VivosX, a disulfide crosslinking method to capture site-specific, protein-protein interactions in yeast and human cells
Figures
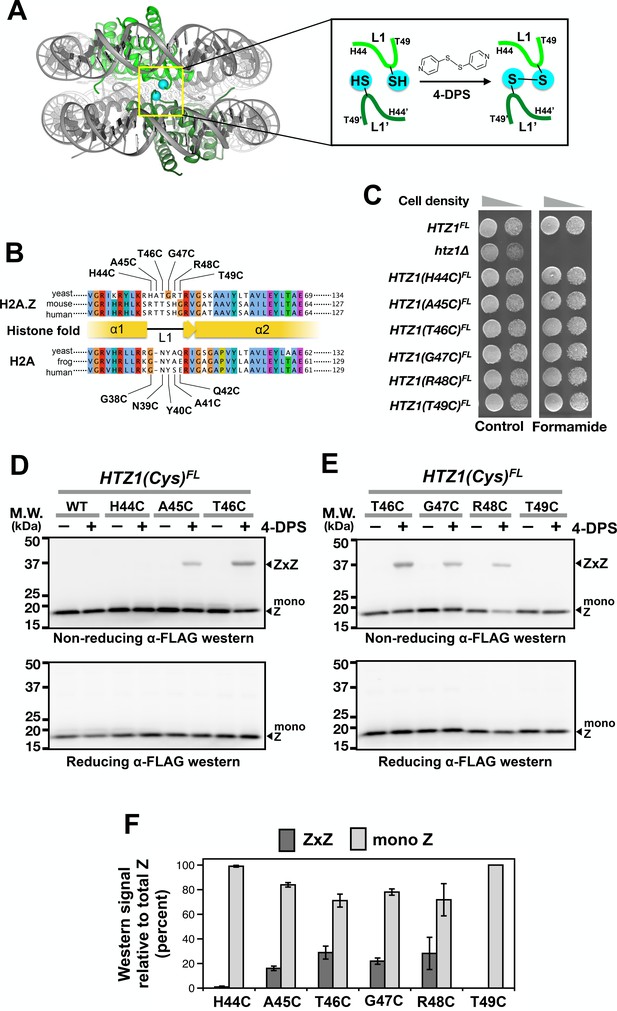
Cysteine substitution at multiple positions of the L1 region of H2A.Z supports Z-to-Z crosslinking in yeast cells.
(A) The L1-L1’ interface of the ZZ nucleosome (PDB: 1F66) is highlighted by a yellow box (Suto et al., 2000). The two nucleosomal H2A.Z-H2B dimers are highlighted in green. Cyan spheres mark the alpha-carbon (Cα) of T41 of mouse H2A.Z, which corresponds to the T46 position of yeast Htz1. Inset depicts the 4-DPS-dependent crosslinking reaction between the L1 cysteines of the two H2A.Z molecules within the ZZ nucleosome. Numbering of the amino acids refers to the yeast Htz1. (B) Sequence alignment analysis in and around the L1 region of H2A.Z and H2A from three different species was performed using the Clustal Omega algorithm in Jalview (Waterhouse et al., 2009). (C) Genetic complementation was performed by spotting equal number of cells (and 10-fold dilution in spots on right) of the indicated strains onto YPD media with and without 2.5% formamide. (D–E) VivosX analysis of yeast H2A.Z. Wild-type and HTZ1(Cys)FL mutants that were treated with 180 µM of 4-DPS (+) or with DMSO (–) were analyzed by non-reducing (top panel) or reducing (bottom panel) SDS-PAGE followed by anti-FLAG immunoblotting. (F) Quantification of Z-to-Z crosslinking efficiency. Bars in dark gray (ZxZ) represent the mean ratios of ZxZ signal over total H2A.Z signal (i.e. ZxZ plus mono Z) in (D) and (E). Bars in light gray (mono Z) represent the mean ratios of mono Z over total H2A.Z. Means and standard deviations (error bars) were calculated from at least three biological replicates. HTZ1(T46C) were performed six times. ZxZ: Z-to-Z cystine adducts. Mono Z: uncrosslinked H2A.Z.
-
Figure 1—source data 1
Values used to plot Figure 1F.
- https://doi.org/10.7554/eLife.36654.007
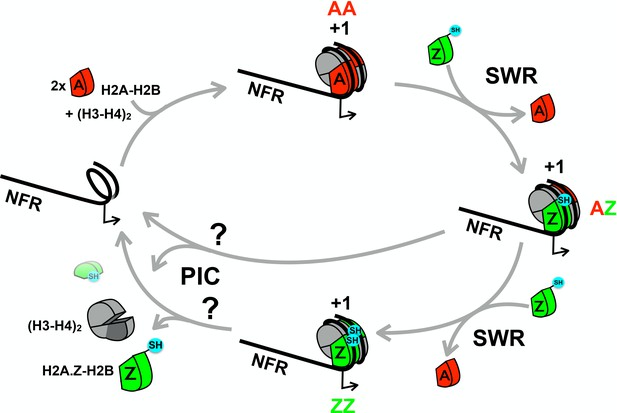
A cartoon depicting the proposed histone cycle.
Z-B dimers are in green, A-B dimers in red and H3-H4 tetramers in gray. NFR: Nucleosome free region. Black arrows: transcription start site. +1: the nucleosome immediately downstream of a promoter. PIC: preinitiation complex. Balls in cyan: cysteine probes. Cysteine probes on H2A are not shown for simplicity.
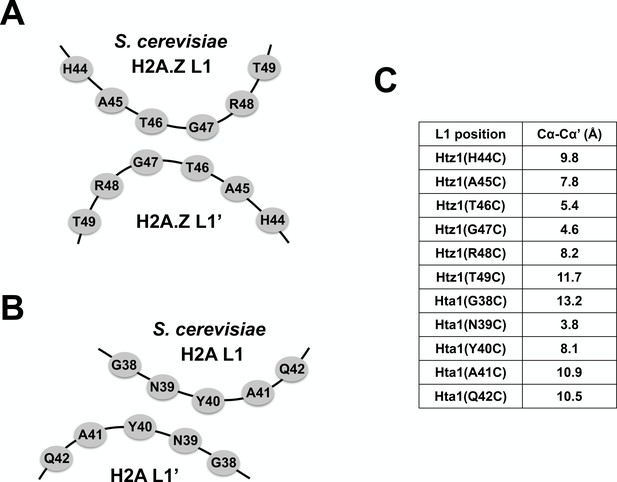
Relative Cα-Cα’ distances at the L1-L1’ interface of AA and ZZ nucleosomes.
(A,B) Cartoons depicting the anti-parallel L1 and L1’ loops of yeast H2A.Z and H2A. (C) A table of Cα-Cα’ distances measured between the indicated amino acid positions in the AA and ZZ nucleosomes. The Cα-Cα’ distances for H2A.Z are based on the published mouse/frog ZZ nucleosome structure (Suto et al., 2000) but the corresponding positions of yeast Htz1 were indicated. The Cα-Cα’ distances are based on the yeast AA nucleosome structure (White et al., 2001).
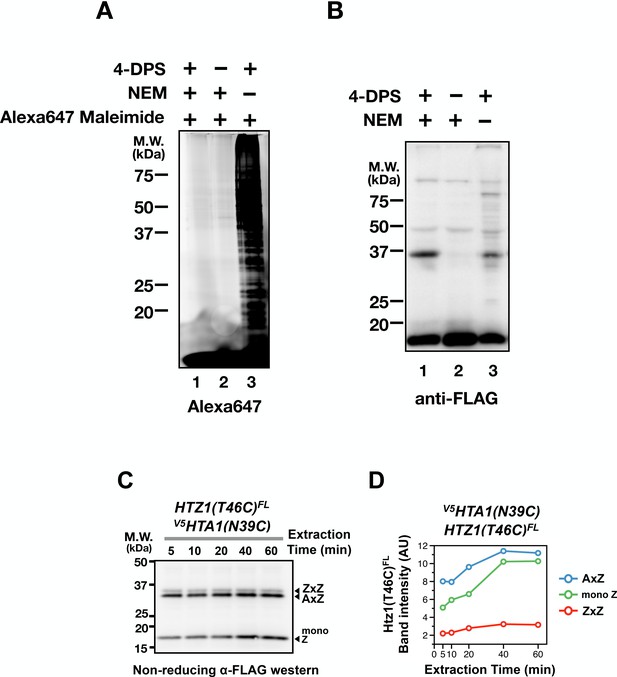
Effects of NEM blocking and cystine linkage stability.
(A) Logarithmically growing HTZ1(T46C)FL cells were incubated with or without 4-DPS (180 µM) before TCA fixation and protein extraction with the TUNES buffer with or without NEM (50 µM). Solubilized total proteins were dialyzed against two changes of 1 L TUNES buffer without NEM using the Slide-A-Lyzer button (10,000 Da MWCO, Thermo Fisher) at room temperature. Alexa647 maleimide (Thermo Fisher) was added to each reaction to a final concentration of 100 µM and was incubated at room temperature for 1 hr before analyzed by reducing SDS-PAGE and fluorescent densitometry. (B) Same as (A), except that the extracted proteins (before the dialysis step) were analyzed by non-reducing SDS-PAGE and anti-FLAG immunoblotting. (C) The V5HTA1(N39C) HTZ1(T46C)FL cells were treated with 4-DPS (180 µM) for 20 min before TCA fixation as described in Figure 1D. But after the addition of the TUNES buffer (with NEM), aliquots of were removed at the indicated times. The extracted proteins were analyzed by non-reducing SDS-PAGE and anti-FLAG western. (D) Band intensities of AxZ, ZxZ, and mono Z in (C) were plotted as a function of extraction time.
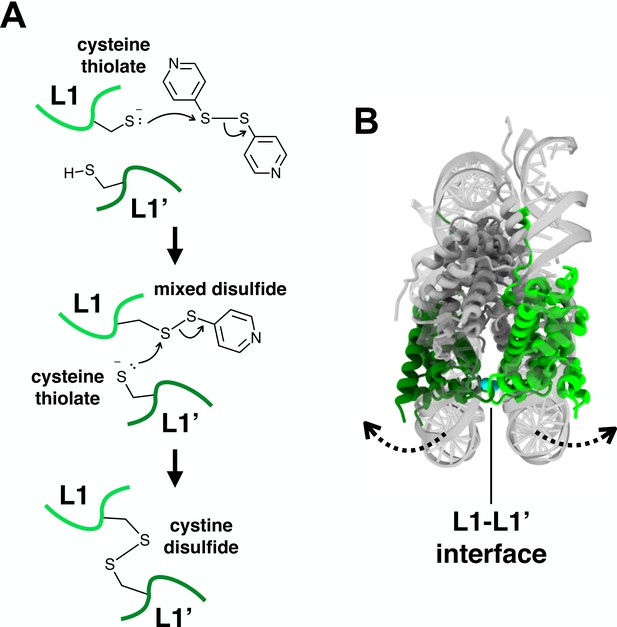
The thiol-disulfide interchange reaction between the cysteine thiols and 4-DPS.
(A) A proposed mechanism of the thiol-disulfide interchange reaction between cysteine thiols and 4-DPS. Light and dark green loops represent the L1 regions of the opposite nucleosomal H2A.Z (or H2A) pair at the L1-L1’ interface. (B) The proposed clamshell opening of the nucleosome structure could facilitate the conformational dynamics of the L1-L1’ interface. Dotted arrows: direction of movement.
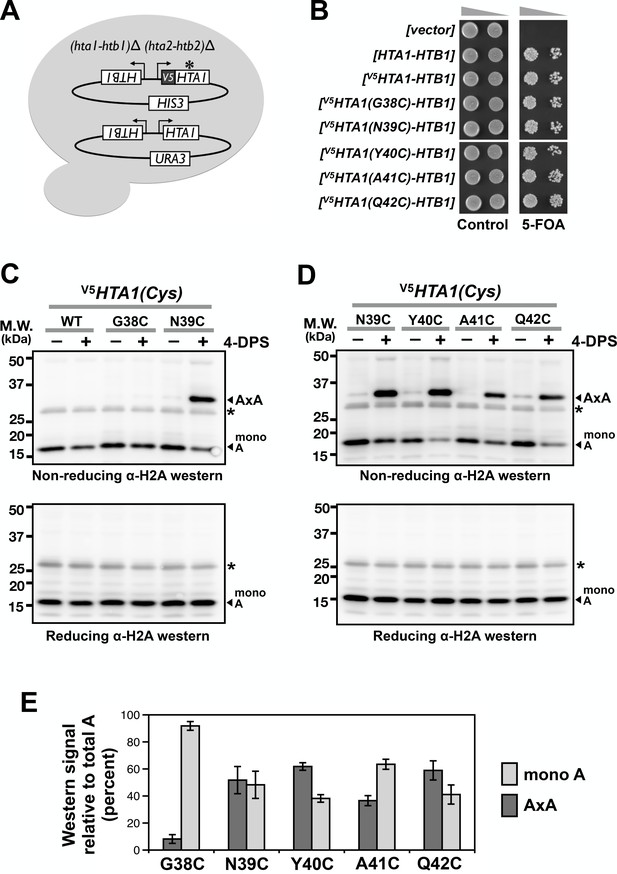
Cysteine substitution at multiple positions of the L1 region of H2A supports A-to-A crosslinking in yeast cells.
(A) The cartoon depicts the plasmid shuffle yeast system used to verify the functionality of the V5HTA1(Cys) mutants. (B) The ability of the HTA1(Cys) mutants to complement the lack of endogenous genes for H2A and H2B was indicated by growth in the presence of 5-FOA, which removes the wild type [HTA1-HTB1-URA3] vector from the cells. (C,D) VivosX analysis of yeast H2A was performed as described in Figure 1D except that the immunoblots were probed with an anti-H2A antibody (Active Motif). Asterisk (*) indicates a non-specific band. (E) Quantification of the AxA adducts and the uncrosslinked H2A (mono A) was performed as described in Figure 1F. Bars and error bars indicate the means and standard deviation of three biological replicates.
-
Figure 2—source data 1
Values used to plot Figure 2E.
- https://doi.org/10.7554/eLife.36654.009
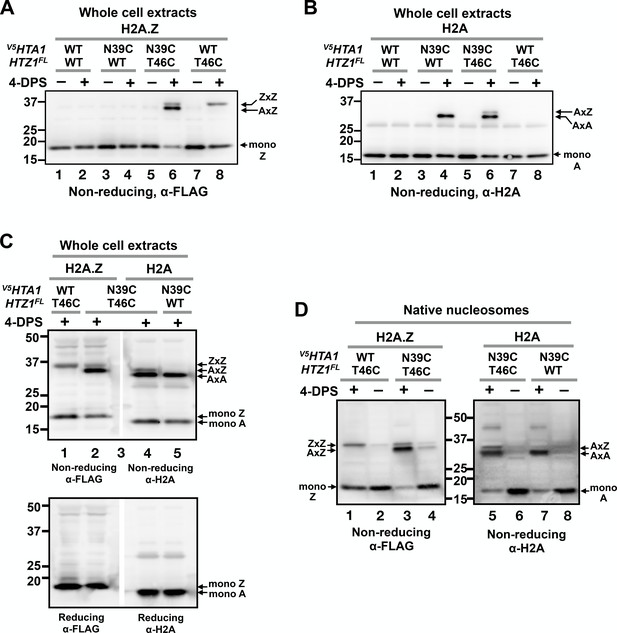
VivosX distinguishes AA, AZ and ZZ nucleosomal species.
(A) Yeast cells expressing V5HTA1(N39C) or V5HTA1 in combination with HTZ1(T46C)FL or HTZ1FL were analyzed by VivosX as described in Figure 1D. An anti-FLAG antibody was used to detect the FLAG-tagged H2A.Z and its crosslink adducts. (B) Same as (A), except that the immunoblot was probed with anti-H2A antibody. (C) The indicated samples were analyzed as in (A) and (B). But after the proteins were transferred to a PVDF membrane, the membrane was cut in the middle of lane 3, which contained the molecular weight markers. The two halves of the membranes were probed with either the anti-FLAG or anti-H2A antibodies. The top and bottom panels were analyzed by non-reducing and reducing SDS-PAGE, respectively. (D) Native nucleosomes released by MNase digestion of the chromatin pellets prepared from the indicated strains were incubated with 4-DPS (+) or DMSO (-). After TCA precipitation and extraction with the TUNES buffer, the histones were analyzed by non-reducing SDS-PAGE and anti-FLAG (left) or anti-H2A (right) immunoblotting.
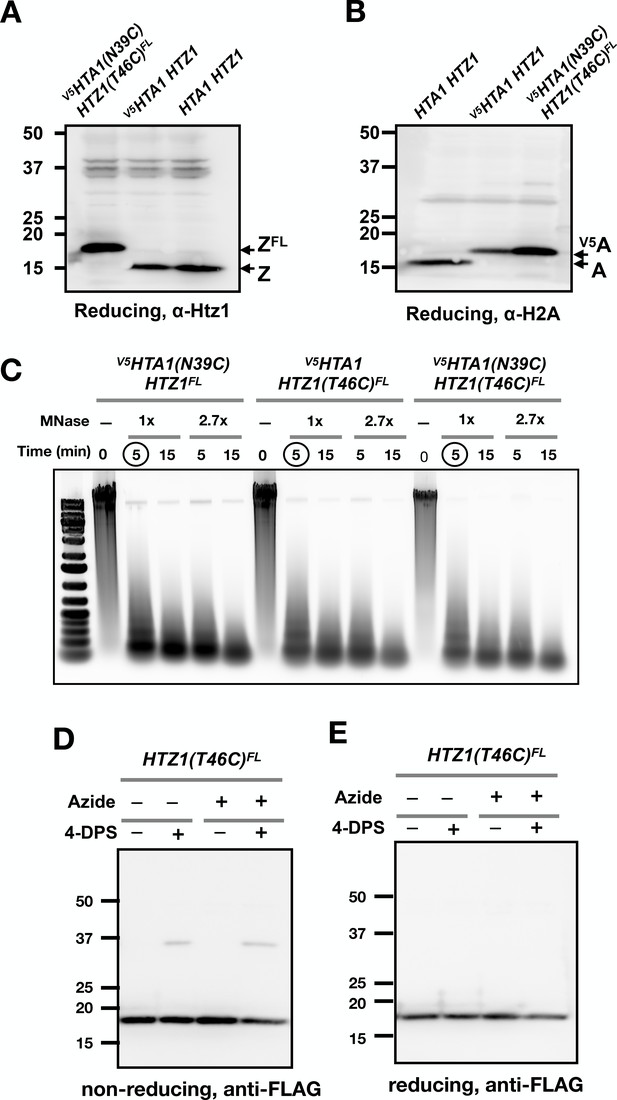
Control experiments for yeast VivosX.
(A,B) Immunoblots showing the different effects of the FLAG and V5 epitope tags on the SDS-PAGE mobility of the H2A.Z and H2A histones. (C) DNA samples purified from chromatin isolated from the indicated strains before and after MNase digestion were analyzed by agarose gel electrophoresis and SYBR Green staining. 1x MNase represents 0.08 U/µL of MNase. Circled lanes: MNase digestion conditions selected for the experiment in Figure 3D. (D,E) Logarithmically growing HTZ1(T46C)FL cells were fixed with sodium azide (0.1%) for 15 min on ice before treatment with and without 4-DPS (180 µM) for 20 min. The cells were then fixed with TCA fixation and the proteins extracted and analyzed as described for Figure 1D.
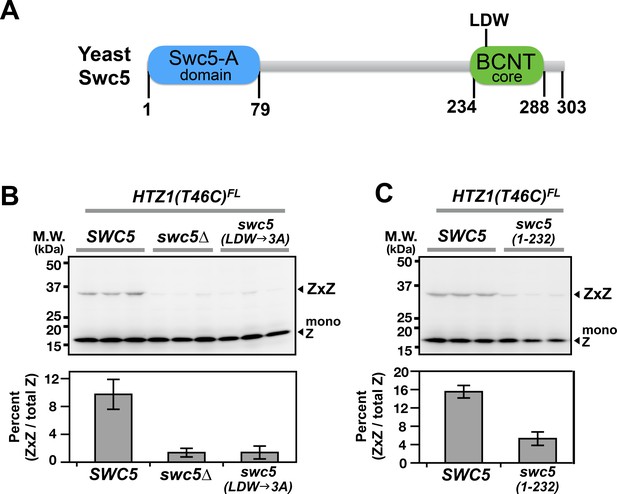
Domain analysis of Swc5 using H2A.Z VivosX.
(A) The cartoon depicts the domain organization of yeast Swc5 (Sun and Luk, 2017). (B,C) The yeast strain, HTZ1(T46C)FL swc5∆, was transformed by a single-copy plasmid containing either the wild-type SWC5 or the indicated SWC5 mutants or by the control vector (swc5∆). Top panels: Each strain was represented by three independent transformants and analyzed by VivosX in parallel. Htz1(T46C)FL and its crosslinked (ZxZ) adducts was detected by anti-FLAG immunoblotting. Bottom panels: Quantification of the immunoblots above. Bars and error bars represent the means and standard deviations of three biological replicates.
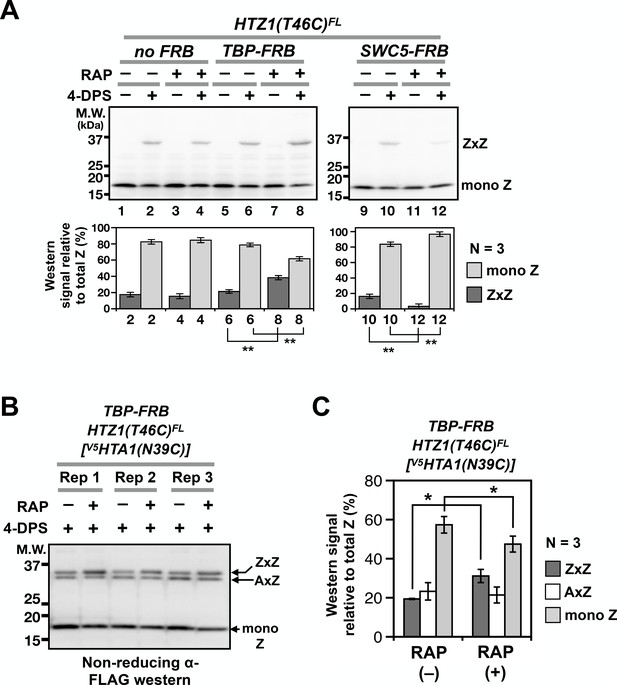
Probing H2A.Z dynamics using VivosX.
(A) HTZ1(T46C)FL strains containing the TBP-FRB or SWC5-FRB alleles or the corresponding wild-type alleles (no FRB) in the Anchor-away genetic background (W303 MAT a tor1-1 fpr1 RPL13A-2xFKBP12) (Haruki et al., 2008) were incubated with rapamycin (+RAP) or without (i.e. DMSO, –RAP) for 1 hr before each culture was divided into halves, where one half was oxidized with 4-DPS (for 20 min) and the other without. Fixation, protein extraction, and immunoblotting analysis were conducted as described for Figure 1D. Bottom panels: Quantification of the ZxZ bands and the mono Z bands was performed as described in Figure 1D. (B) VivosX was performed using HTZ1(T46C)FL TBP-FRB yeast transformed with the [V5HTA1(N39C)-HTB1] plasmid. Rep: biological replicates. (C) Quantification of (B). The immunoblot signals of ZxZ, AxZ, and mono Z were normalized to total H2A.Z. Bars and error bars represent the means and standard deviations of three biological replicates. One asterisk (*) indicates p≤0.05 and two (**) indicates p≤0.01 of t-tests.
-
Figure 5—source data 1
Values used to plot Figure 5A.
- https://doi.org/10.7554/eLife.36654.014
-
Figure 5—source data 2
Values used to plot Figure 5C.
- https://doi.org/10.7554/eLife.36654.015
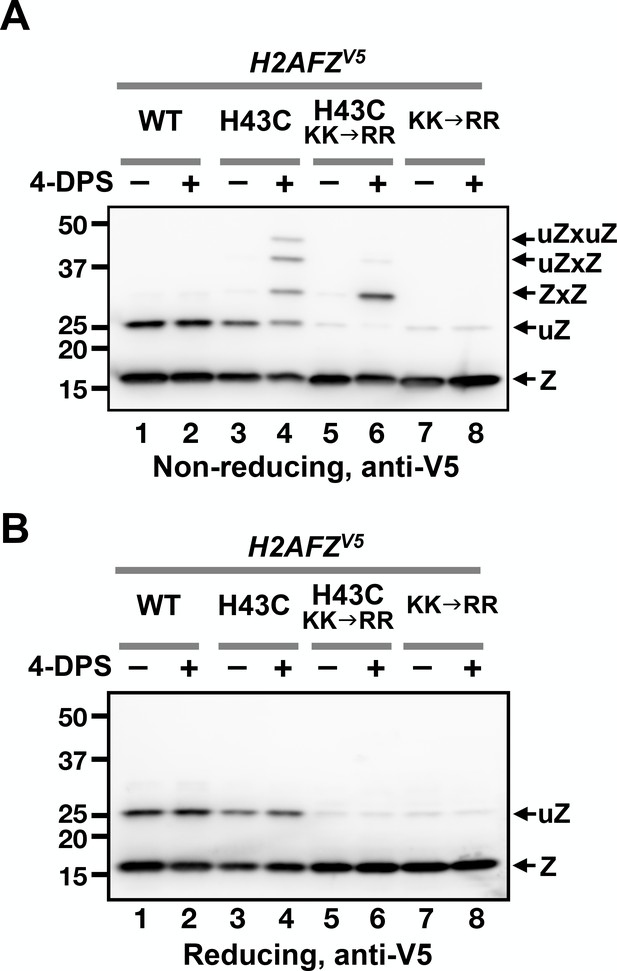
VivosX of H2A.Z in human cells.
(A, B) Human MCF10A cells expressing an ectopic H2AFZ(H43C)V5 gene was incubated with 4-DPS for 20 min before lysis using the TUNES buffer. Total lysates were resolved by non-reducing SDS-PAGE in (A) and reducing SDS-PAGE in (B) before analyzed by anti-V5 immunoblotting. Z: nonubiquitylated H2A.Z; uZ: monoubiquitylated H2A.Z; ZxZ: Z-to-Z crosslink adducts; uZxZ: crosslink adducts with one uZ and one Z; uZxuZ: crosslink adducts of two uZ molecules.
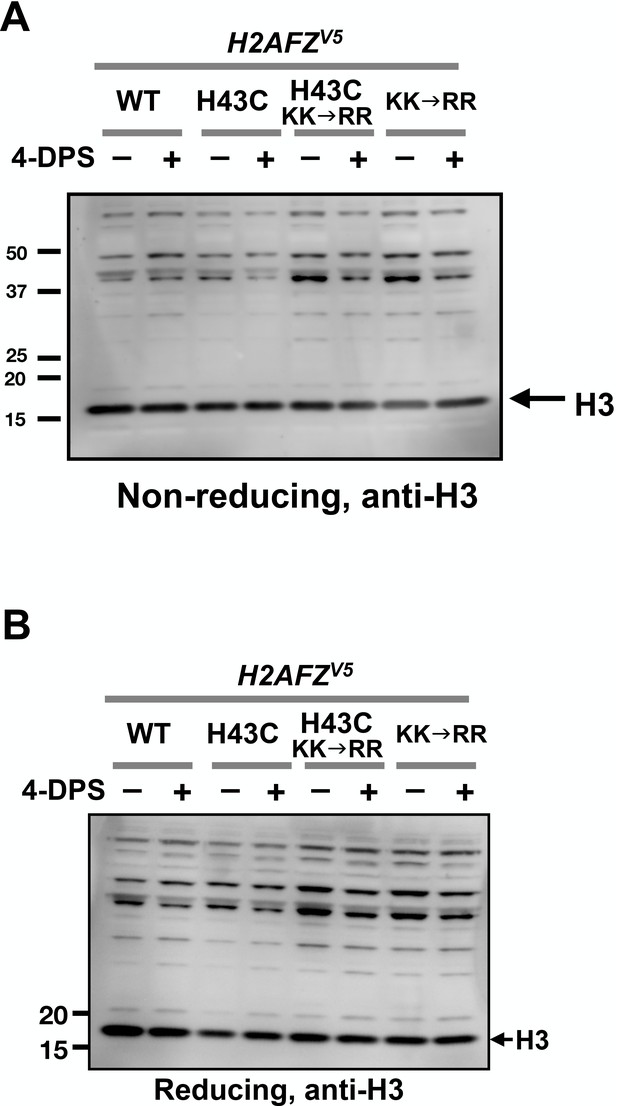
No apparent H3-to-H3 crosslinking observed after 4-DPS treatment.
The protein extracts of MCF10A cells from Figure 6 were analyzed by non-reducing SDS-PAGE in (A) and reducing SDS-PAGE in (B) followed by anti-H3 immunoblotting.
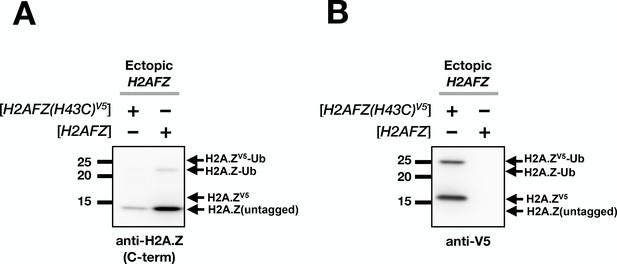
Estimation of ectopic H2A.Z level.
(A) Total lysates of MCF10A cells transfected with H2AFZ(H46C)V5 or H2AFZ (untagged) driven by the same tetracycline-inducible promoter were analyzed by immunoblotting using an antibody directed against the C-terminus of human H2A.Z. (B) The same blot was probed with the anti-V5 antibody. Note that the H2A.Z antibody failed to detect the V5-tagged H2A.Z because the V5 tag at the C-terminus interfered with antibody recognition. In addition, the ubiquitylation sites of H2A.Z overlap the epitope of the anti-H2A.Z antibody. This may explain the disproportionately low H2A.Z-Ub signal in the anti-H2A.Z blot in comparison to the anti-V5 blot. The H2A.Z-Ub recognition bias by the C-terminus H2A.Z antibodies was previously observed (Sarcinella et al., 2007).
Additional files
-
Supplementary file 1
Table of yeast strains used in this study.
- https://doi.org/10.7554/eLife.36654.019
-
Supplementary file 2
Table of plasmids and lentiviral vectors used in this study
- https://doi.org/10.7554/eLife.36654.020
-
Transparent reporting form
- https://doi.org/10.7554/eLife.36654.021





