Control of cyclic oligoadenylate synthesis in a type III CRISPR system
Figures
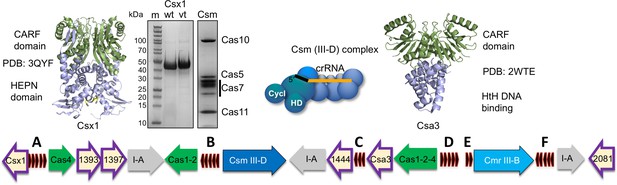
The CRISPR locus and cOA signalling proteins in S. solfataricus.
The six CRISPR loci (A-F) are shown in black. There are three type I-A systems (grey), a Csm/III D system (dark blue) and Cmr/III B system (light blue). Adaptation genes are shown in green. Numbers shown correspond to Sso gene numbers; non-Cas genes are omitted. Genes encoding CARF family proteins are outlined in purple. The structures of two CARF proteins, Sso1389 (Csx1) and Sso1445 (Csa3), are shown. The SDS-PAGE gels show purified Csx1 (Sso1389) wild-type and variant H345N, and the Csm complex.
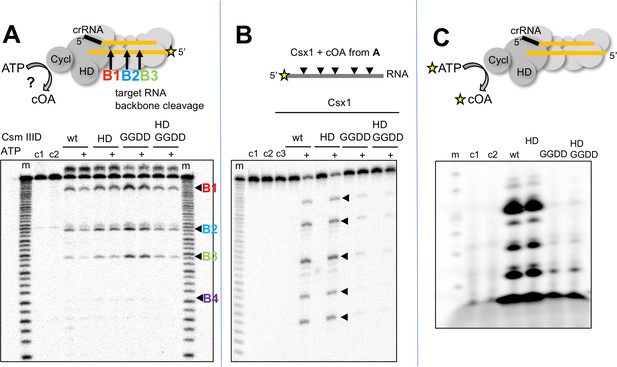
S. solfataricus Csm synthesises cOA on target RNA binding, activating the Csx1 nuclease.
(A) Backbone-mediated cleavage of a labelled target RNA. Three prominent cleavage sites (B1–B3) are indicated. Abrogation of the HD nuclease (HD) or cyclase (GGDD) domain active sites has no effect on this activity. The marker lanes (m) are generated by alkaline hydrolysis of the target RNA. The presence of ATP in the reaction is indicated by a ‘+’ symbol. Control lanes c1 and c2 represent incubation without Csm complex in the presence and absence of ATP, respectively. The diffuse density above the intact RNA is an artefact of electrophoresis. The yellow stars indicate the presence of a radioactive label. (B) Activation of the CARF nuclease Csx1 (Sso1389) by the reaction products of the Csm reaction in A. Csx1 is inactive against the labelled substrate RNA unless activated by the products of the Csm reaction. This activation is dependent on the presence of ATP in the original reaction and an intact cyclase domain. Control lanes c1 and c2 represent incubation of the RNA in buffer at 4 and 50°C, respectively, whereas c3 is a control in presence of Csx1 without added supernatant from Csm reaction. (C) Analysis of the products generated by Csm on addition of target RNA and α−32P-ATP. These products are dependent on the presence of an active cyclase domain. They represent a range of linear and cyclic polyadenosines of varying sizes. Control lanes c1 and c2 respectively represent the reaction in absence of Csm or in presence of Csm without target RNA.
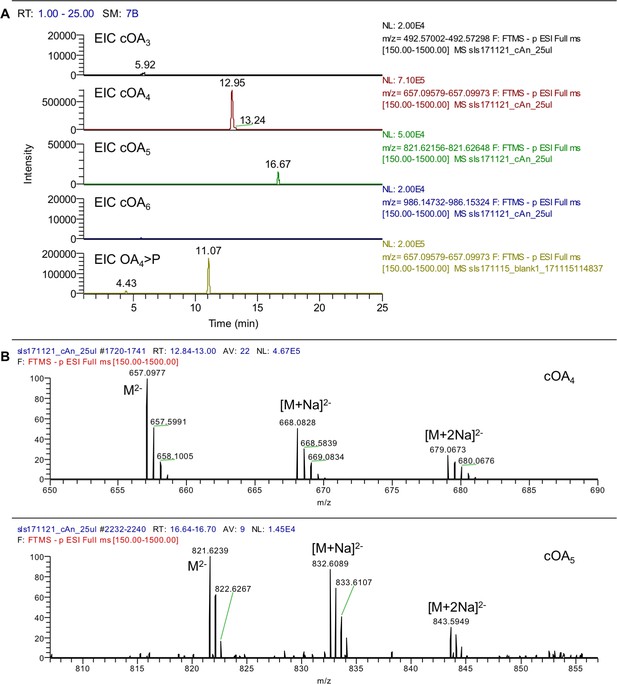
LC-MS analysis of cOA production by S. solfataricus Csm.
(A) Extracted ion chromatograms (EIC) are provided as indicated in the panels. The data confirm that cOA4 is the major product; cOA5 is present at significantly lower concentration. No other oligoadenylate species were present above the MS detection limit. The EIC for linear MazF-derived OA4 >P demonstrates the difference in chromatographic retention for the isomeric species. (B) High-resolution mass spectra for 12.9 min (cOA4) and 16.7 min (cOA5) retention time showing the doubly charged molecular ion peak alongside mono- and disodium adducts. Expected for cOA4 C40H48N20O24P4 1316.2101, found 1316.2100 (-0.1 ppm); expected for cOA5 C50H60N25O30P5 1645.2626, found 1645.2618 (-0.5 ppm).
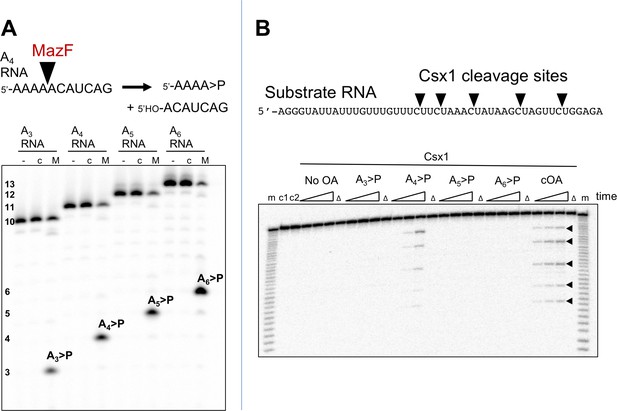
The Csx1 nuclease is activated by linear cOA analogues generated by MazF.
(A) Synthesis of linear A3 – A6 oligoadenylate molecules with cyclic 3’-phosphate termini by MazF. MazF cleaves on the 5’ side of an ACA recognition sequence. For each reaction set, lane ‘-’ is RNA substrate; ‘c’ is the incubation of the RNA in reaction buffer without added MazF in presence of trypsin; ‘M’ is RNA incubated with active MazF for 30 min. (B) Purified linear oligoadenylate species were incubated with Csx1 nuclease along with a 32P-labelled RNA substrate (A1) for 5, 15, 30 min. No Csx1 nuclease activity was detected in the absence of oligoadenylate. When reactions were supplemented by the products of the reaction with Csm, ATP and target RNA (visualised in Figure 2C) the nuclease activity of Csx1 was strongly activated. Screening of linear A3 – A6 >P demonstrated that A4 >P is the cognate activator for Csx1. Lane m shows an alkaline hydrolysis ladder of the substrate RNA; lanes marked ‘Δ’ have the inactive variant of Csx1, H345N, instead of the wild-type enzyme, incubated for 30 min at 50°C; c1 and c2 represent RNA incubated without Csx1 for 30 min at 4 and 50°C, respectively.
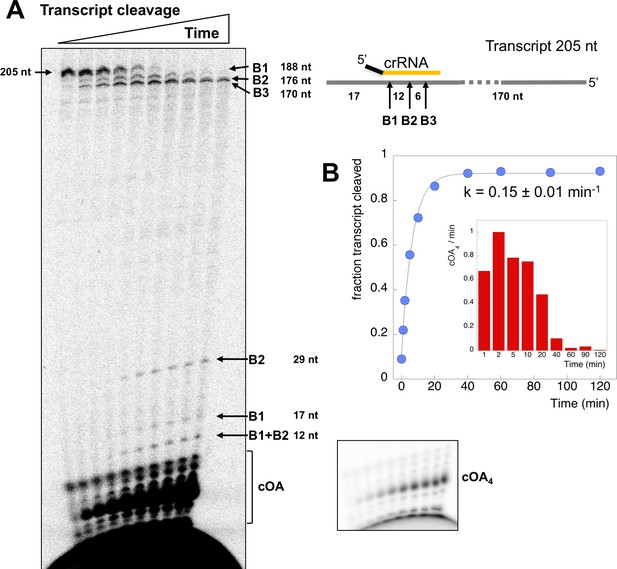
Csm transcript cleavage and cOA production.
(A) Denaturing gel electrophoresis showing cleavage of a 205 nt RNA tanscript target by Csm over time, and cOA synthesis. Products arising from cleavage at sites B1-B3 are indicated. Lanes 1–10 represent time points of 0, 1, 2, 5, 10, 20, 40, 60, 90, 120 min. (B) Quantification of the fraction of cleaved transcript against time, fitted to an exponential equation, yielded a rate of 0.15 min−1. A bar chart showing the change in cOA4 production per minute for each time period (normalised to the value at 2 min) is shown as an insert.
-
Figure 4—source data 1
Raw data for the kinetic analysis and cOA production presented in Figure 4B.
- https://doi.org/10.7554/eLife.36734.009
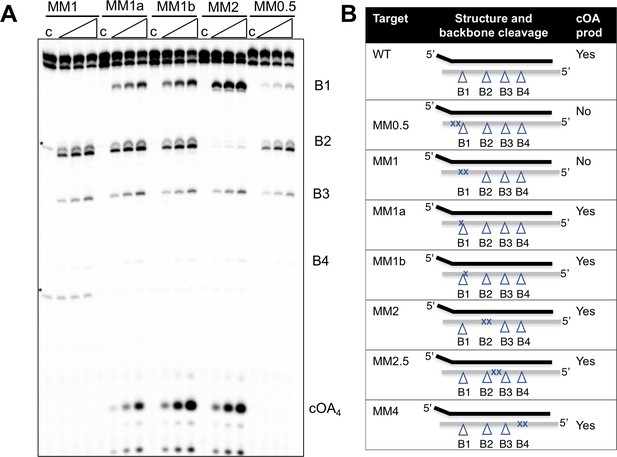
Effect of mismatched targets on cOA production.
Oligonucleotides containing mismatches ranging along the length of the target RNA were used to test both backbone cleavage and cOA synthesis activity by Csm. Backbone cleavage is abolished when two mismatches flank the cleavage site, but is otherwise active. cOA synthesis is not observed for substrates MM0.5 and MM1, which introduce two consecutive mismatches close to the 3’ end of the target RNA. Bands marked with an asterisk are artefactual bands present in the RNA substrate. For all targets, lane c shows the reaction at the zero time point and the three further time points correspond to incubation for 5, 10 and 20 min. Further substrates are shown in Figure 5—figure supplement 1. (B) Table summarising backbone cleavage and cOA production for the mismatch substrates studied in Figure 5A and Figure 5—figure supplement 1. Backbone cleavage sites are indicated with a triangle and mismatches with an ‘X’. The target RNA is represented by the grey line.
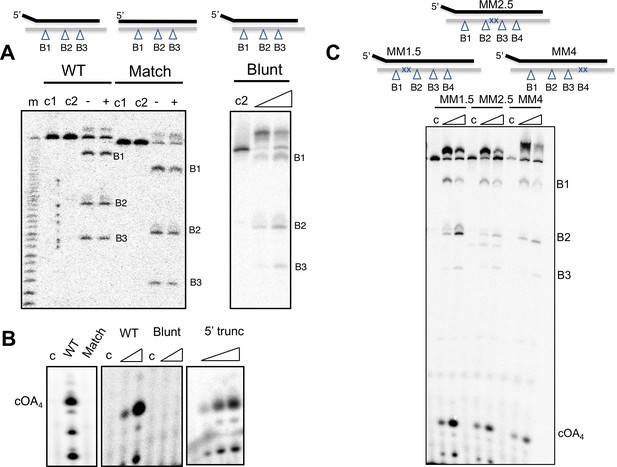
Backbone cleavage and cOA production for target RNAs with altered 5’ and 3’ ends.
(A) Backbone cleavage for Target RNA wild-type, Match (with eight nt match to 5’ handle of crRNA) and 3’-blunt (with no 3’ extension). All three targets are cleaved by the Csm complex. Control c1 and c2 represent incubation of target RNA without Csm for 30 min at 0 or 70°C, respectively. Lanes – and + represent assays in the presence or absence of 1 mM ATP, respectively. For the 3’-blunt target, time points of 5 and 30 min are shown. (B) cOA production is seen for the WT but not the Match or 3’-blunt targets. A control target (5’ -blunt) lacking the 5 nt 5’ overhang present in the WT target behaves like the WT target. This serves as a control for the Match target, which also lacks these 5 nt. Lane c is run in the absence of Csm protein. The time points are 5 and 30 min for WT and 3’-blunt, and 5, 10, 30 min for the 5’-blunt substrate. All oligonucleotide sequences are shown in Table 1. (C) Target RNAs carrying mismatches at sites M1.5, M2.5 and M4 all support backbone cleavage and cOA production. Details as for Figure 5.
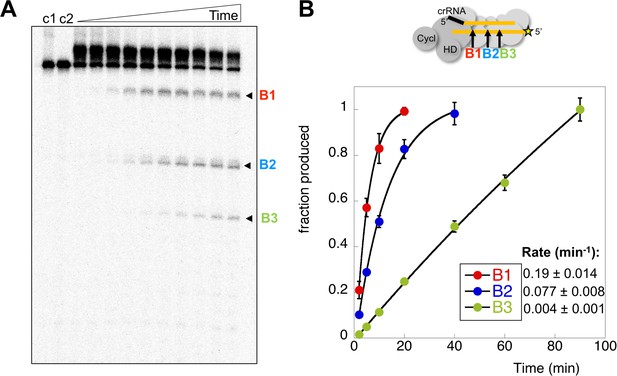
Rates of target RNA cleavage at each position.
(A) Backbone cleavage of target RNA over time (0,1, 2, 5, 10, 20, 40, 60, 90, 120 min). Controls c1 and c2 represent target RNA incubated for 120 min in the absence of Csm at 4 and 70°C, respectively. (B) The rates of target RNA backbone cleavage at sites B1-B3 were determined. Data points represent means of 4 experiments with standard deviation shown. The star represents the radioactive label.
-
Figure 6—source data 1
Raw data for the kinetic analysis presented in Figure 6B.
- https://doi.org/10.7554/eLife.36734.013
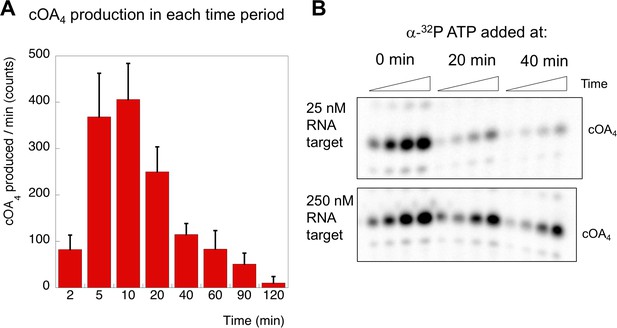
cOA production as a function of time and target RNA concentration.
(A) cOA4 synthesis was quantified using radioactive ATP under the assay conditions described for Figure 6 and plotted as cOA4 synthesised per min (densitometric counts) during the period of each reaction time slot. This was carried out in triplicate: means and standard deviations are shown. (B) In this experiment, α-32P-ATP was added to reactions at 0, 20 or 40 min and the reaction followed for a further 40 min (time points 5, 10, 20 and 40 min). Under standard single turnover conditions (25 nM target RNA), cOA synthesis was largely complete after 20 min. When target RNA was increased ten-fold, cOA synthesis persisted for a longer period.
-
Figure 7—source data 1
Raw data for the kinetic analysis presented in Figure 7A.
- https://doi.org/10.7554/eLife.36734.015
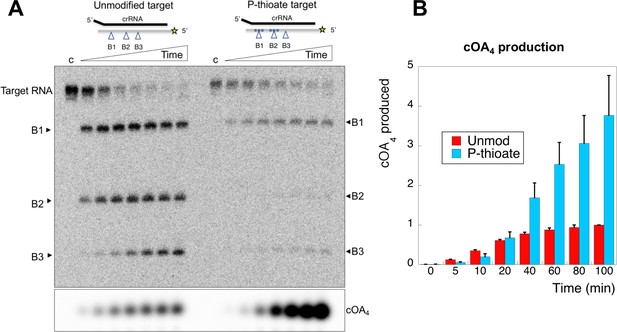
An oligonucleotide with phosphorothioate linkages reduces target RNA cleavage and enhances cOA production.
(A) Comparison of target RNA cleavage for unmodified target RNA (A26) and the same sequence with two sets of three phosphorothioate bonds at sites B1 and B2 (indicated by *). Target RNA cleavage was slowed significantly, with a particular reduction in cleavage at site B2. The synthesis of cOA4, on the other hand, was strongly stimulated over the course of the reaction. Time points are 0 (denoted ‘c’), 5, 10, 20, 40, 60, 80, 100 min. (B) Quantification of cOA4 production, showing four-fold higher levels of cOA4 after 100 min when the P-thioate modified target RNA was used. Values are the means of triplicate experiments, with standard deviation shown, and are normalised to the signal from the unmodified target RNA at 100 min.
-
Figure 8—source data 1
Raw data for the cOA production data, quantified in triplicate.
- https://doi.org/10.7554/eLife.36734.017
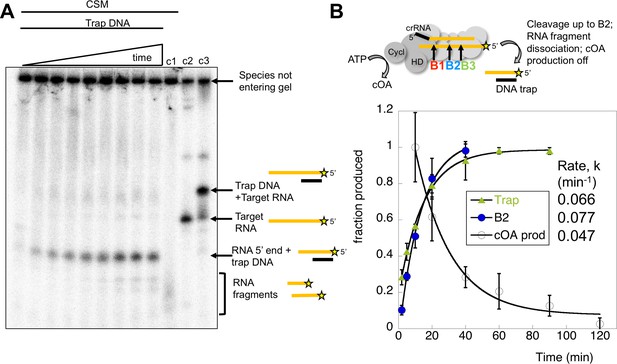
Kinetic analysis of the inactivation of cOA synthesis.
(A) Native polyacrylamide gel following the reaction products generated by Csm bound to radioactively labelled target RNA. A time course of cleavage is shown in the first nine lanes (0, 2, 5, 10, 20, 40, 60, 90, 120 min), where trapping DNA complementary to the 5’ end of the target RNA was added at the end of the reaction. Only species released from the Csm complex enter the gel. A time dependent increase in a species corresponding to a trapped RNA:DNA heteroduplex was observed. Lane c1 shows the reaction run for 2 hr in the absence of trapping DNA – a smear of ssRNA reaction products were observed. Lane c2 shows the position of full length target RNA, and lane c3 shows the full length target RNA bound to the trapping DNA. Figure 9—figure supplement 1 shows the position of a control RNA corresponding to the 5’ end of the B2 cleavage product. (B) The time-dependent accumulation of trapped RNA was quantified in triplicate, normalised and plotted. Means plus standard deviation are shown, yielding a rate koff = 0.066 min−1. The progress of cleavage at site B2 (0.077 min−1) is included for comparison. We also quantified the rate of cOA synthesis in triplicate using target RNA and normalising the rate at 10 min to the value 1.0. This fitted to an exponential with a rate constant kobs = 0.047 min−1, suggesting that product release is correlated with a rapid inactivation of cyclase activity.
-
Figure 9—source data 1
Raw data for the kinetic analysis presented in Figure 9B.
- https://doi.org/10.7554/eLife.36734.020
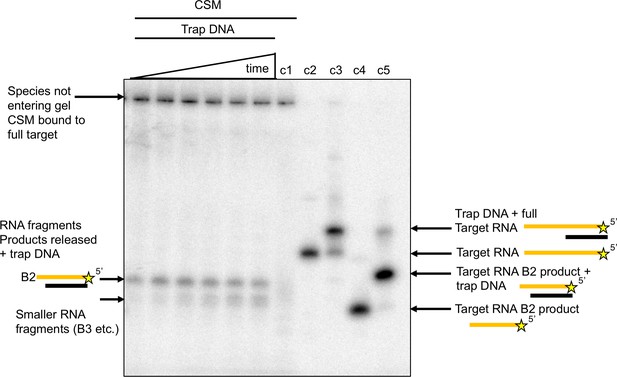
Target RNA cleaved at B2 is the main released product.
Native polyacrylamide gel following the reaction products generated by Csm bound to radioactively labelled target RNA. A time course of cleavage is shown in the first six lanes (10, 20, 40, 60, 90, 120 min), where trapping DNA complementary to the 5’ end of the target RNA was added at the end of the reaction. Only species released from the Csm complex enter the gel. A time dependent increase in a species corresponding to a trapped RNA:DNA heteroduplex was observed. Lane c1 shows the reaction run for 2 hr in the absence of trapping DNA – a smear of ssRNA reaction products can be observed. Lane c2 shows the position of full length target RNA. Lane c3 shows the full length target RNA bound to the trapping DNA. Lane c4 shows the migration position of a labelled RNA oligonucleotide corresponding to cleavage at site B2, and lane c5 shows the same oligonucleotide hybridised to the trap DNA. The gel has a slight ‘smile’ at the edge, but the migration position of the B2 product: trap DNA hybrid fits the major product trapped after Csm cleavage of target RNA.
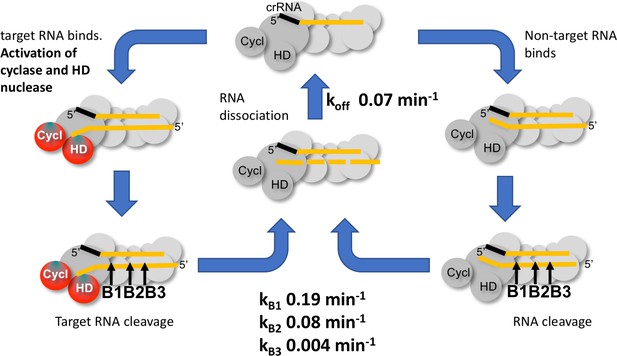
Kinetic scheme for activation and inactivation of cOA production.
In its ground state, Csm adopts a conformation where the HD nuclease and cyclase domains are inactive. On binding cognate target RNA with a non-pairing 3’ end, these domains are allosterically activated, stimulating DNA degradation and cOA synthesis. Backbone cleavage of the target RNA occurs most rapidly at site B1 nearest to the 3’ end, with progressively slower rates of cleavage at sites B2 and B3. Cleaved RNA dissociation is observed with an off rate of 0.07 min−1, suggesting it may be rate limited by cleavage at site B2 and does not require cleavage at site B3. The production of cOA is quickly switched off, consistent with a rapid return to the inactive ground state. Non-target RNAs proceed through the same processing steps, but do not activate the cyclase or HD nuclease domains.
Tables
Oligonucleotides.
Regions complementary to crRNA A26 are italicized and mismatches are shown in bold. crRNA A26 is shown 3’ to 5’ and the 5’ handle is bold. Phosphorothioate linkages are indicated with an asterisk. For substrate A1, Csx1 cleavage sites are in bold.
| crRNA A26 | 3’-GCAACAATTCTTGCTGCAACAATCTTCAACCCATACCAGAAAGUUA |
|---|---|
| Name | Sequence (5’−3’) |
| Target RNA A26 | AGGGUCGUUGUUAAGAACGACGUUGUUAGAAGUUGGGUAUGGUGGAGA |
| 3’-blunt | AGGGUCGUUGUUAAGAACGACGUUGUUAGAAGUUGGGUAUGGU |
| Match | CGUUGUUAAGAACGACGUUGUUAGAAGUUGGGUAUGGUCUUUCAAU |
| 5’-blunt | CGUUGUUAAGAACGACGUUGUUAGAAGUUGGGUAUGGUGGAGA |
| MM0.5 | AGGGUCGUUGUUAAGAACGACGUUGUUAGAAGUUGGGUAUUUUGGAGA |
| MM1 | AGGGUCGUUGUUAAGAACGACGUUGUUAGAAGUUGGGUCGGGUGGAGA |
| MM1a | AGGGUCGUUGUUAAGAACGACGUUGUUAGAAGUUGGGUAGGGUGGAGA |
| MM1b | AGGGUCGUUGUUAAGAACGACGUUGUUAGAAGUUGGGUCUGGUGGAGA |
| MM1.5 | AGGGUCGUUGUUAAGAACGACGUUGUUAGAAGCCGGGUAUGGUGGAGA |
| MM2 | AGGGUCGUUGUUAAGAACGACGUUGUCGGAAGUUGGGUAUGGUGGAGA |
| MM2.5 | AGGGUCGUUGUUAAGAACGACGUUCAUAGAAGUUGGGUAUGGUGGAGA |
| MM4 | AGGGUCGUUGUUAAAGACGACGUUGUUAGAAGUUGGGUAUGGUGGAGA |
| P-thioate A26 | AGGGUCGUUGUUAAGAACGACGUUGU*U*A*GAAGUUGGGU*A*U*GGUGGAGA |
| Substrate (A1) | AGGGUAUUAUUUGUUUGUUUCUUCUAAACUAUAAGCUAGUUCUGGAGA |
| A3 (MazF) | AAAACAUCAG |
| A4 | AAAAACAUCAG |
| A5 | AAAAAACAUCAG |
| A6 | AAAAAAACAUCAG |
| Trap DNA | GTCGTTCTTAACAACGACCCT |
| Target RNA B2 product | AGGGUCGUUGUUAAGAACGACGUUGUU |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Sulfolobus solfataricus) | Csm complex (eight subunits) | PMID: 24119402 | virus expression construct | |
| Gene (Sulfolobus solfataricus) | Csx1/Sso1389 | this paper | UniProtKB - Q97YD5 | plasmid expression construct |
| Gene (Eschericia coli) | MazEF | PMID: 22447587 | plasmid expression construct |





