF2)- EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(∆ZF1/2)- EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(∆RRM1)- EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(∆RRM2)- EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(∆RRM1/2)- EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(Q66K ∆RRM1/2)-EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(A72T ∆RRM1/2)-EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(S85C ∆RRM1/2)-EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(F115C ∆RRM1/2)-EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(P154S ∆RRM1/2)-EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(T622A ∆RRM1/2)-EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(∆pNLS1)- EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(∆pNLS2)- EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(∆pNLS3)- EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(∆pNLS4)- EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(∆pNLS5)- EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(∆pNLS6)- EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(∆pNLS7)- EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(∆pNLS4N)- EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(∆pNLS4C)- EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 pNLS4N-EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 SV40 large T antigen NLS-EGFP | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(WT)- Dendra2 | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(S85C)- Dendra2 | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(F115C)- Dendra2 | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(P154S)- Dendra2 | This paper | ||
| Recombinant DNA reagent | pGW1 MATR3(T622A)- Dendra2 | This paper |
Primer sequences used to generate domain deletion mutants.
https://doi.org/10.7554/eLife.35977.018| Deletion mutation | Amino acids | Primers | Sequences |
|---|---|---|---|
| ∆ZF1 | 288–322 | Forward | 5’-CTT GAA ATC TAC CCA GAA TG-3’ |
| Reverse | 5’-CTT CGG TAA GAG TCC ATG-3’ | ||
| ∆ZF2 | 798–833 | Forward | 5’-CTG AAT AAA TTG GCA GAA GAA C-3’ |
| Reverse | 5’-AGG TAT CAC ATA GTC TAT ACC-3’ | ||
| ∆RRM1 | 398–473 | Forward | 5’-TAT AAA AGA ATA AAG AAA CCT GAA GG-3’ |
| Reverse | 5’-GCT AGT TTC CAC TCT GCC-3’ | ||
| ∆RRM2 | 496–575 | Forward | 5’-GTT CTG AGG ATT CCA AAC AG-3’ |
| Reverse | 5’-TCC AAG CTC TTG CTT TTG-3’ |
Primer sequences used to generate putative NLS deletion mutants.
https://doi.org/10.7554/eLife.35977.019| Deletion mutation | Amino acids | Primers | Sequences |
|---|---|---|---|
| ∆pNLS1 | 146–171 | Forward | 5’-AGA GTA CCT AGG GAT GAT TG-3’ |
| Reverse | 5’-AAG CTG TAG AAG GAT TTG G-3’ | ||
| ∆pNLS2 | 473–479 | Forward | 5’-CCT GAA GGA AAG CCA GAT C-3’ |
| Reverse | 5’-CTG GGA TAA ATG AAC TCT CAC-3’ | ||
| ∆pNLS3 | 571–574 | Forward | 5’-CTG GTT CTG AGG ATT CCA ACC-3’ |
| Reverse | 5’-CTC AGA CAG GTC AAC CTT C-3’ | ||
| ∆pNLS4 | 588–611 | Forward | 5’-ACT GAT GGT TCC CAG AAG-3’ |
| Reverse | 5’-CAG TAA ATC AAT GCC TCT G-3’ | ||
| ∆pNLS5 | 701–720 | Forward | 5’-GAG GAA CTT GAT CAA GAA AAC-3’ |
| Reverse | 5’-CAC AGC TTT ATC TGA TGG TTC-3’ | ||
| ∆pNLS6 | 780–784 | Forward | 5’-CAG CCC AAT GTT CCT GTT G-3’ |
| Reverse | 5’-ATA CTC ATC TGG GAT TGT ATA G-3’ | ||
| ∆pNLS7 | 798–833 | Forward | 5’-GAA ACT ATG ACT AGT ACT AGA G-3’ |
| Reverse | 5’-CTG ATA ATG AGG AAG GCT G-3’ | ||
| ∆pNLS4N | 588–595 | Forward | 5’-TCT TAC TCT CCA GAT GGC-3’ |
| Reverse | 5’-CAG TAA ATC AAT GCC TCT G-3’ | ||
| ∆pNLS4C | 608–611 | Forward | 5’-ACT GAT GGT TCC CAG AAG-3’ |
| Reverse | 5’-ATC ACT TGG AGA TTC TTT GC-3’ |
Primer sequences used to generate point mutations.
https://doi.org/10.7554/eLife.35977.020| Mutation | Primers | Sequences |
|---|---|---|
| Q66K | Forward | 5’-TTC TTC ATT GAA TAA ACA AGG AGC TC-3’ |
| Reverse | 5’-GAG CTC CTT GTT TAT TCA ATG AAG AA-3’ | |
| A72T | Forward | 5’-AAG GAG CTC ATA GTA CAC TGT CT-3’ |
| Reverse | 5’-AGA CAG TGT ACT ATG AGC TCC TT-3’ | |
| S85C | Forward | 5’-AAT TTG CAG TGT ATA TTT AAC ATT GG-3’ |
| Reverse | 5’-ATG GGA AGA AGT ACT AGC AGA-3’ | |
| F115C | Forward | 5’-ATT TTG GCC AGC TGT GGT CTG TCT GCT-3’ |
| Reverse | 5’-GTT ACT GGC CTG GTC TGC ATC-3’ | |
| P154S | Forward | 5’-GAA GAA GGC TCT ACC TTG AGT TAT GG-3’ |
| Reverse | 5’-AGT TCT CCT CCT TTT AAG CTG-3’ | |
| T622A | Forward | 5’-GAG AGT TCA GCC GAA GGT AAA GAA C-3’ |
| Reverse | 5’-AGT CTT CTG GGA ACC ATC AGT-3’ |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.35977.021





