PDF-1 neuropeptide signaling regulates sexually dimorphic gene expression in shared sensory neurons of C. elegans
Figures
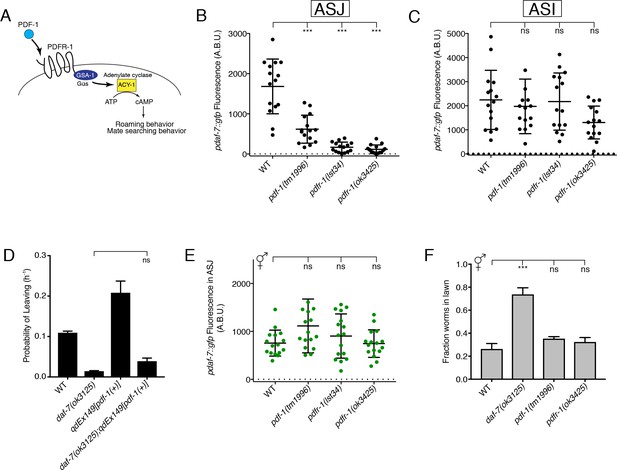
The PDF-1 pathway is required for the male-specific expression of daf-7 in the ASJ neurons and its effects on male mate-searching behavior.
(A) PDF-1 signaling activates cAMP production and regulates both roaming behavior and male mate-searching behavior in C. elegans. (B–C) Maximum fluorescence values of pdaf-7::gfp in the ASJ (B) and ASI (C) neurons of adult male animals. ***p<0.001 as determined by ordinary one-way ANOVA followed by Dunnett’s multiple comparisons test. Error bars represent standard deviation (SD). ns, not significant. n = 15 animals for all genotypes. (D) Probability of leaving values for epistasis experiment between daf-7(ok3125) and a PDF-1 overexpressing line. Values plotted are the mean +SEM for three independent experiments. Significance determined by unpaired t-test with Welch’s correction. ns, not significant. n = 60 total animals for all genotypes except the daf-7(ok3125); qdEx149 strain where n = 48. (E) Maximum fluorescence values of pdaf-7::gfp in the ASJ neurons of hermaphrodites after 16 hr on P. aeruginosa. Significance determined by ordinary one-way ANOVA followed by Dunnett’s multiple comparisons test. Error bars represent SD. ns, not significant. n = 15 animals for all genotypes. (F) Lawn occupancy of animals on P. aeruginosa after 16 hr. ***p<0.001 as determined by ordinary one-way ANOVA followed by Dunnett’s multiple comparisons test. Values plotted indicate the mean + SD for three replicates. Number of animals assayed are as follows: WT (n = 89), daf-7 (n = 66), pdf-1 (n = 117), pdfr-1 (n = 105).
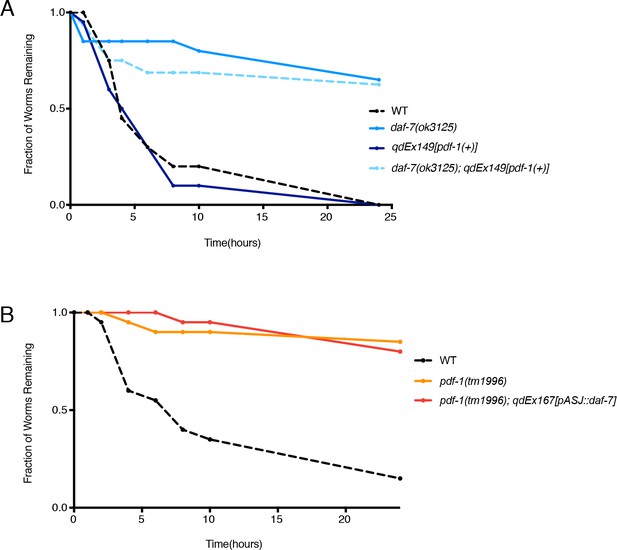
The PDF-1 pathway regulates mate-searching behavior via male-specific modulation of daf-7 expression in ASJ and through parallel mechanisms.
(A) Representative mate-searching data curves for WT (dashed black), daf-7 mutant (light blue), pdf-1(+) transgenics (dark blue), and the daf-7; pdf-1(+) double (light blue dashed) males. n = 20 animals for each curve shown except for daf-7;pdf-1(+) where n = 16 animals. (B) Representative mate searching data curves for WT (dashed black), pdf-1 mutant (orange), and pdf-1 mutant males with daf-7 overexpressed from ASJ (red). n = 20 for all genotypes.
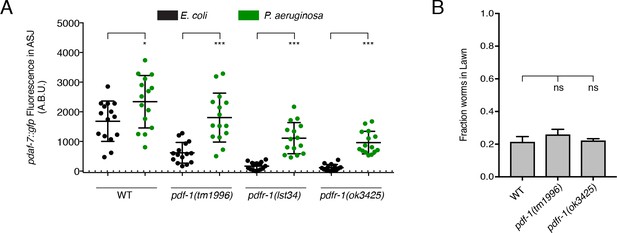
PDF-1 pathway mutant males can upregulate daf-7 expression in ASJ in response to P. aeruginosa exposure and show intact pathogen avoidance behavior.
(A) Maximum fluorescence values of pdaf-7::gfp expression in ASJ for males exposed to E. coli (black) or P. aeruginosa (green) for 16 hr. ***p<0.001, *p<0.05 as determined by unpaired t-tests with Welch’s correction. n = 15 animals for all genotypes and conditions. (B) Lawn occupancy of animals on P. aeruginosa after 16 hr. Significance as determined by ordinary one-way ANOVA followed by Dunnett’s multiple comparisons test. ns, not significant. Values plotted indicate the mean + SEM for three independent experiments. 30 animals were assayed per experiment (n = 90 across all experiments for all genotypes).
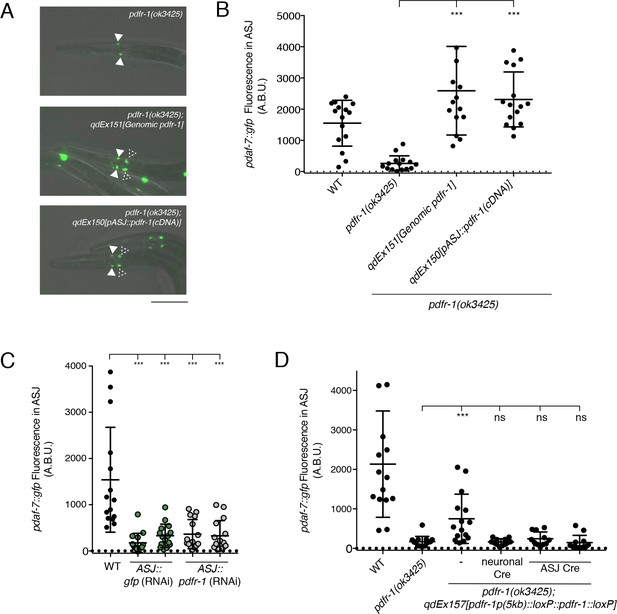
PDF-1 signaling is necessary and sufficient in the ASJ neurons for the regulation of daf-7 expression in male C. elegans.
(A) pdaf-7::gfp expression in pdfr-1(ok3425) mutant (top), genomic rescue (middle), and ASJ-specific rescue (bottom) male animals. Filled arrowheads indicate the ASI neurons; dashed arrowheads indicate the ASJ neurons. Scale bar indicates 50 μm. (B) Maximum fluorescence values of pdaf-7::gfp in the ASJ neurons of pdfr-1 rescue males. ***p<0.001 as determined by ordinary one-way ANOVA followed by Dunnett’s multiple comparisons test. Error bars represent SD. n = 15 animals for all genotypes. (C) Maximum fluorescence values of pdaf-7::gfp in the ASJ neurons of WT males (black) and animals with ASJ-specific RNAi of either GFP (green) or pdfr-1(gray). ***p<0.001 as determined by ordinary one-way ANOVA followed by Dunnett’s multiple comparisons test. Error bars indicate SD. n = 15 animals for all conditions. (D) Maximum fluorescence values of pdaf-7::gfp in the ASJ neurons of WT, pdfr-1(ok3425) mutants, and animals with floxed pdfr-1 rescued under the control of a 5 kb distal reporter as reported in Flavell et al. (2013) (left three columns). pdfr-1 function was removed either in all neurons or specifically in ASJ using cell-specific expression of Cre recombinase (right three columns). ***p<0.001 as determined by ordinary one-way ANOVA followed by Dunnett’s multiple comparisons test. ns, not significant. Error bars indicate SD. n = 12–15 animals for each condition.
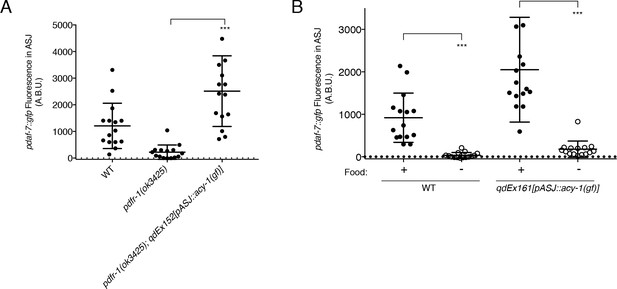
ACY-1 acts downstream of PDFR-1 to regulate daf-7 expression in male ASJ neurons.
(A) Maximum fluorescence values of pdaf-7::gfp in the ASJ neurons of males expressing the gain-of-function ACY-1(P260S) cDNA specifically in ASJ. ***p<0.001 as determined by unpaired t-test with Welch’s correction. Error bars represent SD. n = 15 animals for all genotypes. (B) Maximum fluorescence values of pdaf-7::gfp in the ASJ neurons of WT and ASJ-specific acy-1(gf) expressing fed (filled circles) and starved (open circles) males. ***p<0.001 as determined by unpaired t-test with Welch’s correction. Error bars indicate SD. n = 15 animals for all conditions.
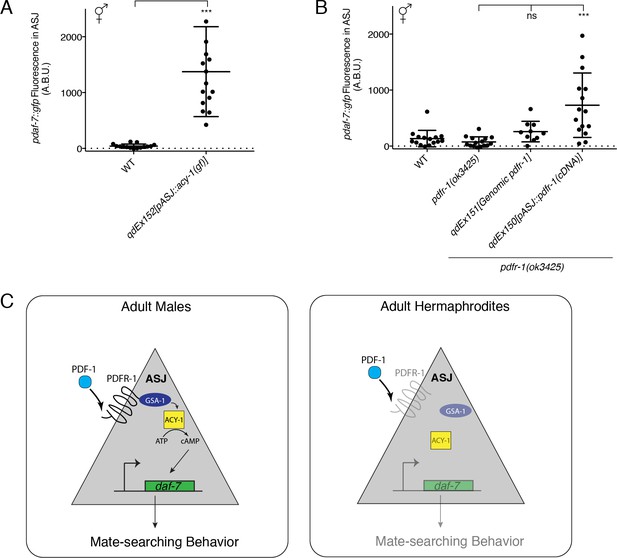
Heterologous activation of the PDF-1 pathway in ASJ is sufficient to activate daf-7 transcription in adult hermaphrodites.
(A) Maximum fluorescence values of pdaf-7::gfp in the ASJ neurons of WT hermaphrodites and hermaphrodites where ACY-1 has been activated (via the gain of function P260S mutant) specifically in ASJ. ***p<0.001 as determined by unpaired t-test with Welch’s correction. Error bars represent SD. n = 15 animals for both genotypes. (B) Maximum fluorescence values of pdaf-7::gfp in the ASJ neurons of hermaphrodites overexpressing pdfr-1 from either a genomic fragment or under the control of a heterologous ASJ-specific promoter. ***p<0.001 as determined by ordinary one-way ANOVA followed by Dunnett’s multiple comparisons test. Error bars represent SD. ns, not significant. n = 15 animals for all genotypes except pdfr-1(ok3425); qdEx151 where n = 10 animals. (C) Model for the sex-specific regulation of daf-7 expression in the ASJ chemosensory neurons by the PDF-1 signaling pathway.
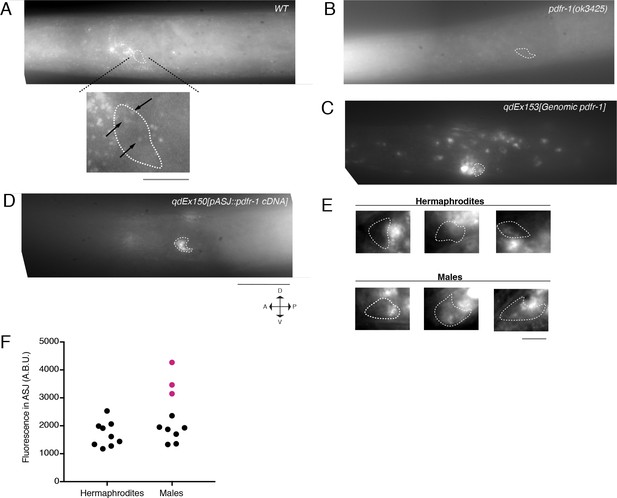
FISH imaging of endogenous pdfr-1 mRNA transcripts.
(A) pdfr-1 transcripts in a WT male animal. mRNA can be observed in neurons as well as diffusely in muscle and throughout the body. Magnified image of an ASJ neuron is shown on bottom. Arrows indicate pdfr-1 transcripts. Scale bar represents 5 µm. (B) pdfr-1 mRNA in a pdfr-1(ok3425) mutant male. No mRNA can be visualized indicating the specificity of the designed probe set. (C) pdfr-1 transcripts in a male carrying a genomic fragment including the pdfr-1 locus on an extrachromosomal transgenic array. Animals carrying this array have increased pdfr-1 expression but with all the endogenous regulatory sequence, facilitating easier analysis of expression in specific neurons, such as ASJ. (D) pdfr-1 mRNA in a pdfr-1(ok3425) male with heterologous expression of pdfr-1 cDNA in the ASJ neurons. No transcripts can be observed outside of the ASJ neurons. All images presented in (A)-(D) are maximum intensity z-projections of 28 stacked exposures of pdfr-1 mRNA. ASJ neurons are outlined in each image by a white dotted line as localized by either ptrx-1::gfp or pdaf-7::gfp (for the ASJ rescue line). Animals are representative samples from mixed stage populations. All images were taken at 100x magnification. Scale bar indicates 50 µm. (E) Representative FISH images of endogenous pdfr-1 mRNA molecules in adult hermaphrodite (top) and male (bottom) ASJ neurons. White dotted lines indicate outline of ASJ cell body as localized by ptrx-1::gfp. Animals imaged carried a transgene with the pdfr-1 genomic locus to increase expression (see Figure 4—figure supplement 1C). Male images are representative of animals where puncta can be observed. Scale bar represents 5 µm. (F) Quantification of pdfr-1 probe fluorescence in ASJ of 9 hermaphrodite and 10 male ASJ neurons. Magenta data points indicate animals where pdfr-1 mRNA puncta were visible in the ASJ neurons.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Caenorhabditis elegans) | pdf-1 | NA | WBGene00020317 | |
| Gene (C. elegans) | pdfr-1 | NA | WBGene00015735 | |
| Gene (C. elegans) | daf-7 | NA | WBGene00000903 | |
| Gene (C. elegans) | acy-1 | NA | WBGene00000068 | |
| Genetic reagent (C. elegans) | ksIs2 | PMID: 11677050 | WBTransgene00000788 | pdaf-7::gfp |
| Recombinant DNA reagent | Moerman Fosmid Library | Source Bioscience | WRM0629dH07, WRM0627cG01, WRM0641dA07, WRM068aD11 | |
| Recombinant DNA reagent | pPD95.75 | Fire Lab C. elegans Vector Kit | Addgene plasmid # 1494 | |
| Recombinant DNA reagent | pCFJ90 | PMID:18953339 | Addgene plasmid # 19327 | pmyo-2::mCherry, used as co-injection marker |
| Recombinant DNA reagent | pZH42 | this paper | pdf-1 genomic DNA in pUC19 | |
| Recombinant DNA reagent | pZH48 | this paper | ptrx-1::pdfr-1(cDNA, b isoform, no STOP codon)::F2A::mCherry:: unc-54 3'UTR | |
| Recombinant DNA reagent | pZH53 | this paper | ptrx-1::ACY-1(P260S)::unc-54 3'UTR | |
| Recombinant DNA reagent | pZH58 | this paper | pdfr-1p(distal, 5 kb)::loxP::pdfr-1 cDNA(B isoform)::loxP::unc-54 3'UTR | |
| Recombinant DNA reagent | pZH59 | this paper | ptrx-1::nCre | |
| Recombinant DNA reagent | pJDM30 | PMID: 25303524 | ptrx-1::daf-7 | |
| Recombinant DNA reagent | pSF11 | PMID: 23972393 | ptag-168::nCre, gift of C. Bargmann and S. Flavell | |
| Commercial assay or kit | NEBuilder HiFi DNA Assembly Master Mix | NEB | E2621 | |
| Software, algorithm | GraphPad Prism | GraphPad | RRID:SCR_002798 | |
| Other | Alexa Fluor 647 DHS ester | Invitrogen/Thermo Fisher Scientific | A20006 | dye used for conjugation of FISH probes |
Additional files
-
Supplementary file 1
C. elegans strains used in this study.
A comprehensive list of the strains used in this study. Strain source (this study or others) is indicated.
- https://doi.org/10.7554/eLife.36547.009
-
Supplementary file 2
Oligos used for transgenic strain construction and pdfr-1 FISH
A comprehensive list of the oligos created for this study. All sequences are listed in the 5’ to 3’ direction.
- https://doi.org/10.7554/eLife.36547.010
-
Transparent reporting form
- https://doi.org/10.7554/eLife.36547.011





