Glutathione de novo synthesis but not recycling process coordinates with glutamine catabolism to control redox homeostasis and directs murine T cell differentiation
Figures
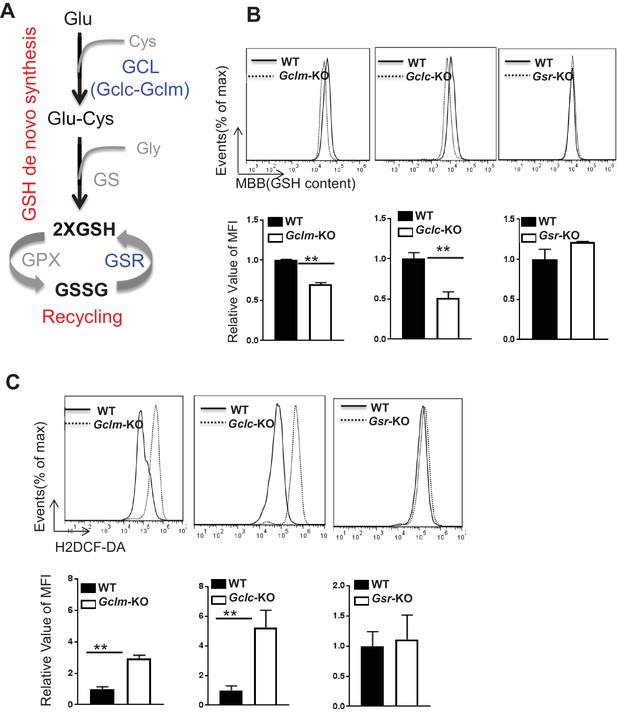
De novo synthesis but not recycling of GSSG is required for producing GSH and fine-tuning ROS upon TCR stimulation.
(A) Diagram of GSH biosynthesis, with metabolic pathways highlighted in red and enzymes highlighted in blue. (B) Naive CD4+T cells from WT and Gclm KO (left), or WT (CD4-Cre-, Gclcfl/fl) and Gclc KO (CD4-Cre+, Gclcfl/fl, (middle), or WT and Gsr KO (right) were activated by plate-bound anti-CD3 plus anti-CD28 for 24 hr, followed by the measurement of GSH levels. (C) Naive CD4+T cells from WT and Gclm KO (left), or WT (CD4-Cre-, Gclcfl/fl) and Gclc KO (CD4-Cre+, Gclcfl/fl, (middle), or WT and Gsr KO (right) were activated by plate-bound anti-CD3 plus anti-CD28 for 24 hr, followed by the measurement of ROS levels. Data in Figure 1B–C are representative of two independent experiments. Data represent the mean ± S.D.
-
Figure 1—source data 1
Source data for B and C.
- https://doi.org/10.7554/eLife.36158.005
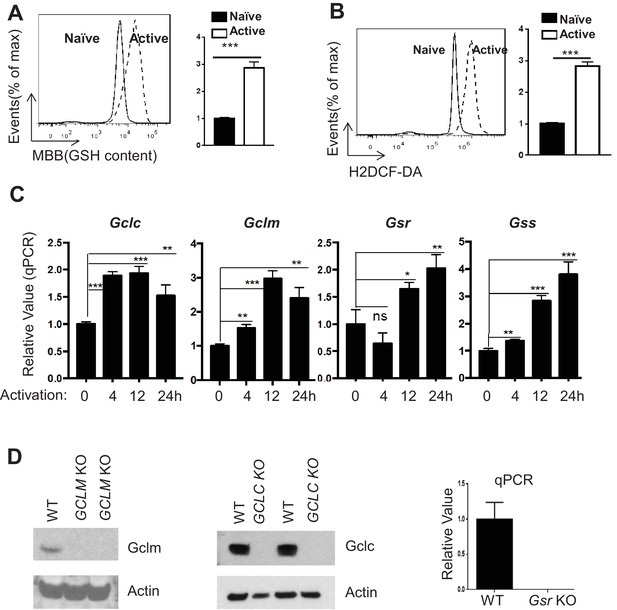
TCR stimulation drives GSH and ROS production in T cells.
(A–B) Naive CD4 +T cells from C57BL/6 mice were either cultured in the presence of IL-7 (naive) or activated by plate-bound anti-CD3 and anti-CD28 for 24 hr, followed by measuring intracellular GSH (A) and ROS (B) by FACS. (C) RNAs were isolated from naïve or activated T cells for indicated times, and used for real-time qPCR analyses of indicated genes. Expression levels in naive cells were set to 1. (D) The protein levels of Gclm (left) or Gclc (middle) in total T cells from mice with indicated genotypes were determined by western blot. RNAs were isolated from WT or Gsr KO T cells and used for real-time qPCR analyses of Gsr gene. Expression levels in WT sample were set to 1. Data in Figure A-C are representative of two independent experiments. Data are represented the mean ± S.D.
-
Figure 1—figure supplement 1—source data 1
Source data for A, B, C and D.
- https://doi.org/10.7554/eLife.36158.004
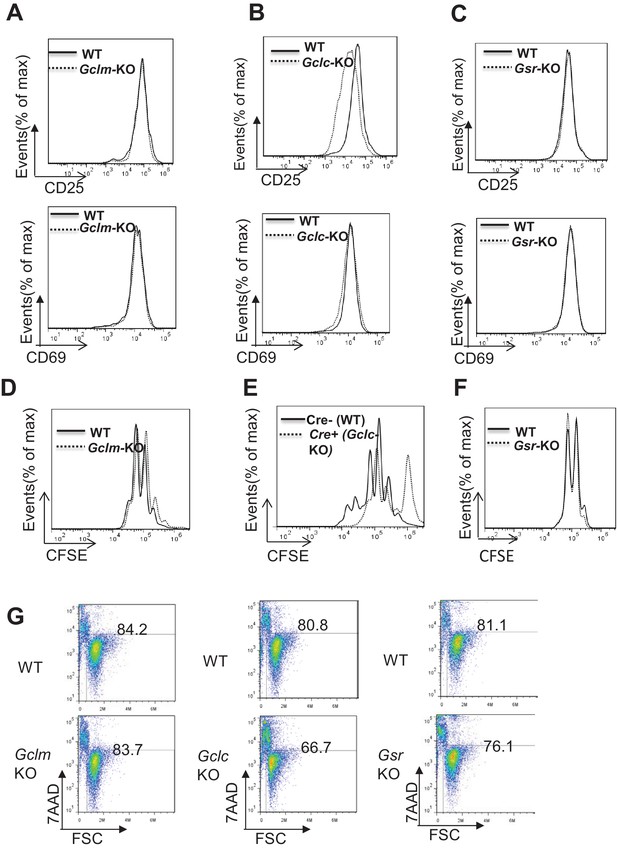
Severe depletion of GSH by blocking de novo synthesis suppresses T cell activation and proliferation.
(A–C) Naive CD4 +T cells from WT and Gclm KO (A), or WT (CD4-Cre-, Gclcfl/fl) and Gclc KO (CD4-Cre+, Gclcfl/fl) (B), or WT and Gsr KO (C) mice were activated by plate-bound anti-CD3 plus anti-CD28 for 24 hr, followed by cell surface expression of CD25 (upper panel) and CD69 (lower panel). (D–F) Cell proliferation of active CD4 +T cells (72 hr) with indicated genotypes was determined by CFSE dilution. (G) Naive CD4 +T cells isolated from mice with indicated genotypes were activated by plate-bound anti-CD3 and anti-CD28 for 24 hr. Cell viability was determined by FACS. Figure 2A–G are representative of three independent experiments.
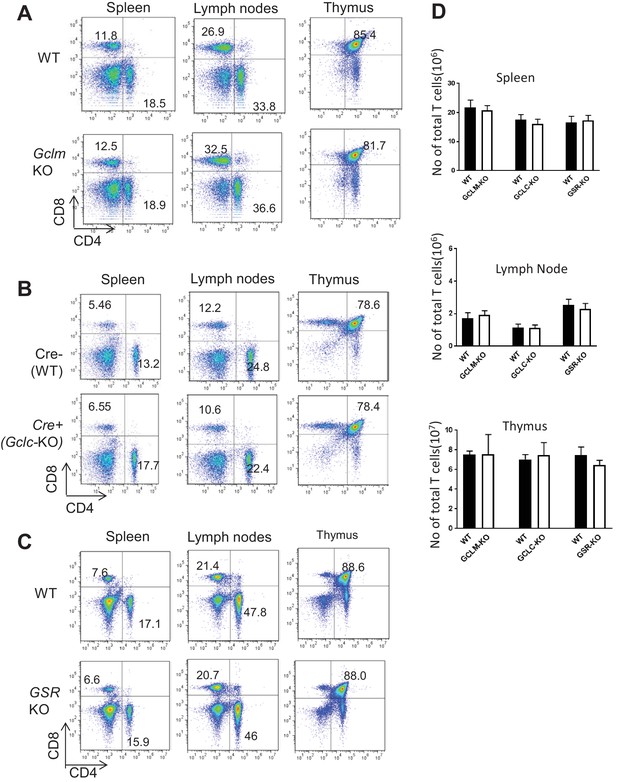
Severe depletion of GSH by blocking de novo synthesis suppresses T cell activation and proliferation.
(A–C) Distribution of CD4+ and CD8+ cells in the spleen, lymph nodes or thymus from mice with indicated genotypes were determined by FACS. Figure 2A -D are representative of two independent experiments (D) Bar graph represents the number of T cells calculated from total splenocytes, lymph node cells and thymocytes.
-
Figure 2—figure supplement 1—source data 1
Source data for A, B, C and D.
- https://doi.org/10.7554/eLife.36158.008
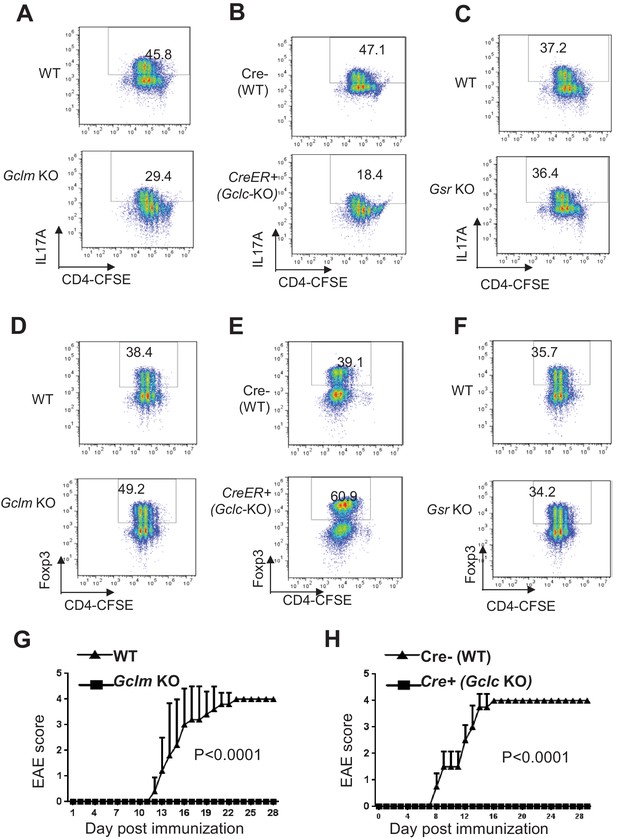
Ablation of de novo synthesis but not recycling of GSSG reciprocally alters TH17 and iTreg cell differentiation.
(A–F) Naive CD4+ T cells from WT and Gclm KO, or WT (Cre-,) and CreER+ (Gclc-KO- in the presence of 100 nM of 4-hydroxytamoxifen (4OHT)), or WT and Gsr KO mice were stained with 4 µm CFSE and differentiated under TH17 or iTreg -inducing conditions for 5 days, followed by intracellular staining of IL-17 and Foxp3. (G–H) mice with indicated genotypes were immunized with MOG to induce EAE and pathological progressions were scored daily. Data in Figure 4A–H are representative of two-three independent experiments.
-
Figure 3—source data 1
Source data for G and H.
- https://doi.org/10.7554/eLife.36158.012
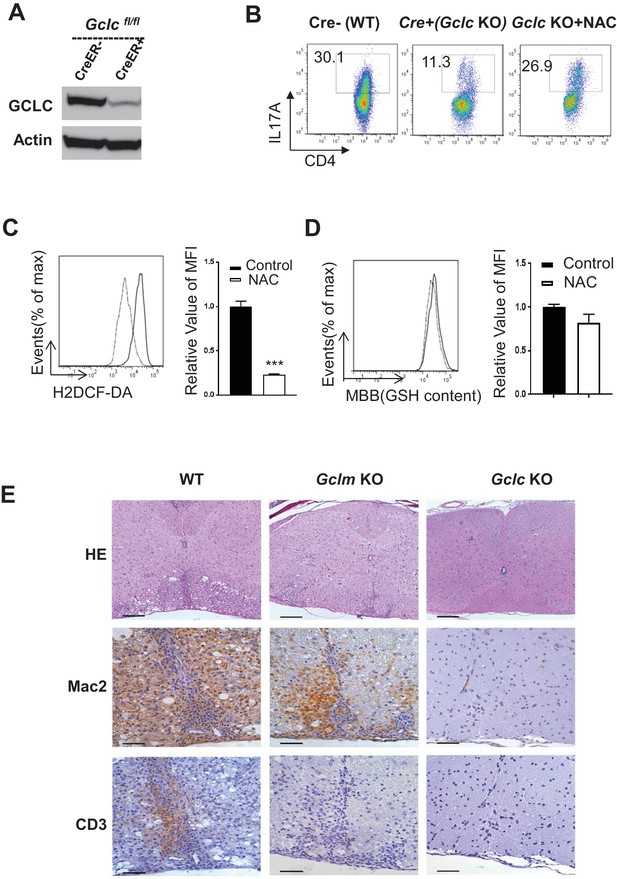
Ablation of de novo synthesis but not recycling of GSSG reciprocally alters TH17 and iTreg cell differentiation.
(A) The protein levels of Gclc in cells collected two days following 4OHT treatment were determined by western blot. (B) Naïve CD4+ T cells from WT (CD4-Cre-, Gclcfl/fl) and Gclc KO (CD4-Cre+, Gclcfl/fl) mice were differentiated under TH17-inducing conditions with or without 5 mM N-acetyl cysteine (NAC) for 5 days, followed by intracellular staining of IL-17. (C) Naive CD4+ T cells were activated for 24 hr with or without 5 mM NAC, followed by measuring ROS and (D) GSH by FACS. (E) WT, Gclm-/- (KO) and CD4-Cre, Gclcfl/fl (KO) mice were immunized with MOG/CFA. After 20 days, cervical spinal cord sections were analyzed by H and E (bars, 200 μm) and anti-Mac2 and anti-CD3 immunohistochemistry (bars, 50 μm).Data are representative of two independent trials. Data in Figure A-D are representative of two-three independent experiments.
-
Figure 3—figure supplement 1—source data 1
Source data for C and D.
- https://doi.org/10.7554/eLife.36158.011
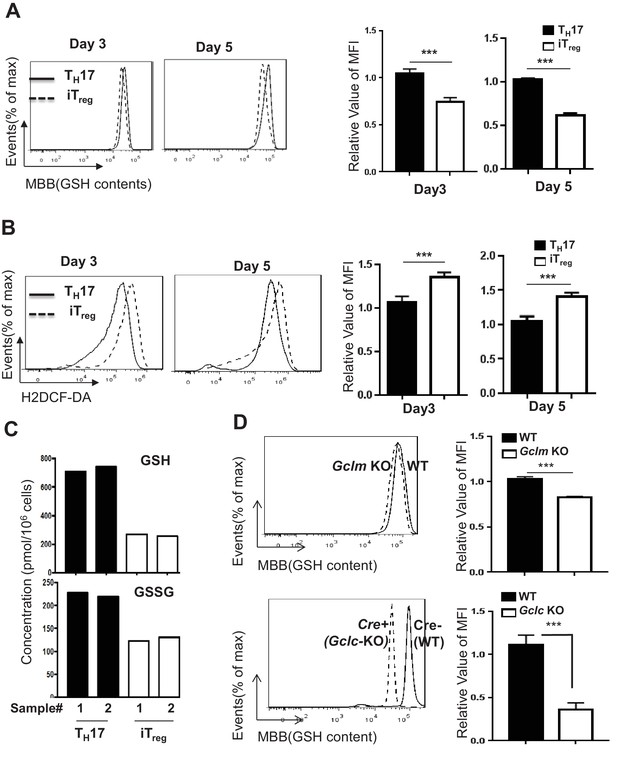
TH17 cells preferentially maintain higher level of GSH than iTreg cells.
(A–B) Naive CD4+ T cells from C57BL/6 mice were differentiated under iTreg or TH17–inducing conditions and cells were collected at indicated times, followed by measuring intracellular GSH (A) and ROS (B) by FACS. (C) Naive CD4+ T cells from C57BL/6 mice were differentiated under TH17 or iTreg–inducing conditions for 5 days. The intracellular levels of GSH and GSSG were determined by mass spectrometry. (D) Naive CD4+T cells from WT and Gclm KO (top) or WT (CD4-Cre-, Gclcfl/fl) and Gclc KO (CD4-Cre+, Gclcfl/fl, (bottom) were differentiated under TH17-inducing conditions for 5 days, followed by the measurement of GSH levels. Data in Figure 4A–D are representative of three independent experiments. Data represent the mean ± S.D.
-
Figure 4—source data 1
Source data for A, B, C and D.
- https://doi.org/10.7554/eLife.36158.016
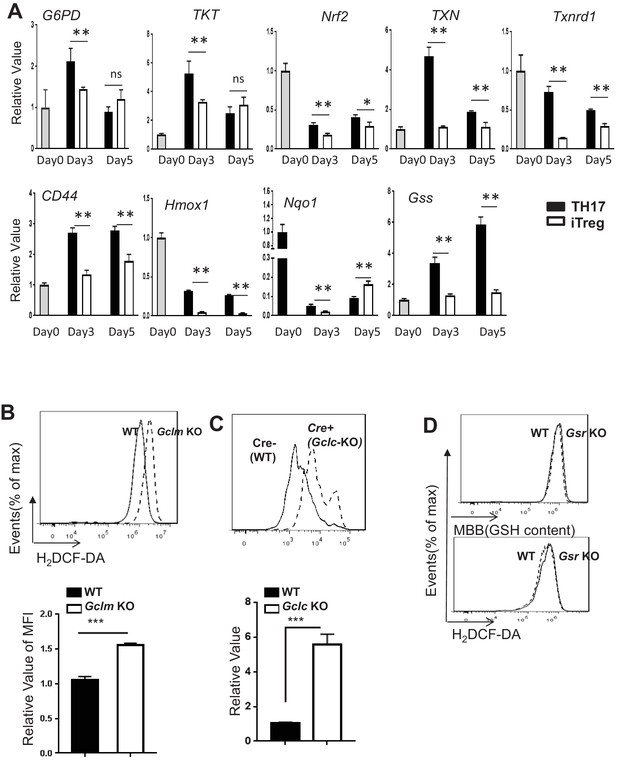
De novo synthesis but not recycling of GSSG is required for providing GSH and suppressing ROS during TH17 cell differentiation.
(A) RNAs were isolated from cells differentiated under TH17 or iTreg-inducing conditions for indicated times, and used for real-time qPCR analyses of indicated genes. Expression levels in day 0 were set to 1 (B–D) Naive CD4+ T cells from WT and Gclm KO, or WT (CD4-Cre-, Gclcfl/fl) and Gclc KO (CD4-Cre+, Gclcfl/fl), or WT and Gsr KO mice were differentiated under TH17-inducing conditions for 5 days, followed by measuring ROS by FACS. Data in Figure B-D are representative of two independent experiments. Data represent the mean ±S.D.
-
Figure 4—figure supplement 1—source data 1
Source data for A, B and C.
- https://doi.org/10.7554/eLife.36158.015
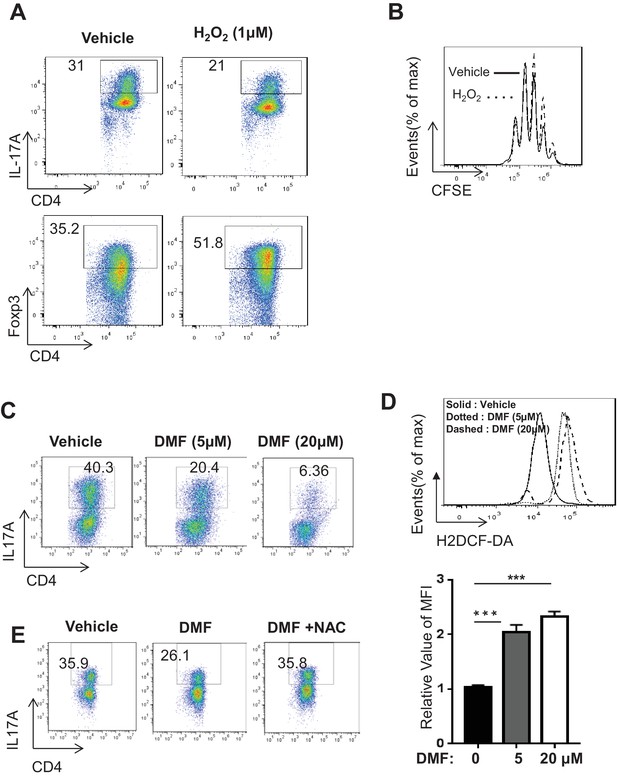
DMF suppresses TH17 differentiation by augmenting ROS generation.
(A) Naive CD4+ T cells from C57BL/6 mice were differentiated under TH17 or iTreg-inducing conditions with or without H2O2 (1 µM) for 5 days, followed by intracellular staining of IL-17 and Foxp3. (B) Cell proliferation of active CD4+ T cells (72 hr) with or without H2O2 (1 µM) was determined as CFSE dilution. (C–D) Naive CD4+ T cells from C57BL/6 mice were differentiated under TH17-inducing conditions with indicated dose of DMF for 5 days, followed by intracellular staining of IL-17 (C) and ROS (D). (E) Naive CD4+ T cells from C57BL/6 mice were differentiated under TH17-inducing conditions with indicated treatment for 5 days, followed by intracellular staining of IL-17. Data in Figure 5 are representative of two-three independent experiments. Data represent the mean ±S.D.
-
Figure 5—source data 1
Source data for D.
- https://doi.org/10.7554/eLife.36158.019
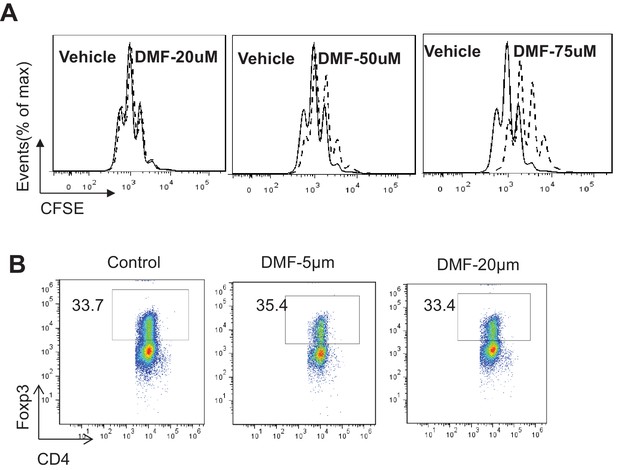
DMF suppresses TH17 differentiation through augmenting ROS generation.
(A) Cell proliferation of active CD4 +T cells (72 hr) with indicated treatments was determined as CFSE dilution (B). Naive CD4 +T cells from C57BL/6 mice were differentiated with 5 and 20 µM of DMF under iTreg cell -inducing conditions for 5 days, followed by intracellular staining of Foxp3.
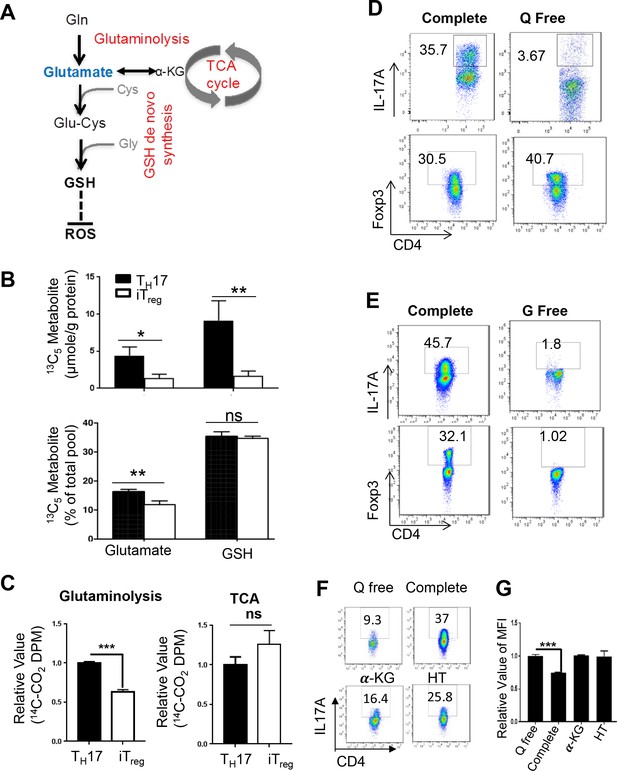
Glutamate that fuels GSH de novo synthesis is partially derived from glutamine in T cells.
(A) Diagram of metabolic steps linked to the GSH production, with metabolic pathways highlighted in red. (B) Naive CD4+ T cells from C57BL/6 mice were differentiated under TH17 and iTreg–inducing conditions for 5 days, followed by culturing in media containing 13C515N2-glutamine. The intracellular levels of Glutamate and GSH including 13C-, 13C,15N-, and 12C-unlabeled forms were determined by IC-UHRFTMS. (C) Naive CD4+ T cells from C57BL/6 mice were differentiated under TH17 or iTreg cell–inducing conditions for 5 days, were used for measuring the generation of 14CO2 from [U-14C]-glutamine (glutaminolysis), from [2-14C]-pyrvuate (TCA). (D–E) Naive CD4+ T cells from C57BL/6 mice were differentiated in completed, glutamine-free (Q free) or glucose-free (G free) medium under TH17 or iTreg cell-inducing conditions for 5 days, followed by intracellular staining of IL-17 and Foxp3. (F) Naive CD4+ T cells from C57BL/6 mice were activated in complete medium for 24 hr and cells were washed with PBS and switch to conditional medium in presence or absence of glutamine, 3 mM αKG or 100 μM hypoxanthine and 16 μM thymidine (HT) for 5 days followed by intracellular staining of IL-17 and (G) and ROS. Data represent the mean ±S.D.
-
Figure 6—source data 1
Source data for B, C and G.
- https://doi.org/10.7554/eLife.36158.024
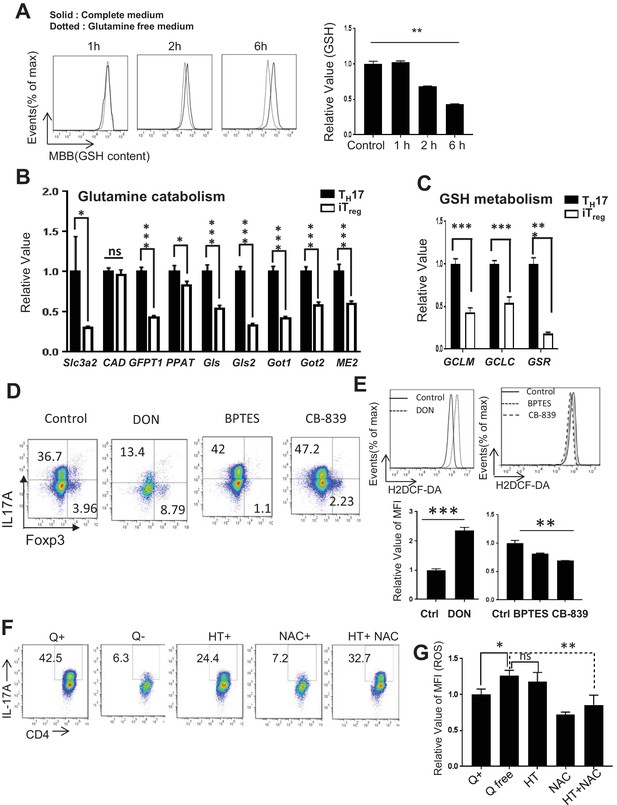
Glutamine catabolism is required for driving TH17 and iTreg cell differentiation.
(A) Naive CD4 +T cells from C57BL/6 mice were differentiated with TH17 cell -inducing conditions for 5 days in complete medium and cells were washed and cultured in Q-free medium as indicated time followed by the measurement of GSH. (B–C) RNAs were isolated from cells differentiated under TH17 or iTreg-inducing conditions for 3–5 days, and used for real-time qPCR analyses of indicated metabolic genes. Expression levels in TH17 group were set to 1. (D) Naive CD4+ T cells from C57BL/6 mice were differentiated in presence of 30 µM DON, 25 µM BPTES and 25 µM CB-839 under TH17 cell -inducing conditions for 5 days, followed by intracellular staining of IL-17 and Foxp3 (E) ROS production by measuring the DCF-fluorescence intensity and the data was represented by histogram (upper panel) and relative intensity by bar graph (lower panel). (F) Naive CD4+ T cells from C57BL/6 mice were differentiated in presence and absence of glutamine and 5 mM NAC, 100 μM hypoxanthine and 16 μM thymidine (HT) and in combination of HT and NAC under TH17 cell-inducing conditions for 5 days, followed by intracellular staining of IL-17 (G). Bar graph shows the relative value of mean fluorescence intensity of ROS measurement. Data represent the mean ±S.D of two-three independent experiments.
-
Figure 6—figure supplement 1—source data 1
Source data for A, B, C, E and G.
- https://doi.org/10.7554/eLife.36158.022
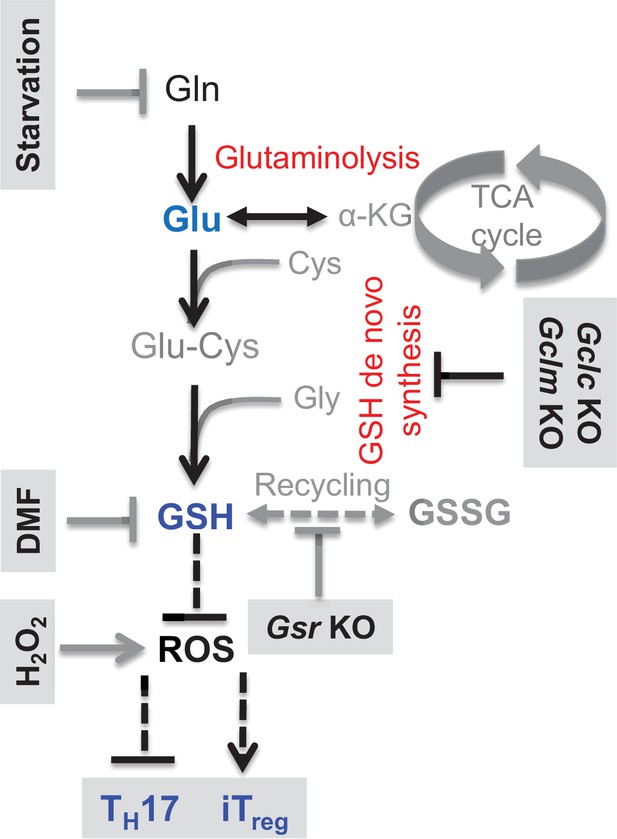
Glutamine catabolism coordinates with GSH metabolism in modulating ROS homeostasis and T cell differentiation.
Modulation of GSH biosynthesis and ROS production in directing T cell differentiation.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | C57BL/6 (B6) mice | Taconic | ||
| Strain, strain background (Mus musculus) | C3H/HeN | Envigo | ||
| Genetic reagent (Mus musculus) | CD4-Cre Gclcflox/flox | PMID:23226398 | ||
| Genetic reagent (Mus musculus) | Gclm-KO | PMID:12384496 | ||
| Genetic reagent (Mus musculus) | ROSA26CreERT2 | RRID: IMSR_JAX:008463 | The Jackson Laboratory | |
| Genetic reagent (Mus musculus) | GSR-KO | PMID: 10218442 | ||
| Antibody | Mouse anti- CD3 mAb | Cat. #:BE0001-1, RRID:AB_1107634 | BioXcell | |
| Antibody | Mouse anti- CD28 mAb | Cat. #BE0015-1 RRID:AB_1107624 | BioXcell | |
| Antibody | Mouse anti -IL2 mAb | Cat.#BE0043 RRID::AB_1107702 | BioXcell | |
| Antibody | Mouse anti- IL4 mAb | Cat. #BE0045 RRID:AB_1107707 | BioXcell | |
| Antibody | Mouse anti- IFNγ mAb | Cat. #BE0055 RRID:AB_1107694 | BioXcell | |
| Antibody | Anti mouse CD4-FITC | Cat. #11–0042 RRID:AB_464897 | eBioscience | (1:200) |
| Antibody | Anti mouse CD4-APC | Cat. #17-0041-81 RRID:AB_469319 | eBioscience | (1:200) |
| Antibody | Anti mouse CD8-APC-Cy7 | Cat. #100714 RRID:AB_312753 | Biolegend | (1:200) |
| Antibody | Anti mouse Foxp3-APC | Cat. # RRID:AB_469456 | eBioscience | (1:200) |
| Antibody | Anti mouse IL-17A-PECy7 | Cat. #25-7177-82 RRID:AB_10732356 | eBioscience | (1:200) |
| Antibody | Anti GCLC antibody (rabbit monoclonal) | Cat. #ab190685 RRID:AB_10975474 | Abcam | WB (1:1000) |
| Antibody | Anti GCLM antibody (rabbit monoclonal) | Cat. #ab124827 RRID:AB_10975474 | Abcam | WB (1:1000) |
| Antibody | anti-mouse CD25 -PE | Cat. #101904 RRID:AB_312847 | Biolegend | (1:200) |
| Antibody | anti-mouse CD69-PECy7 | Cat. #552879 RRID:AB_394508 | BD Bioscience | (1:200) |
| Antibody | Anti mouse monoclonal CD3 | Cat. #sc-101442 RRID:AB_1120355 | Santa Cruz | IHC (1:50) |
| Antibody | Anti mouse monoclonal galectin-3 | Cat. #sc-32790, RRID:AB_627657 | Santa Cruz | IHC (1:50) |
| Peptide, recombinant protein | MOG35-55 peptide | synthesized and HPLC-purified | St. Jude Hartwell Center for Biotechnology | |
| Peptide, recombinant protein | Recombinant mouse IL-6 | 216–16 | Peprotech | |
| Peptide, recombinant protein | Recombinant human TGFb | 100–21 c | Peprotech | |
| Peptide, recombinant protein | Recombinant human or mouse IL-2 | 200–02 | Peprotech | |
| Commercial assay or kit | Foxp3/Transcription Factor Staining Buffer Set | 00-5523-00 | e-Bioscience | |
| Commercial assay or kit | Naive CD4 + T cell isolation kit,mouse | 5160725186 | Miltenyi Biotec | |
| Commercial assay or kit | CD45R(B220) microbeads, mouse | 5150309030 | Miltenyi Biotec | |
| Commercial assay or kit | ABC kit | PK-7200 | Vector laboratories | |
| Commercial assay or kit | MojoSort Mouse naive CD4 T Cell Isolation Kit | 480031 | Biolegend | |
| Chemical compound, drug | Diethly Fumerate | Sigma Aldrich | D95654 | |
| Chemical compound, drug | N-Acetyl-L-cysteine | Sigma-Aldrich | A7250 | |
| Chemical compound, drug | Tamofixen | Sigma-Aldrich | T5648 | |
| Chemical compound, drug | 4-hydroxytamoxifen | Sigma | H7904 | |
| Chemical compound, drug | Dimethy a-keto glutarate/aKG | Sigma-Aldrich | 34963–1 | |
| Chemical compound, drug | Hypoxathine | Sigma-Aldrich | H9377 | |
| Chemical compound, drug | Thymidine | Sigma | T9250 | |
| Chemical compound, drug | H2O2 | Sigma-Aldrich | 7722-84-1 | |
| Chemical compound, drug | carboxyfluorescein diacetate succinimidyl ester(CFSE) | Invitrogen | C1157 | |
| Chemical compound, drug | DM-H2DCFDA | Invitrogen | C6827 | |
| Chemical compound, drug | DAB | Vector Laboratories | SK-4100 | |
| Chemical compound, drug | Monobromobimane | Invitrogen | M1378 | |
| Chemical compound, drug | 7-amino- actinomycin D(7AAD) | Biolegend | 420404 | |
| Chemical compound, drug | Pertussis toxin | 181 | List Biological Laboratories | |
| Chemical compound, drug | Mycobacterium tuberculosum | 231141 | Difco | |
| Chemical compound, drug | Incomplete Freund’s Adjuvant | 263910 | Difco | |
| Chemical compound, drug | [U-14C]-glutamine | MC 1124 | Moravek | |
| Chemical compound, drug | [2–14C]-pyruvate | ARC 0222 | American Radiolabeled Chemicals | |
| Chemical compound, drug | Cell Stimulation Cocktail (plus protein transport inhibitors) (500X) | 00-4975-93 | eBioscience | |
| Chemical compound, drug | Iscove's Modified Dulbecco's Media - Glucose free conditional medium | ME17058P1 | Thermo Fisher Scientific | |
| Chemical compound, drug | Iscove's Modified Dulbecco's Media - without L-glutamine | 12–726 f | Lonza | |
| Chemical compound, drug | RPMI 1640 Medium, No Glucose | 11-879-020 | Gibco | |
| Chemical compound, drug | Hyclone RPMI 1640 Medium, no glutamine | sh30096.10 | Thermo Fisher Scientific | |
| Chemical compound, drug | U-13C6-glutamine | CNLM-1275–0.1 | Cambridge Isotope Lab | |
| Chemical compound, drug | 6-Diazo-5-oxo-L -norleucine | D2141-5MG | Sigma-aldrich | |
| Chemical compound, drug | Bis-2- (5-phenylacetamido -1,3,4-thiadiazol-2-yl) ethyl sulfide (BPTES) | SML0601 | Sigma-aldrich | |
| Chemical compound, drug | CB-839 | 22038 | Cayman | |
| Software, algorithm | Graphpad Prism | RRID:SCR_002798 | ||
| Software, algorithm | FlowJo | RRID:SCR_008520 |
Additional files
-
Supplementary file 1
List of primer sequences used for RT-PCR analysis.
- https://doi.org/10.7554/eLife.36158.025
-
Transparent reporting form
- https://doi.org/10.7554/eLife.36158.026





