Unravelling the history of hepatitis B virus genotypes A and D infection using a full-genome phylogenetic and phylogeographic approach
Figures
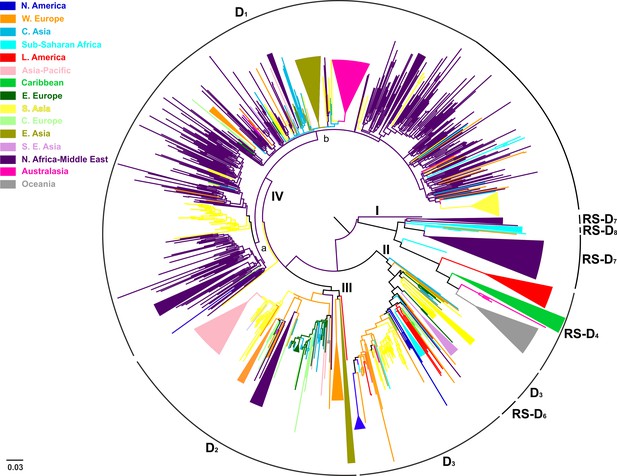
Midpoint rooted phylogeographic tree estimated by RAxML v8.0.20.
HBV genotype D sequences (N = 916) categorized according to the geographic region of sampling were used in the analysis. Monophyletic clusters are indicated as triangles. Major clusters are indicated in Roman numerals.
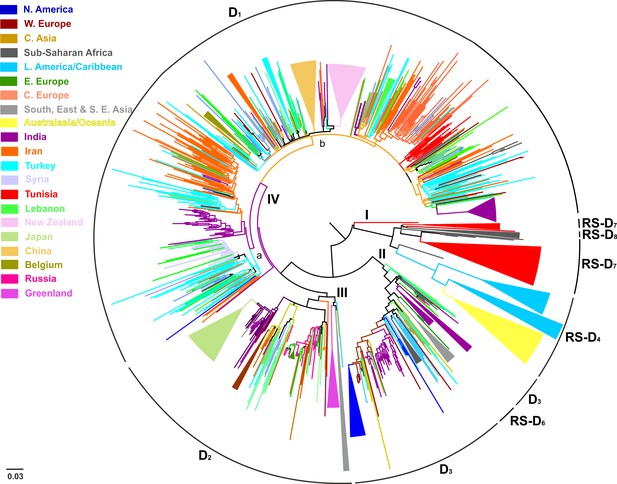
Midpoint rooted phylogeographic tree on 916 HBV genotype D sequences: categorized according to the country/geographic region of sampling.
https://doi.org/10.7554/eLife.36709.005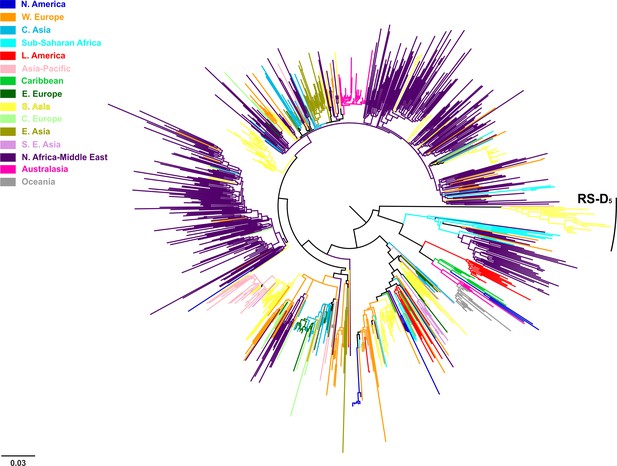
Midpoint rooted phylogeographic tree on HBV genotype D sequences after the inclusion of RS-D5.
https://doi.org/10.7554/eLife.36709.006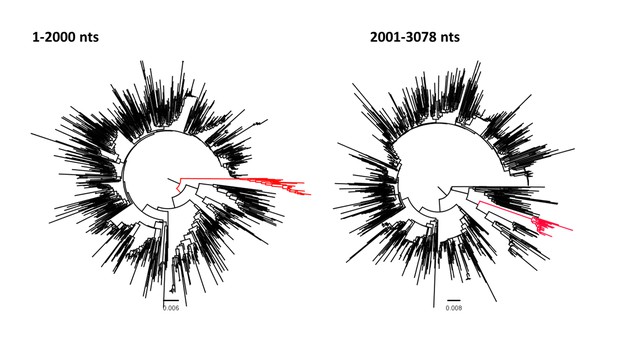
Phylogenetic analysis of genotype D including the RS-D5 in two non-overlapping subgenomic areas (1–2000 nts) and (2001–3078 nts). RS-D5 sequences are shown in red.
https://doi.org/10.7554/eLife.36709.007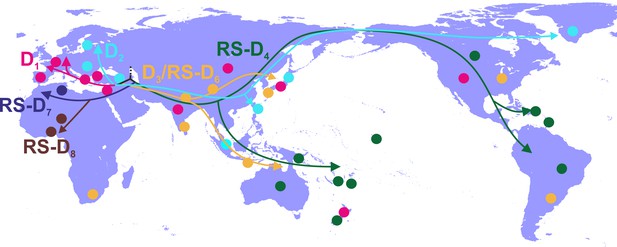
Putative major dispersal pathways of genotype D as estimated by phylogeographic analysis.
The origin is depicted with dotted line. Subgenotypes and their corresponding dispersal routes are shown with different colors. Colored circles depict the geographic areas where subgenotypes are the most prevalent.
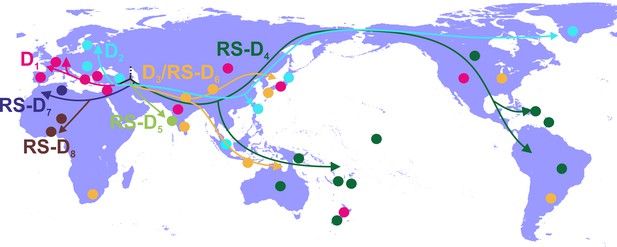
Putative major dispersal pathways of genotype D as estimated by phylogeographic analysis after the inclusion of RS-D5.
https://doi.org/10.7554/eLife.36709.011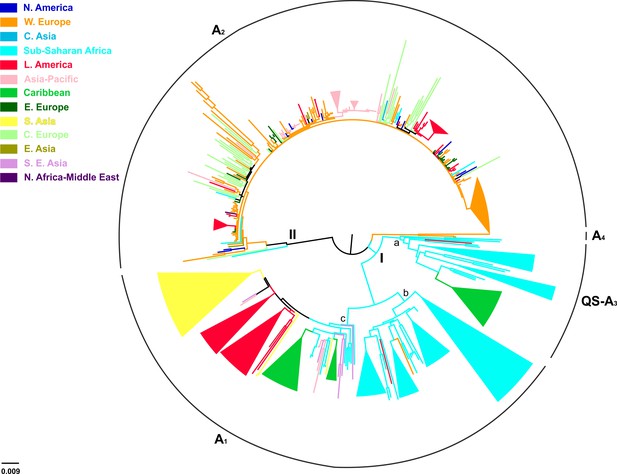
Midpoint rooted phylogeographic tree estimated by RAxML v8.0.20.
HBV genotype A sequences (N = 493) categorized according to the geographic region of sampling were used in the analysis. Monophyletic clusters are indicated as triangles. Major clusters are indicated in Roman numerals.
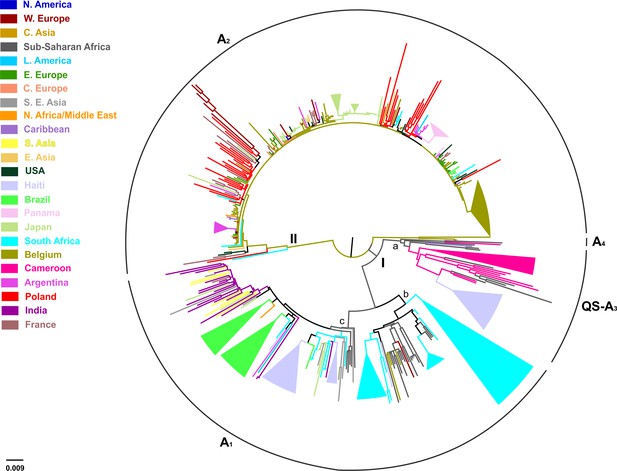
Midpoint rooted phylogeographic tree on 493 HBV genotype A sequences categorized according the country/geographic region of sampling.
https://doi.org/10.7554/eLife.36709.015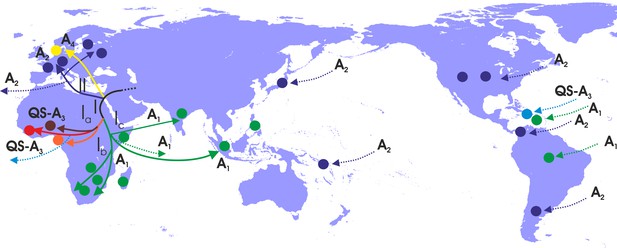
Putative major dispersal pathways for genotype A as revealed by the phylogeographic analysis.
The putative geographic origin of genotype A is in Middle East/Central Asia. Subgenotypes and their corresponding dispersal routes are shown with different colors. Colored circles depict the geographic areas where subgenotypes are the most prevalent. Dotted lines represent the source and sinks for distant dispersal pathways.
Tables
Sampling of HBV* genotype D sequences from different geographic regions.
https://doi.org/10.7554/eLife.36709.003| Region of sampling | Countries (N) | Sequences (N, %) |
|---|---|---|
| North Africa/Middle East | 5 | 394 (43.0) |
| South Asia | 2 | 151 (16.5) |
| Western Europe | 9 | 70 (7.6) |
| Central Asia | 4 | 44 (4.8) |
| Australasia | 2 | 40 (4.4) |
| Asia-Pacific | 1 | 36 (3.9) |
| East Asia | 2 | 35 (3.8) |
| Eastern Europe | 3 | 27 (3.0) |
| Latin America | 2 | 25 (2.7) |
| Oceania | 6 | 23 (2.5) |
| Sub-Saharan Africa | 8 | 20 (2.2) |
| North America | 2 | 16 (1.8) |
| Central Europe | 2 | 14 (1.5) |
| Caribbean | 2 | 11 (1.2) |
| Southeast Asia | 2 | 10 (1.1) |
| Total | 52 | 916 (100) |
-
* HBV, hepatitis B virus.
HBV* genotype D percentages of clustering based on the geographic region of sampling.
https://doi.org/10.7554/eLife.36709.008| Region of sampling | Monophyletic clusters (N) | Clustered sequences (N, %) |
|---|---|---|
| Australasia | 1 | 36 (90) |
| Oceania | 1 | 19 (83) |
| Asia-Pacific | 1 | 30 (83) |
| Southeast Asia | 1 | 8 (80) |
| East Asia | 2 | 24 (69) |
| Caribbean | 1 | 7 (64) |
| Latin America | 1 | 14 (56) |
| Sub-Saharan Africa | 2 | 11 (55) |
| North America | 1 | 8 (50) |
| Western Europe | 3 | 21 (30) |
| South Asia | 3 | 34 (23) |
| Eastern Europe | 1 | 5 (19) |
| North Africa/Middle East | 6 | 63 (16) |
| Central Asia | 0 | 0 (0) |
| Central Europe | 0 | 0 (0) |
| Total | 24 | 279 (31) |
-
* HBV, hepatitis B virus.
Sampling of HBV* genotype A sequences from different geographic regions.
https://doi.org/10.7554/eLife.36709.012| Region of sampling | Countries (N) | Sequences (N, %) |
|---|---|---|
| Sub-Saharan Africa | 14 | 112 (22.7) |
| Western Europe | 8 | 108 (21.9) |
| Latin America | 5 | 62 (12.6) |
| Asia-Pacific | 1 | 53 (10.8) |
| Caribbean | 2 | 52 (10.5) |
| Central Europe | 2 | 37 (7.5) |
| South Asia | 3 | 26 (5.3) |
| Eastern Europe | 4 | 17 (3.5) |
| North America | 2 | 11 (2.2) |
| Southeast Asia | 2 | 8 (1.6) |
| East Asia | 2 | 4 (0.8) |
| North Africa/Middle East | 2 | 2 (0.4) |
| Central Asia | 1 | 1 (0.2) |
| Total | 48 | 493 (100) |
-
* HBV, hepatitis B virus.
HBV* genotype A percentages of clustering based on the geographic region of sampling.
https://doi.org/10.7554/eLife.36709.013| Region of sampling | Monophyletic clusters (N) | Clustered sequences (N, %) |
|---|---|---|
| South Asia | 1 | 23 (88) |
| Caribbean | 3 | 44 (85) |
| Latin America | 4 | 44 (71) |
| Sub-Saharan Africa | 6 | 70 (63) |
| Western Europe | 1 | 33 (31) |
| Asia-Pacific | 2 | 11 (21) |
| Central Europe | 0 | 0 (0) |
| Eastern Europe | 0 | 0 (0) |
| Southeast Asia | 0 | 0 (0) |
| East Asia | 0 | 0 (0) |
| North America | 0 | 0 (0) |
| North Africa/Middle East | 0 | 0 (0) |
| Central Asia | 0 | 0 (0) |
| Total | 17 | 255 (52) |
-
* HBV, hepatitis B virus.
Additional files
-
Supplementary file 1
Sampling and percentages of clustering of HBV genotype D sequences from different countries and geographic regions.
- https://doi.org/10.7554/eLife.36709.009
-
Supplementary file 2
Sampling and percentages of clustering of HBV genotype A sequences from different countries and geographic regions.
- https://doi.org/10.7554/eLife.36709.017
-
Supplementary file 3
List of papers for HBV genotype D sequences included in the analysis.
- https://doi.org/10.7554/eLife.36709.018
-
Supplementary file 4
List of papers for HBV genotype A sequences included in the analysis.
- https://doi.org/10.7554/eLife.36709.019
-
Supplementary file 5
List of countries within each geographic region (as defined by the Global Burden of Disease classification system), in which HBV genotype D sequences were included in the analysis.
- https://doi.org/10.7554/eLife.36709.020
-
Supplementary file 6
List of countries within each geographic region (as defined by the Global Burden of Disease classification system), in which HBV genotype A sequences were included in the analysis.
- https://doi.org/10.7554/eLife.36709.021
-
Transparent reporting form
- https://doi.org/10.7554/eLife.36709.022





