Recruitment of two dyneins to an mRNA-dependent Bicaudal D transport complex
Figures
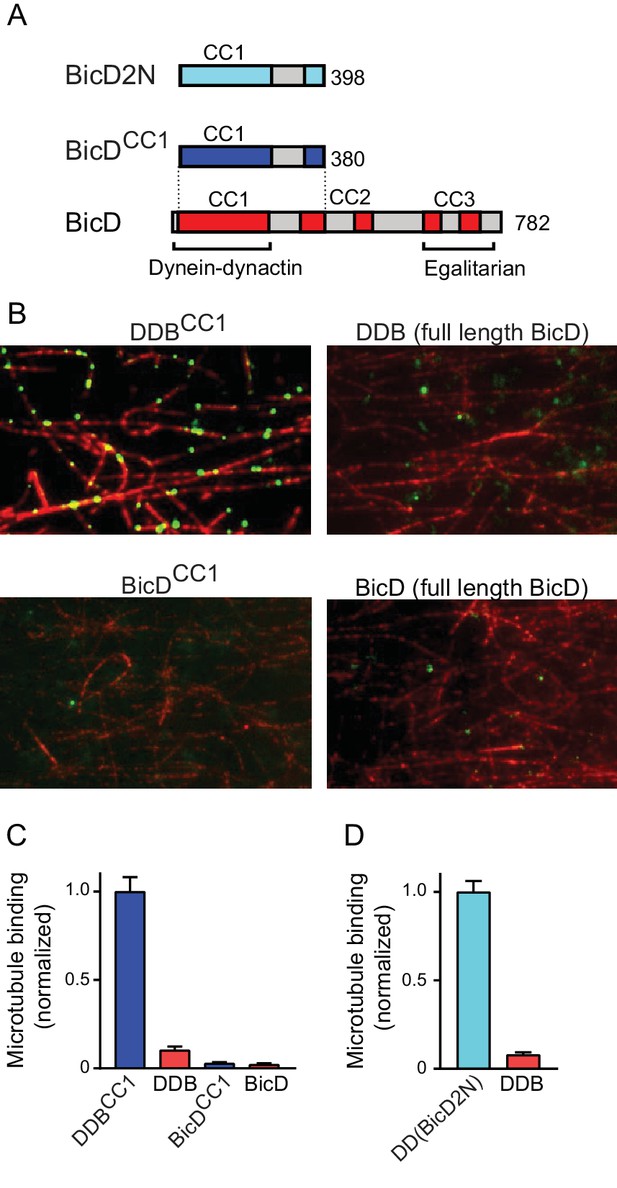
Full-length BicD is auto-inhibited and does not bind dynein-dynactin.
(A) Schematic of BicD constructs. Full-length Drosophila BicD contains three coiled-coil (CC) regions that are designated CC1, CC2, and CC3 (red). CC2 and CC3 are each interrupted by non-coiled-coil regions (gray). Regions predicted to form coiled-coil domains were determined from Paircoil2 analysis (McDonnell et al., 2006). Truncated versions of Drosophila BicD (BicDCC1) and mammalian BicD2 (BicD2N) are also shown. CC1 is the minimal fragment that activates dynein-dynactin, but does not bind cargo adaptor proteins, which occurs via CC3. (B) Single-molecule pulldowns on microtubules (red) show that dynein-dynactin associates with BicDCC1 (green) but not with full-length Drosophila BicD (green) in the presence of AMP-PNP (upper panels). Controls show that the two BicD constructs alone do not bind non-specifically to microtubules (lower panels). BicD was visualized with a 525 nm-streptavidin Qdot bound to an N-terminal biotin tag on BicD. (C) Quantification of the number of dynein-dependent associations of complexes containing Drosophila BicDCC1 (blue) versus full-length Drosophila BicD (red) to microtubules, normalized to microtubule length and dynein concentration (average ± SEM from 5 to 15 fields). Controls for non-specific binding are also quantified. (D) Quantification of the number of dynein-dependent associations of complexes containing mammalian BicD2N (cyan) or full-length Drosophila BicD (red) to microtubules, normalized to microtubule length and dynein concentration (average ± SEM from 5 to 15 fields), from independent experiments. See Figure 1—source data 1.
-
Figure 1—source data 1
Dataset for Figure 1.
- https://doi.org/10.7554/eLife.36306.005
SDS-PAGE gels of BicD constructs and tissue-purified bovine brain dynein and dynactin.
(A) (lane 1) Drosophila BicDCC1, (lane 2) full-length Drosophila BicD, (lane 3) human BicD2N, and (lane 4) molecular mass markers. 4–12% SDS-PAGE gel, MOPS buffer. (B) dynein and dynactin purified from bovine brain, and molecular mass markers. 4–12% SDS-PAGE gel, MOPS buffer.
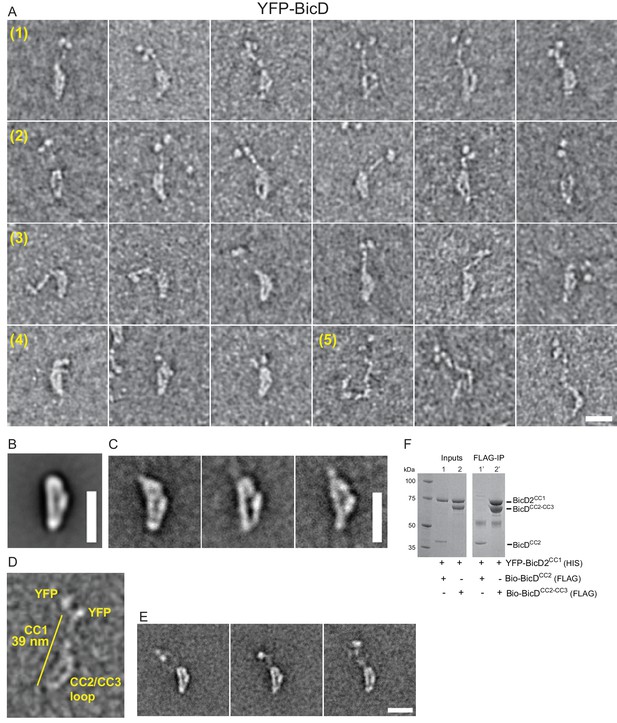
Electron micrographs of the auto-inhibited full-length YFP-BicD.
(A) A montage of negatively stained images showing (row 1) the most common ‘b’ orientation, (row 2) a less common ‘d’ orientation, (row 3) the range over which the molecule can flex, (row 4) compact molecules, and (row 5) rare open molecules. (B) Global average of all aligned YFP-BicD images. (C) Example class averages of BicD with classification focused on the loop. (D) One YFP-BicD molecule illustrating the interpretation of the EM image, with the length of the long straight section indicated. (E) Example class averages of YFP-BicD with classification focused on the protruding YFP-CC1 region. See Figure 2—figure supplement 1 for full classifications. (F) Pulldown showing that CC1 interacts with CC2-CC3 but not CC2. Scale bars = 20 nm.
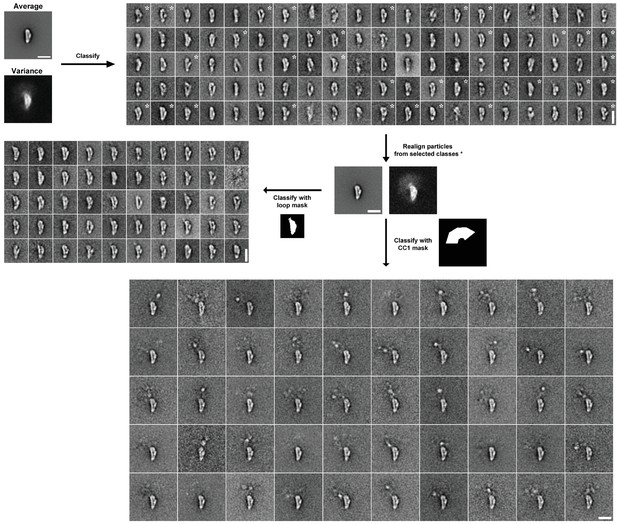
Image processing scheme for YFP-BicD EM data.
Following initial alignment of the full dataset (n = 4205), images were classified into 100 classes using a variance mask. Classes in similar orientations were selected (based on the appearance of class averages and class variances, asterisks show selected classes). Particles from selected classes were re-aligned (n = 1513). Aligned images were classified into 50 classes using masks around the loop or the protruding YFP-CC1 region. Scale bars = 20 nm.
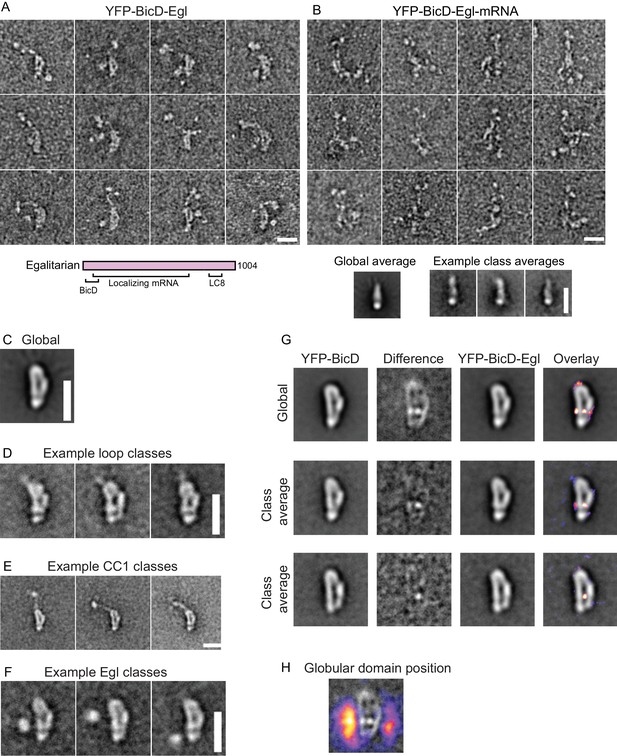
Electron micrographs of the YFP-BicD-Egl and YFP-BicD-Egl-mRNA complex.
(A) Montage of images showing that BicD retains the auto-inhibited looped conformation in the presence of bound Egl. A schematic of Egl (below) shows that the N-terminal domain binds BicD, the C-terminus binds dynein light chain (LC8), and mRNA binding is mediated through a large number (~800) of amino acids. (B) Montage of images showing that YFP-BicD no longer retains the auto-inhibited looped conformation in the presence of bound Egl and mRNA (K10min). Global and class averages are shown below the montage. (C) Average of all YFP-BicD-Egl molecules. (D) Example class averages of YFP-BicD-Egl with classification focused on the loop. (E) Example class averages of YFP-BicD-Egl with classification focused on the protruding YFP-CC1 region. (F) Example class averages of YFP-BicD-Egl with classification focused adjacent to the loop, to reveal the globular domain of Egl. (G) Co-alignment and classification of YFP-BicD and YFP-BicD-Egl. Upper row shows the average of YFP-BicD and YFP-BicD-Egl images (after an initial alignment, classification and selection of particles in similar orientations). Difference maps are shown in the second column and the difference map is overlaid as a heatmap onto the average of YFP-BicD-Egl in the fourth column, to show clearly the locations of greatest difference. Second and third rows show the averages and differences for individual classes. See Figure 3—figure supplement 5 for the full classification. (H) Heatmap showing the position of the globular Egl domain. Scale bars = 20 nm.
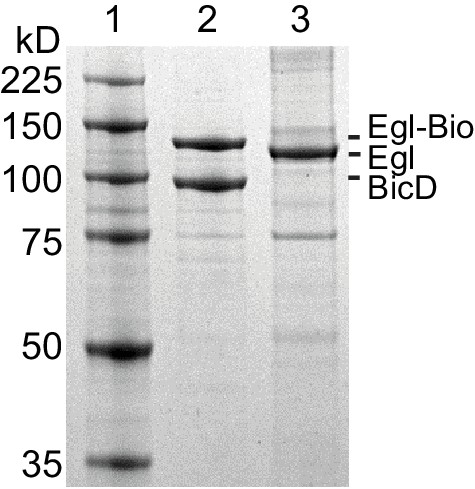
SDS-PAGE gels of Egl.
(lane 1) Molecular mass markers, (lane 2) BicD co-expressed with Bio-tagged Egl, (lane 3) Egl expressed alone. 4–12% SDS-gel, MOPS buffer.
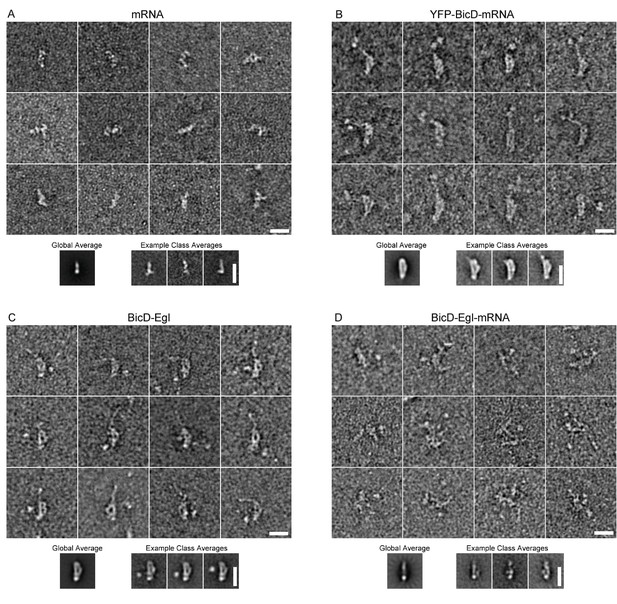
EM of other conditions imaged in this study.
(A) Montage showing example raw images of mRNA (K10min). Global average and example class averages are shown below the montage (n = 2039). (B) Montage showing example raw images of YFP-BicD-mRNA. Global average and example class averages are shown below the montage (n = 1393). (C) Montage showing example raw images of BicD-Egl. Global average and example class averages are shown below the montage (n = 3697). (D) Montage showing example raw images of BicD-Egl-mRNA. Global average and example class averages are shown below the montage (n = 3526).
Fields of view of all conditions imaged in this study.
Scale bars = 50 nm.
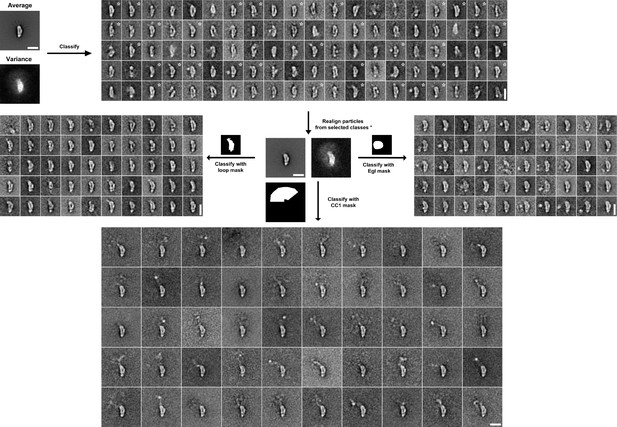
Image processing scheme for YFP-BicD-Egl EM data.
Following initial alignment of the full dataset (n = 4117), images were classified into 100 classes using a variance mask. Classes in similar orientations were selected (based on the appearance of class averages and class variances, asterisks show selected classes). Particles from selected classes were re-aligned (n = 1488). Aligned images were classified into 50 classes using masks around the loop, the protruding YFP-CC1 region or the region adjacent to the loop, in order to reveal the Egl globular domain. Scale bars = 20 nm.
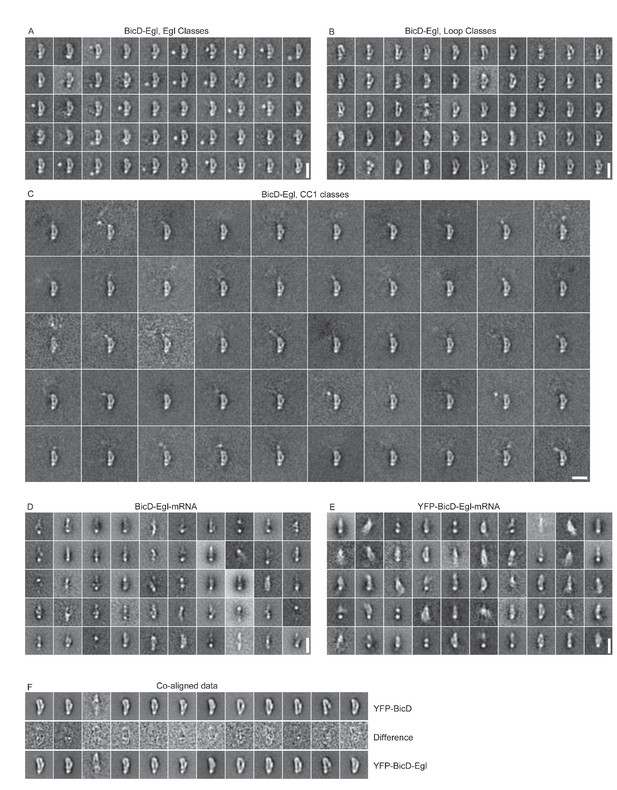
Other classifications used in this study.
(A) Classification of BicD-Egl using a mask adjacent to the loop to reveal the globular domain of Egl. (B) Classification of BicD-Egl using a mask around the loop. (C) Classification of BicD-Egl using a mask around the protruding CC1 region. (A), (B) and (C) are classifications of re-aligned images (n = 1765) following an initial alignment (n = 3697) and selection of classes in the most common orientation. (D) Classification of BicD-Egl-mRNA (n = 3526). Scale bars = 20 nm. (E) Classification of YFP-BicD-Egl-mRNA (n = 3345). (F) Classification of combined YFP-BicD and YFP-BicD-Egl datasets (most common orientation only). Central row shows difference images between the two datasets. Scale bars = 20 nm.
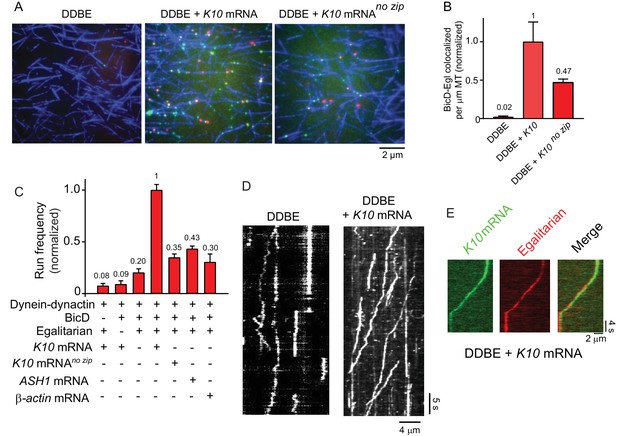
K10 mRNA is needed for BicD-Egl to recruit dynein-dynactin for motility.
(A) Single-molecule pulldowns of YFP-BicD (green) and Qdot-Egl (red) by dynein-dynactin (DDBE) bound to microtubules (blue) in the absence or presence of K10 mRNA. (B) Quantification of the single-molecule pulldown data showing the normalized number of colocalized BicD-Egl complexes bound to dynein-dynactin per µm microtubule length in the absence of mRNA (DDBE), in the presence of K10 mRNA (DDBE + K10), or K10 without the TLS zip code (DDBE + K10 no zip). (C) Normalized run frequencies (per μM dynein per μm microtubule length per time) of motile mRNPs. (Bars, left to right) Little movement of labeled K10 mRNA in the absence of either BicD or Egl. Few events are also observed for mRNPs lacking mRNA (movement was visualized with a Qdot bound to Egl). Fully reconstituted mRNPs with K10 mRNA have the highest run frequency, while mRNPs reconstituted with K10 mRNA lacking the TLS zip code showed reduced run frequencies. Two unrelated mRNA constructs, S. cerevisiae ASH1 mRNA and mammalian β-actin mRNA showed run frequencies similar to that of K10 mRNA lacking the TLS zip code. Error bars, sem, n ≥ 4 movies per condition, two independent experiments. (D) Kymographs illustrating motion of dynein-dynactin-BicD-Egl (DDBE) in the absence or presence of K10 mRNA. (E) Kymograph showing moving DDBE-K10 mRNA complexes dual labeled with Egl bound to a Qdot (red) and K10 mRNA labeled with Alexa Fluor 488-UTP (green). See Figure 4—source data 1.
-
Figure 4—source data 1
Dataset for Figure 4.
- https://doi.org/10.7554/eLife.36306.016
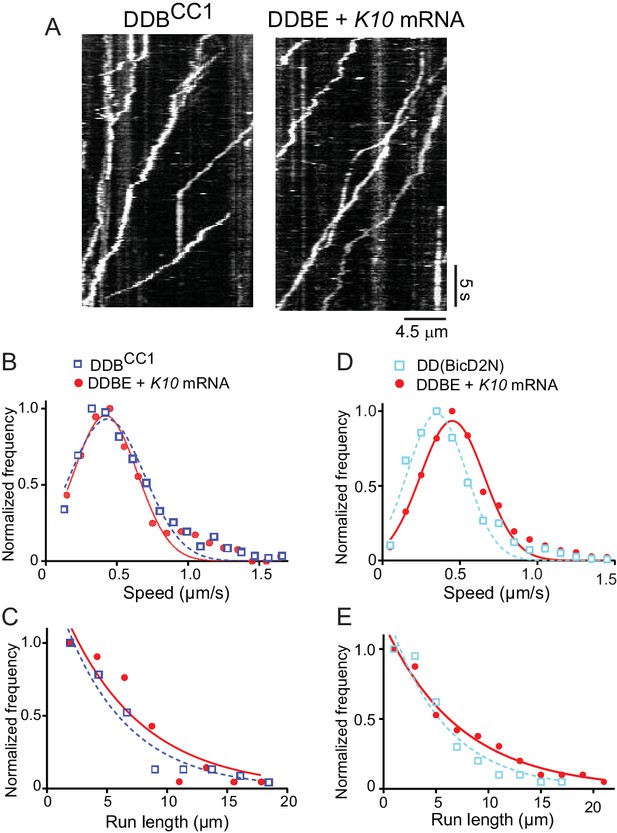
The motile properties of the fully reconstituted mRNP are similar to dynein-dynactin complexes reconstituted with Drosophila DDBCC1 or mammalian BicD2N.
(A) Kymograph of (left panel) a minimal dynein-dynactin-BicDCC1 (DDBCC1) complex, and (right panel) a complex of dynein, dynactin, full-length Drosophila BicD, Egl and K10 mRNA, visualized with a Qdot on BicD (B) Speed distributions of DDBE + K10 mRNA (red circles) (0.42 ± 0.22 μm/s, n = 505) and DDBCC1 (blue squares) (0.43 ± 0.26 μm/s, n = 516, p=0.25, t-test, mean ± SD). (C) Run length distributions of DDBE + K10 mRNA (red circles) (6.4 ± 0.25 μm, n = 71) and DDBCC1 (blue squares) (5.3 ± 0.17 μm, n = 66, p=0.8, Kolmogorov–Smirnov test, mean ±SE of the fit). (D) Speed distributions of DDBE + K10 mRNA (red circles) (0.45 ± 0.21 µm/s, n = 1126) and DD(BicD2N) (blue squares) (0.35 ± 0.19 µm/s, n = 1147; p<0.05, t-test, from 50 representative run trajectories for each condition, mean ± SD). (E) Run length distributions of DDBE + K10 mRNA (red circles) (7.2 ± 0.5 μm, n = 142) and DD(BicD2N) (blue squares) (5.4 ± 0.7 μm, n = 137, p=0.053, Kolmogorov–Smirnov test, mean ± SE of the fit). See Figure 5—source data 1.
-
Figure 5—source data 1
Dataset for Figure 5.
- https://doi.org/10.7554/eLife.36306.019

Kymograph of (left panel) a minimal dynein-dynactin-BicD2N complex, visualized with a Qdot bound to BicD2N, and (right panel), a complex of dynein, dynactin, full-length Drosophila BicD, Egl and K10 mRNA labeled with Alexa Fluor 488-UTP.
The straight vertical line at the left of both kymographs shows complex accumulation at the minus-end of the microtubule.
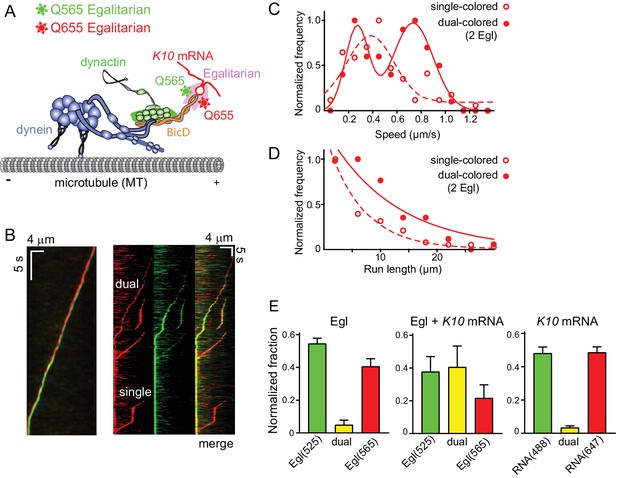
Motile properties of complexes containing one versus two Egl molecules.
(A) Schematic of the two-color experiment in which Egl is labeled with either a 565 or 655 nm QDot (figure adapted from [Reck-Peterson et al., 2018]). (B) Kymographs of dual-color runs. (Left panel) trajectories of a run with both a 565 nm (green) and a 655 nm (red) Qdot-labeled Egl bound to the moving complex. The red trajectory is shifted horizontally for presentation purposes. (Right panels) Red, green and combined channels of several processive runs. The top three trajectories are dual-color runs, while the bottom one is a single-color run. (C) Speeds of single-color complexes (open red circles) were best fit to a single Gaussian distribution with a speed of 0.38 ± 0.20 μm/s (n = 81). In contrast, the dual-colored complex (filled red circles) was best fit with a double Gaussian distribution with a slow speed of 0.27 ± 0.1 μm/s, and a faster speed of 0.73 ± 0.16 μm/s (n = 62). (D) Run length histogram of single-color events (open red circles) (6.4 ± 0.7 μm, n = 81) are significantly shorter than dual-color runs (filled red circles) (12.1 ± 2.4 μm, n = 62; p=0.045, Kolmogorov–Smirnov test, mean ± SE of the fit). The curves are exponential fits to the data. (E) Dual-colored Egl (525 or 565 Qdots), dual-colored Egl plus mRNA, or dual-colored mRNA (Alexa 488-UTP or Andy 647-UTP) mixed in solution and adhered to a glass coverslip. The percent single- and dual-colored complexes are indicated. After correction for complexes with two copies of the same color Egl (see Materials and methods), 83% of complexes were determined to contain two molecules of Egl. See Figure 6—source data 1.
-
Figure 6—source data 1
Dataset for Figure 6.
- https://doi.org/10.7554/eLife.36306.021
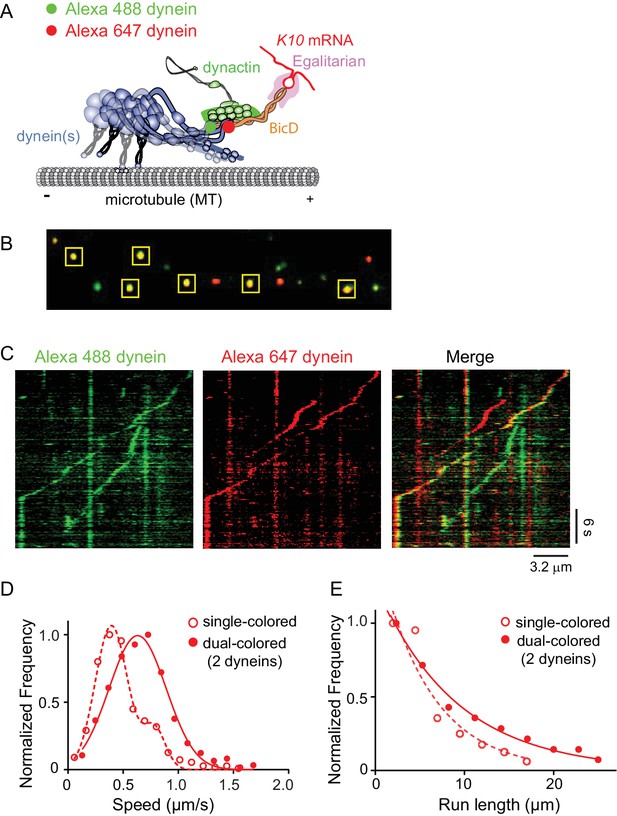
Recruitment of two dynein motors to the mRNP results in faster and longer runs.
(A) Schematic of the two-color experiment. Dynein was either labeled with Alexa 488 (green) or Alexa 647 (red) for the single-molecule pulldowns shown in panel B. mRNPs were formed by incubating with two dyneins per dynactin (figure adapted from [Reck-Peterson et al., 2018]). (B) Single-molecule pulldowns in the presence of AMP-PNP showed that 22% of complexes were dual-colored, implying that 44% of complexes had two dynein motors bound. (C) Kymographs showing single- and dual-colored runs, with dynein labeled with either a 525 or a 655 nm Qdot. Speed and run length are quantified in panels D and E. (D) The speed of dual-colored complexes (filled red circles) (0.63 ± 0.26 μm/s, n = 40), were compared to that of single-colored complexes (open red circles) (0.40 ± 0.14 μm/s and 0.74 ± 0.13 μm/s, n = 44). (E) Run lengths of dual-colored runs (filled red circles) (8.9 ± 0.5 μm, n = 40) were 53% longer than the single-colored complexes (open red circles) (5.8 ± 1.1 μm, n = 44, p=0.036, Kolmogorov–Smirnov test, mean ± SE of the fit). See Figure 7—source data 1.
-
Figure 7—source data 1
Dataset for Figure 7.
- https://doi.org/10.7554/eLife.36306.023
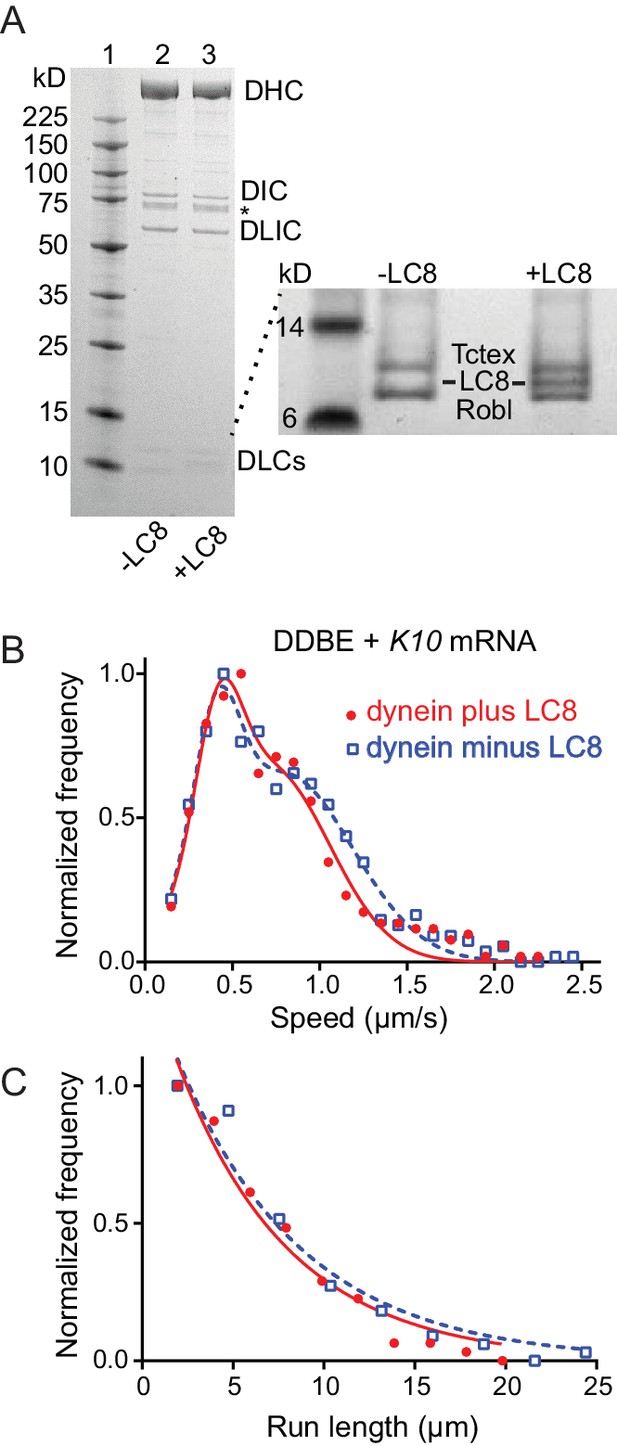
Comparison of motile mRNPS reconstituted with WT dynein or dynein without LC8.
(A) SDS-PAGE of (lane 1) molecular mass markers, (lane 2) expressed WT dynein, (lane 3) dynein expressed without LC8. The identity of the bands labeled DIC and DLIC was confirmed by mass spectrometry. The band marked with an asterisk is a FLAG-reactive fragment that was shown by mass spectrometry to be derived from the heavy chain. DHC, dynein heavy chain; DIC, dynein intermediate chain; DLIC, dynein light intermediate chain. Higher loads of the same samples are shown in the inset so that the light chains can be visualized. 4–12% SDS-PAGE, MES buffer. (B) WT dynein speeds (filled red circles) (0.42 ± 0.13 μm/s and 0.71 ± 0.34 μm/s, n = 396) were not statistically different from dynein minus the LC8 light chain (open blue squares) (0.40 ± 0.13 μm/s and 0.79 ± 0.39 μm/s, n = 488; p=0.57, t-test, mean ± SD). (C) WT dynein run lengths (6.2 ± 0.11 μm, n = 113) were the same as dynein without LC8 (6.9 ± 0.14 μm, n = 102 r; p=0.46, Kolmogorov–Smirnov test, mean ± SE). Complexes were reconstituted with two dyneins per dynactin. See Figure 8—source data 1.
-
Figure 8—source data 1
Dataset for Figure 8.
- https://doi.org/10.7554/eLife.36306.026
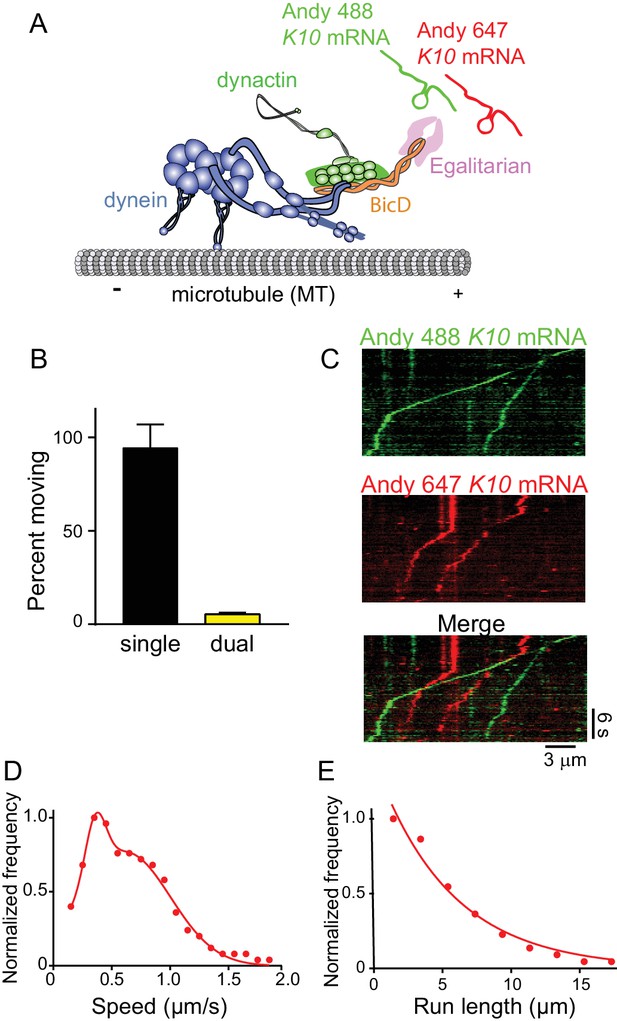
The mRNP predominantly contains one K10 mRNA.
(A) Schematic of the two-color mRNA experiment. mRNA was either labeled with Andy 488-UTP (green) or Andy 647-UTP (red) (figure adapted from [Reck-Peterson et al., 2018]). (B) 5.5% of moving complexes were dual-labeled, implying that 11% of the complexes contained 2 mRNAs, and 89% contained one mRNA. (C) Representative kymographs highlighting the predominance of single-colored moving complexes. (D) The speed pattern was bimodal (0.36 ± 0.09 μm/s and 0.62 ± 0.38 μm/s, n = 187) because the complex was reconstituted with two dyneins per dynactin. (E) Run lengths were 5.5 ± 0.09 μm (n = 67). See Figure 9—source data 1.
-
Figure 9—source data 1
Dataset for Figure 9.
- https://doi.org/10.7554/eLife.36306.028
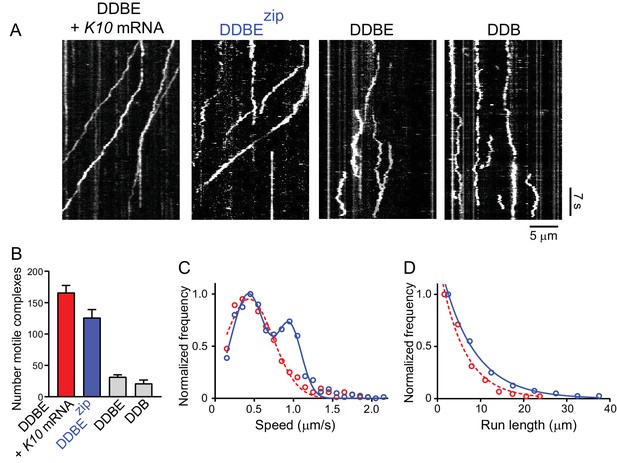
A truncated Egl-leucine zipper construct supports motility in the absence of mRNA.
(A) Kymographs illustrating motion of mRNPS reconstituted from dynein-dynactin-BicD-Egl-K10 mRNA, dynein-dynactin-BicD-Eglzip, dynein-dynactin-BicD-Egl, or dynein-dynactin-BicD. (B) Run frequency normalized to dynein concentration and time for the same four scenarios illustrated in panel A. (C) Bimodal speeds of dynein-dynactin-BicD-Eglzip (blue circles), with the slower population moving at 0.43 ± 0.21 μm/s and the faster population at 0.95 ± 0.15 μm/s (n = 633). For comparison, dynein-dynactin-BicD-Egl-K10 mRNA (red circles) moved at 0.44 ± 0.31 μm/s (n = 560) (p<0.001, t-test, mean ± SD) (D) Run lengths of dynein-dynactin-BicD-Eglzip (blue circles) were longer than those of dynein-dynactin-BicD-Egl-K10 mRNA (red circles) (7.6 ± 0.03 μm, n = 84 vs. 5.7 ± 0.09 μm, n = 106; p=0.03, Kolmogorov–Smirnov test, mean ± SE). See Figure 10—source data 1.
-
Figure 10—source data 1
Dataset for Figure 10.
- https://doi.org/10.7554/eLife.36306.030
Videos
Low-resolution 3D map of negative stain EM data (related to Figure 3).
The video shows the size comparison of the apparent loop (EM volume depicted in gray mesh) with existing structures for parts of the YFP-BicD-Egl complex. The map has not been validated, and serves only as a guide. YFP molecules at the N-terminus of BicD are depicted in green (PDB ID: 2Y0G). The blue molecule is the exonuclease domain of RNAseD (PDB ID:1YT3), which serves as a proxy for the proposed exonuclease-like domain of Egl (Mach and Lehmann, 1997; Moser et al., 1997; Navarro et al., 2004). The white coiled-coil is the BicD2 N-terminal fragment from the structure of the dynein-dynactin-BicD2 complex (Urnavicius et al., 2015) (PDB ID:5AFU), fitted into the straighter part of the looped structure. In this interpretation, the remaining loop bulge must consist of CC2 and CC3. Given that CC2 is contiguous in sequence with the end of CC1, CC2 is likely to be the bottom part of the ‘b’ structure, while CC3 binds to a region near the middle of CC1.
mRNPs with two dyneins move faster and longer (related to Figure 7).
The DDBE plus K10 mRNA complex containing two dimeric dyneins (yellow due to colocalization of a red and green Qdot) moved 12.2 µm in 20.4 s at a speed of 0.6 µm/s on a microtubule track (unlabeled). For comparison, a single dynein (red Qdot) moved a shorter distance (4.5 µm) at a slower speed (0.36 µm/s). The image was magnified twofold and played at 6x real time.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Chemical compound, drug | Alexa Fluor 488–5-UTP | Molecular Probes | C11403 | |
| Chemical compound, drug | Andy Fluor 488-X-UTP | GeneCopoeia | C410A | |
| Chemical compound, drug | Andy Fluor 647-X-UTP | GeneCopoeia | C418A | |
| Chemical compound, drug | Rnase Inhibitor | Promega | N261B | |
| Chemical compound, drug | Q-dot 525 streptavidin conjugate | Invitrogen | Q10141MP | |
| Chemical compound, drug | Q-dot 565 streptavidin conjugate | Invitrogen | Q10131MP | |
| Chemical compound, drug | Q-dot 655 streptavidin conjugate | Invitrogen | Q10121MP | |
| Chemical compound, drug | SNAP-Surface Alexa Fluor 488 | New England BioLabs | S9129S | |
| Chemical compound, drug | SNAP-Surface Alexa Fluor 647 | New England BioLabs | S9136S | |
| Chemical compound, drug | SNAP-Biotin | New England BioLabs | S9110S | |
| Chemical compound, drug | Ribonucleic acid, transfer from Escherichia coli | Sigma-Aldrich | R1753 | |
| Chemical compound, drug | Tubulin protein (X-rhodamine): bovine brain | Cytoskeleton, Inc | TL620M-A | |
| Chemical compound, drug | paclitaxel | Cytoskeleton, Inc | TXD01 | |
| Commercial kit | RiboMAX Large Scale RNA Production Systems | Promega | P1280 | |
| Recombinant DNA | pDyn1 (SNAPf-His-ZZ-LTLT- DYNC1H1 in pACEBac1) (Homo sapiens) | Schlager et al. (2014) | NCBI:NP_001367.2 | Expression plasmids for dynein in Sf9 cells. See details in Materials and methods. |
| Recombinant DNA | pDyn3 (ZZ-SNAPf-DYNC1H1, DYNC1I2, DYNC1LI2, DYNLT1, DYNLRB1,DYNLL1 in pDynBac1) (Homo sapiens) | Schlager et al. (2014) | NCBI:NP_001367.2 | Expression plasmids for dynein in Sf9 cells. See details in Materials and methods. |
| Recombinant DNA | dynein nBiotin tag (Homo sapiens) | This paper | NCBI:NP_001367.2 | Expression plasmids for dynein in Sf9 cells. See details in Materials and methods. |
| Recombinant DNA | Bicaudal D, isoform A (Drosophila melanogaster) | This paper | NCBI:NP_724056.1 | Expression plasmids for BicD, YFP-BicD in Sf9 cells, and BicDCC1 in E. coli. see details in Materials and methods. |
| Recombinant DNA | Bicaudal D homolog 2 isoform 2 (Homo sapiens) | This paper | NCBI:NP_056065.1 | Expression plasmids for BicD2N in E. coli. See details in Materials and methods. |
| Recombinant DNA | Egalitarian (Drosophila melanogaster) | This paper | NCBI:AAB49975.2 | Expression plasmids for Egl in Sf9 cells, and Egl-ZIP in E. coli. See details in Materials and methods. |
| Recombinant DNA | K10 mRNA (Drosophila melanogaster, f) | This paper | NCBI:NM_058143.3 | Expression plasmids for K10 mRNA, K10 no zip mRNA. See details in Materials and methods. |
| Recombinant DNA | β-actin mRNA (Rattus norvegicus) | This paper | NCBI:NM_031144.3 | Expression plasmids for b actin mRNA. See details in Materials and methods. |
| Recombinant DNA | ASH1 mRNA (Saccharomyces cerevisiae) | Sladewski et al. (2013) | NCBI:NM_001179751.1 | Expression plasmids for Ash1 mRNA. See details in Materials and methods. |
| Recombinant DNA | kinesin G235A (Mus musculus) | This paper | NCBI:NM_008449.2 | Expression plasmids for rigor kinesin in E. coli. See details in Materials and methods. |
| Biological sample | dynein - dynactin | Bovine brain | See details in Materials and methods. | |
| Biological sample | tubulin | Bovine brain | See details in Materials and methods. | |
| Software, algorithm | Nikon ECLIPSE Ti microscope | Nikon | ||
| Software, algorithm | Nikon NIS Elements | Nikon | ||
| Software, algorithm | Andor EMCCD Camera | Andor Technology USA | ||
| Software, algorithm | Prism | GraphPad | v7; RRID:SCR_002798 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.36306.031





