Structures reveal opening of the store-operated calcium channel Orai
Figures
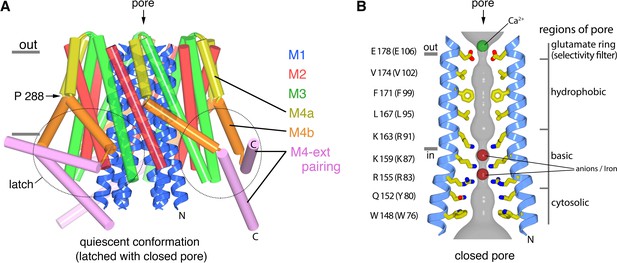
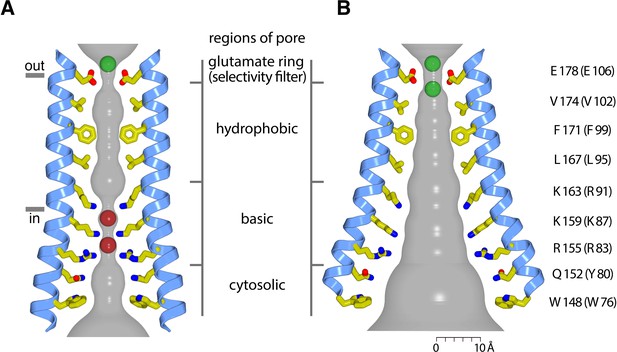
The quiescent conformation –latched with a closed pore.
(a) Overall structure of Orai, from PDB ID 4HKR, in a ‘quiescent’ conformation (Hou et al., 2012). The pore is closed; M4-ext helices pair with one another. The M1 helices are depicted as blue ribbons; other helices are cylinders. The approximate boundaries of the plasma membrane are shown as gray bars. Regions referred to as ‘latches’ in this study are indicated as dashed ovals. (b) Close-up view of the closed pore. Two M1 helices are drawn as ribbons (four M1 helices are omitted for clarity). The pore is a depicted as a gray surface indicating the minimal radial distance to the nearest van der Waals contact. Amino acid side chains that form the walls of the pore are drawn as sticks and colored (yellow for carbon, blue for nitrogen, and red for oxygen). Amino acid numbering is shown for Drosophila melanogaster Orai without parentheses and for human Orai in parentheses. Sections of the pore are indicated. Horizontal gray bars correspond to the approximate boundaries of the membrane, although the M1 helices are shielded from the membrane by M2 and M3. A Ca2+ ion is indicated. Red spheres mark the location of anomalous difference electron density attributed to iron, perhaps bound as (FeCl6)3- (Hou et al., 2012). The complex anion (IrCl6)3- also binds in these sites (Hou et al., 2012).
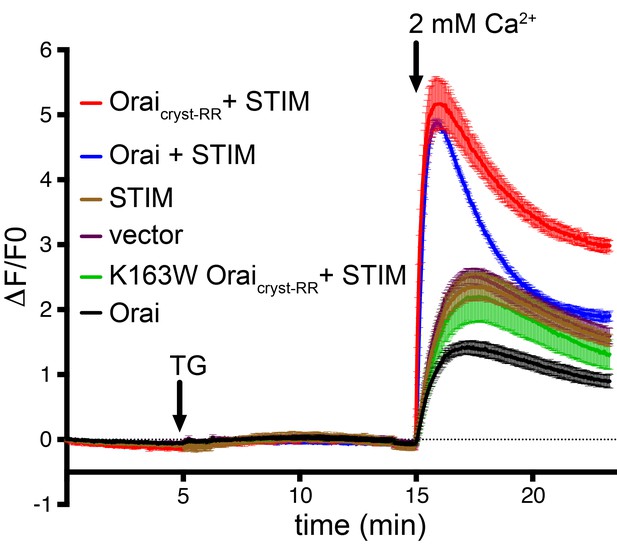
Ca2+ influx measurements show that Drosophila Orai constructs function as CRAC channels when co-expressed with STIM.
HEK293 cells were transfected with Orai and/or STIM as indicated (also see Materials and methods). Cytosolic [Ca2+] levels were detected using the genetically-encoded fluorescent Ca2+ indicator GCaMP6s (Chen et al., 2013); data are plotted as the change in fluorescence intensity relative to the initial value (ΔF/F0) versus time. Thapsigargin (TG), which is used to deplete ER calcium stores, and 2 mM CaCl2 were added at the indicated times (arrows). Ca2+ influx above the background level (vector alone) was observed for the co-expression of STIM with Oraicryst-RR or wild-type Orai, but not for the constitutively closed K163W mutant or for Orai or STIM alone. Signal from endogenous CRAC channels is apparent from the vector control (empty expression vector). Standard error, derived from three independent measurements, is shown.
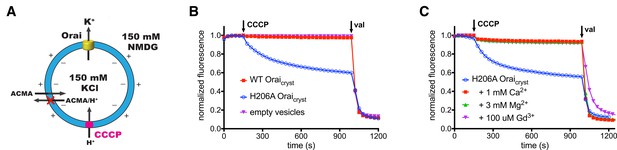
Ion flux through H206A Oraicryst in liposomes.
(a) Schematic of the fluorescence-based flux assay. Vesicles containing WT or H206A Oraicryst or those prepared without protein (empty vesicles) were loaded with 150 mM KCl and were diluted 50-fold into flux buffer containing a fluorescent pH indicator (ACMA) and 150 mM N-methyl-D-glucamine (NMDG) to establish a K+ gradient (Materials and methods). After stabilization of the fluorescence signal (150 s), a proton ionophore (CCCP) was added. An electric potential arising from K+ efflux was used to drive the uptake of protons, which quenches the fluorescence of ACMA. A red ‘X’ indicates that ACMA is not membrane-permeable in the protonated form. (b) K+ flux measurements for WT and H206A Oraicryst. The time-dependent decrease in fluorescence observed for H206A Oraicryst after the addition of CCCP is indicative of K+ flux. Valinomycin (val) was added after 990 s to render all vesicles permeable to K+ and establish a baseline fluorescence. Traces were normalized by dividing by the initial fluorescence value, which was within ±10% for each experiment. (c) K+ flux through H206A Oraicryst is inhibited by Ca2+, Mg2+ and Gd3+.
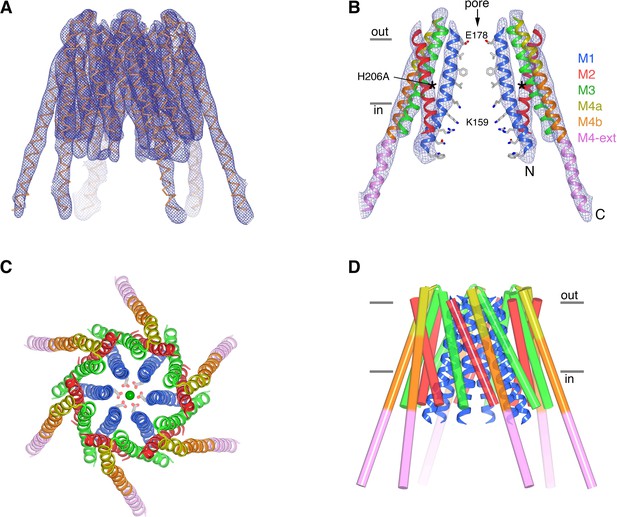
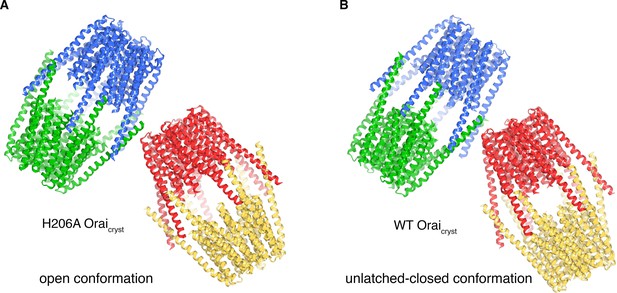
The structure of H206A Oraicryst reveals an open conformation.
(a) Electron density map of H206A Oraicryst. The map (blue mesh, contoured at 1.4 σ, and covering one channel) was calculated from 20 to 6.7 Å using native-sharpened amplitudes and phases that were improved by 24-fold non-crystallographic symmetry (NCS) averaging, solvent flattening and histogram matching (Materials and methods). The atomic model is shown in Cα representation. Figure 4—video 1 shows a video of this Figure. (b) Side view showing two opposing subunits of H206A Oraicryst and the same electron density map. Asterisks mark the location of the H206A substitution. Amino acid side chains on the pore are shown only for reference (sticks). Approximate boundaries of the membrane are shown as horizontal bars. Helices are depicted as ribbons and colored as indicated. (c) Extracellular view showing the hexameric architecture. Helices are depicted as ribbons, with Glu178 side chains (sticks) and Ca2+ ion (green sphere) shown for reference. (d) Overall structure, shown in the same orientation as (a). The M1 helices are drawn as blue ribbons and the other helices are shown as cylinders.
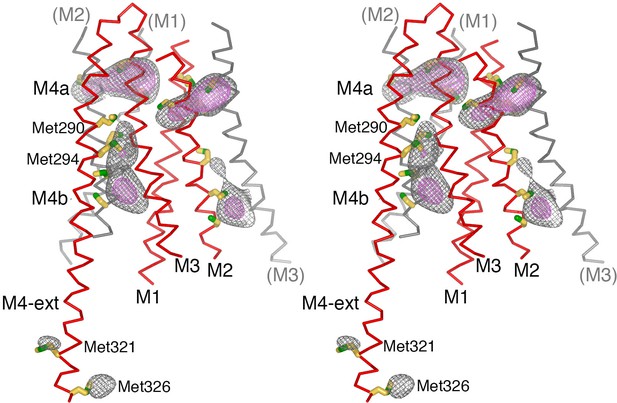
Anomalous-difference electron-density at cysteine and methionine residues in the final model of H206A Oraicryst, depicted in stereo.
An anomalous-difference electron-density map was calculated from 25 to 10 Å resolution from highly redundant diffraction data collected with λ = 1.7085 Å X-rays (NaI experiment, Table 1) using anomalous differences as amplitudes and phases that were determined by MR-SAD, 24-fold NCS averaging, solvent flattening and histogram matching (Materials and methods). This map was then averaged in real-space according to the 24-fold NCS symmetry to yield the map shown. The map is contoured at 5.5 σ (gray mesh) and 8.5 σ (pink mesh) and shown in the vicinity of a subunit of Orai (red Cα trace). Methionine and cysteine residues are shown as sticks (colored yellow for carbon and green for sulfur atoms). Methionine residues on M4b and M4-ext are labeled. Portions of neighboring Orai subunits (gray Cα traces) are shown for reference with their helices labeled in parentheses. While their side chain conformations are hypothetical on account of the limited resolution of the diffraction data, anomalous-difference electron-density peaks for methionine and/or cysteine residues on each of the M1-M4 helices and on the M4-ext helix confirm the amino acid register of the atomic model.
Electron density for H206A Oraicryst from Figure 4A.
https://doi.org/10.7554/eLife.36758.007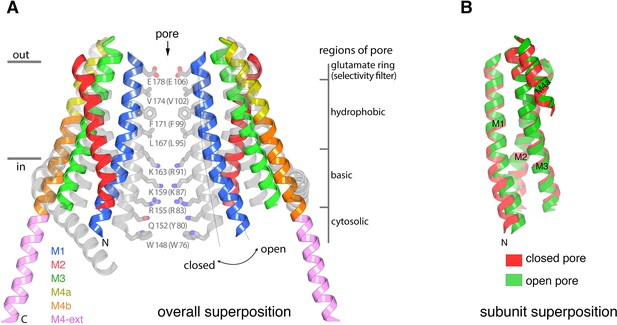
Conformational changes between the quiescent and open conformations.
(a) Superimposed structures of the quiescent (PDB ID: 4HKR) and open (H206A Oraicryst) conformations are drawn in ribbon representation. Two opposing subunits are shown, surrounding the pore, with the open conformation colored as indicated and the closed conformation in gray. Thin lines and a curved arrow highlight the outward rotation of subunits (with its fulcrum near Glu178) and the slight additional bend in M1. Conformational changes of M4/M4-ext are also apparent. Amino acids forming the walls of the closed pore (from the quiescent conformation) are shown as sticks, with corresponding regions of the pore indicated. Amino acids in parentheses denote human Orai1 counterparts. Horizontal bars indicate approximate boundaries of the plasma membrane. (b) Comparison of M1-M4a from individual subunits between the quiescent and open conformations. The region of an Orai subunit spanning M1 through M4a was superimposed between the quiescent (red ribbons, PDB ID 4HKR) and open (green ribbons, H206A Oraicryst) conformations. The slight additional bend in M1 of the open conformation is apparent at its N-terminal end. Otherwise the M1-to-M4b region of the two subunits superimpose within the error of the coordinates of the open conformation (the root-mean-squared deviation for the Cα positions of residues 163 to 288 superimposed in this manner is 1.1 Å).
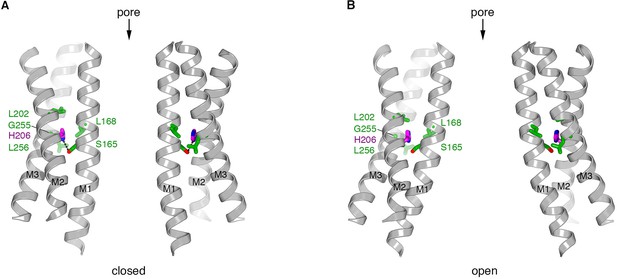
Residue 206 in closed and open pores.
(a) Depiction of the closed pore (PDB ID 4HKR). M1 through M3 are drawn as ribbons for two opposing subunits. His206 (H206, pink) and the amino acid side chains within van der Waals distance (green) are drawn as sticks. A hydrogen bond made between His206 and Ser165 is shown as a dashed line. (b) Depiction of the open conformation (H206A Oraicryst), showing the corresponding regions as in (a). The conformations of the amino acid side chains in the atomic model are shown for reference to indicate plausibility despite the limited resolution of the diffraction data. Amino acid 206 is depicted as the wild-type histidine to indicate that this amino acid could be accommodated in the observed conformation of the channel without steric hindrance.
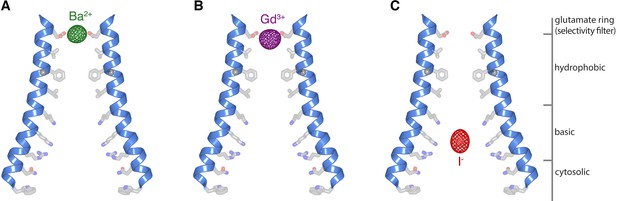
Ion binding in the open pore.
(a–c) Anomalous-difference electron density maps (mesh) for crystals of H206A Oraicryst with Ba2+ (a), Gd3+ (b), and I- (c). M1 helices of two opposing subunits are shown as ribbons. Side chains proposed to line the pore (sticks) are drawn for reference; their conformations are hypothetical. The maps are contoured at 10 σ and calculated from 25 to 9 Å for (a–b), and at 7 σ and calculated from 25 to 10 Å for (c).
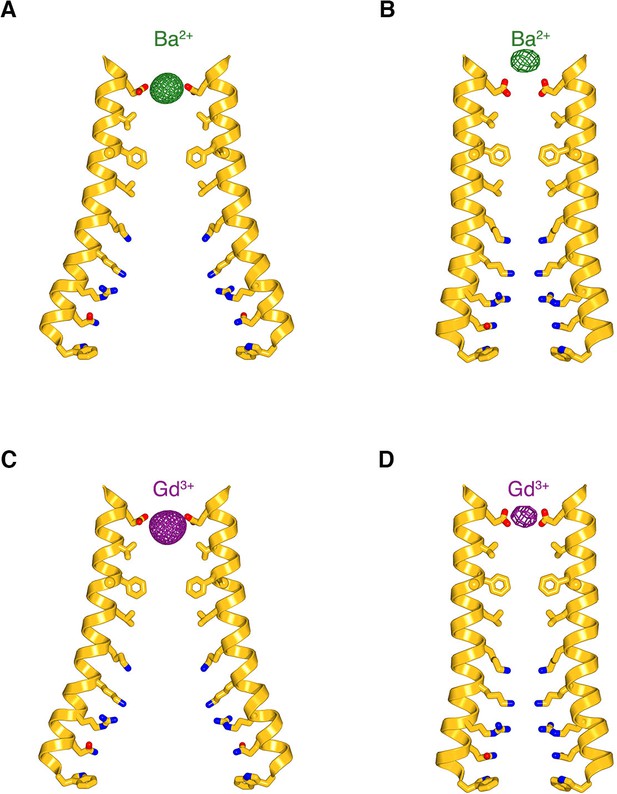
Comparison of the anomalous-difference electron-density peaks for Ba2+ and Gd3+ in the open and closed pores.
(a–b) Anomalous-difference electron density for Ba2+ (green mesh) in open pore of H206A Oraicryst (from Figure 5A) and in the closed pore of the quiescent conformation (from Hou et al., 2012), respectively. M1 helices (amino acids 148 to 180) are depicted from two opposite subunits as ribbon representations. Pore-lining side chains (sticks) are drawn for reference in (a) since their conformations cannot be determined due to the limits of the diffraction data, and they are shown in (b) according to their observed conformations (Hou et al., 2012). (c–d) Analogous depictions for the anomalous-difference electron-density peaks for Gd3+ in the open pore (c), from Figure 5B and closed pore (d), from Hou et al. (2012).
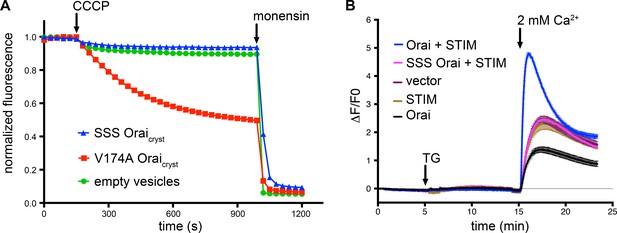
SSS Oraicryst is neither constitutively active nor forms functional CRAC channels when expressed with STIM.
(a) Proteoliposome-based sodium (Na+) flux assay. Purified proteins were reconstituted into liposomes to assay for Na+ flux under divalent-free conditions (Materials and methods) as described previously (Hou et al., 2012). After stabilization of the fluorescence signal (150 s), the proton ionophore CCCP was added to the sample. A decrease in fluorescence is indicative of Na+ flux out of the proteoliposomes. The Na+ ionophore monensin was added after 990 s to render all vesicles permeable to Na+ and establish the minimum baseline fluorescence. The traces were normalized to the initial fluorescence value, which was within ±10% in the experiments. The signal for SSS Oraicryst (Oraicryst with the R155S, K159S, and K163S mutations) is comparable to what is observed for liposomes without protein (‘empty vesicles’). As a control for the assay, we observed Na+ flux through purified channels containing the V174A mutation of the hydrophobic region of the pore, which has previously been shown to produce leaky channels with diminished selectivity for Ca2+ (Hou et al., 2012; McNally et al., 2012). (b) Ca2+ influx measurements in HEK293 cells expressing indicated Orai and STIM constructs. Methods are identical to those for Figure 2. SSS Orai does not show Ca2+ uptake above the background level (vector control).
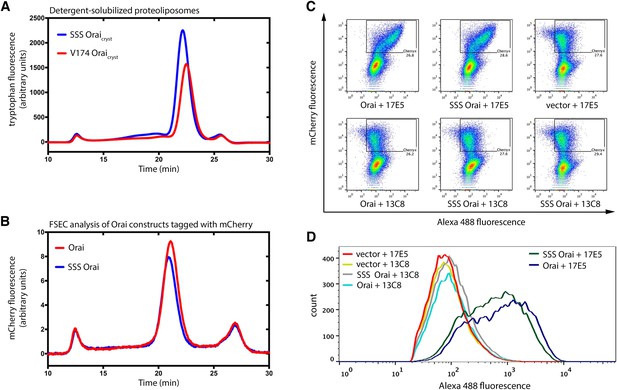
SSS Orai is properly folded and expressed on the plasma membrane.
(a) SEC analysis of detergent (DDM)-solubilized proteoliposomes that were used for the assay show in Figure 7a. A monodisperse peak for SSS Oraicryst is observed at the appropriate elution volume. (b) FSEC analysis of Orai constructs transfected into HEK293 cells (e.g. used for the assay shown in Figure 7b). The leftward shift relative to (a) is due to the added mass of the mCherry tag. (c–d) Flow cytometry indicates that SSS Orai is expressed at the plasma membrane. A monoclonal antibody (17E5) that targets the M1-M2 extracellular loop of Drosophila Orai was used to detect surface expression. A monoclonal antibody (13C8) that targets the cytosolic region of Drosophila Orai was used as negative control. (c) Orai expression is indicated by mCherry fluorescence on the Y-axis. The cell-surface binding of primary antibodies was detected using an Alexa Fluor 488 secondary antibody (X-axis). (d) Histogram overlay, indicating cell-surface expression of Orai and SSS Orai. Histogram plots are shown for the boxed regions from (c). The rightward-shifts for Orai and SSS Orai in the presence of 17E5 indicate surface expression of these constructs. The percentages of cells that had strong mCherry fluorescence and were selected for the histogram plots are indicated below the boxed regions from (c).
Molecular packing in the crystals of H206A Oraicryst and WT Oraicryst.
(a) Crystal packing of H206A Oraicryst. The contents of the asymmetric unit, consisting of four complete channels, is shown. Each channel is colored a unique color and shown in ribbon representation. The channels interact with one another via coiled-coil interactions between their M4-ext helices. (b) Packing of WT Oraicryst in the crystal, showing the contents of the asymmetric unit, depicted analogously to a).
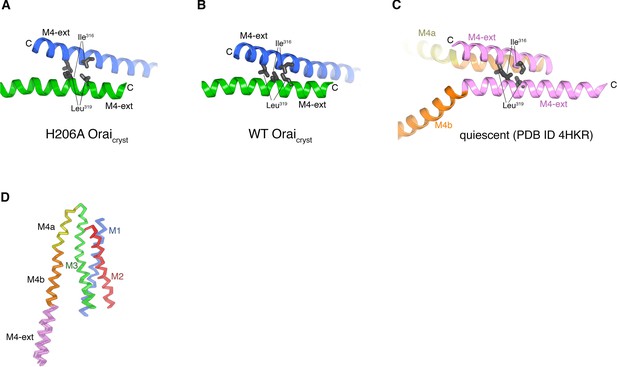
M4-ext helices and subunit comparisons.
(a) Close-up view of a coiled-coil interaction between two M4-ext helices of the blue- and green-colored channels in the asymmetric unit of H206A Oraicryst (from Figure 7A). Ile316 and Leu319, which form the hydrophobic interface of the coiled-coil interaction on each of the M4-ext helices, are drawn as gray sticks for reference. (b) Analogous coiled-coil interaction between two M4-ext helices of the blue- and green-colored channels in the asymmetric unit of WT Oraicryst (from Figure 7B). (c) Coiled-coil interaction of two paired M4-ext helices observed in the quiescent conformation, which occurs between adjacent subunits of the same channel (from Figure 1, PDB ID 4HKR). (d) Superposition of the 24 individual subunits (from four hexameric channels) of H206A Oraicryst within the asymmetric unit, shown in Cα representation.
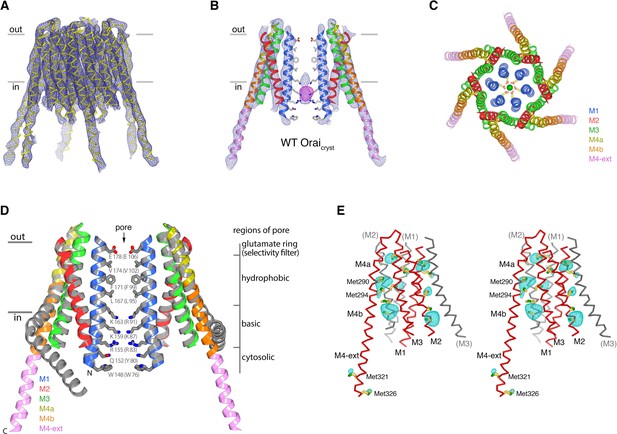
The structure of WT Oraicryst reveals an unlatched-closed conformation.
(a) Electron density for WT Oraicryst, shown as blue mesh covering the channel (Cα representation). The map (contoured at 1.3 σ) was calculated from 20 to 6.9 Å using native sharpened amplitudes and phases that were determined by MR-SAD and improved by 24 fold NCS averaging, solvent flattening and histogram matching (Materials and methods). (b) Electron density, from (a) (blue mesh), covering two opposing subunits of WT Oraicryst (cartoon representation, colored as indicated in c). Anomalous-difference electron-density (from iron) in the basic region of the pore is shown as magenta mesh (map calculated from 25 to 9 Å and contoured at 5 σ). Conformations of pore residues are based on the quiescent conformation (PDB ID 4HKR). (c) Extracellular view of WT Oraicryst. Helices are drawn as ribbons and colored as indicated, with Glu178 side chains (sticks) and Ca2+ ion (green sphere) shown for reference. (d) Superposition of the quiescent conformation (PDB ID 4HKR) and the structure of the unlatched-closed conformation (WT Oraicryst). Two subunits of each channel are shown. The quiescent conformation is gray; the structure of WT Oraicryst is shown in colors. Amino acids lining the pore of the quiescent conformation are shown as sticks. A slight outward displacement of the intracellular side of M3 is observed in the structure of WT Oraicryst; otherwise the conformations of M1-M4a are indistinguishable within the resolution limits of the diffraction data (RMSD for Cα positions 148 to 288 is 0.9 Å). (e) Anomalous-difference electron-density peaks at cysteine and methionine residues confirms the amino acid register of the WT Oraicryst structure (stereo representation). An anomalous-difference electron-density map was calculated from 25 to 9 Å resolution from data collected with λ = 1.738 Å X-rays (Extended Data Table 2) using anomalous differences as amplitudes and phases from (a). This map was then averaged in real-space according to the 24-fold NCS symmetry to yield the map shown. The map is contoured at 5 σ (cyan mesh) and shown in the vicinity of a subunit of Orai (red Cα trace). Methionine and cysteine residues are shown as sticks (colored yellow for carbon and green for sulfur atoms). Methionine residues on M4b and M4-ext are labeled. Portions of neighboring Orai subunits (gray Cα traces) are shown for reference with their helices labeled in parentheses. Side chain conformations are hypothetical.
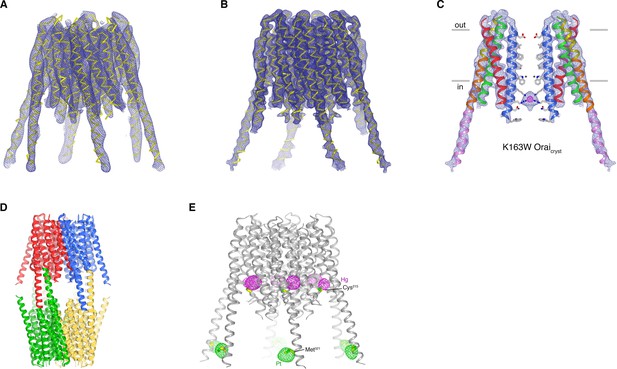
Structures of K163W Oraicryst in I41 and P42212 crystal forms (unlatched-closed conformation).
(a) Structure of K163W Oraicryst from the I41 crystal form. The map (contoured at 1.3 σ) was calculated from 20 to 6.1 Å using native-sharpened amplitudes and experimental phases that were determined by MIRAS and were improved by 24-fold NCS averaging, solvent flattening and histogram matching (Materials and methods). The atomic model is shown in Cα representation (yellow). The crystal form is analogous to crystals of H206A Oraicryst and WT Oraicryst, and has analogous crystal packing. The root-mean-squared deviation (RMSD) for Cα positions between the structures of WT Oraicryst and K163W Oraicryst is 0.5 Å. (b) 4.35 Å resolution structure of K163W Oraicryst in the P42212 crystal form. Electron density (mesh) covering the channel is shown. The map (contoured at 1.3 σ) was calculated from 20 to 4.35 Å using sharpened amplitudes and phases that were determined by MR and were improved by threefold non-crystallographic symmetry (NCS) averaging, solvent flattening and histogram matching (Materials and methods). (c) Two opposing subunits of K163W Oraicryst (P42212 crystal form), showing the pore, with electron density from (b). Amino acids on the pore are depicted as sticks (conformations based on PDB 4HKS; Materials and methods). Anomalous-difference electron density in the pore is shown as magenta mesh (calculated from 30 to 8 Å resolution using anomalous differences as amplitudes, and contoured at 3.8 σ). Asterisks mark the locations of K163W substitutions. (d) Packing of K163W Oraicryst in the P42212 crystal form. The contents of each asymmetric unit (three Orai subunits) are colored a unique color. Two asymmetric units (e.g. blue and red) form a complete channel. The channels interact with one another in the crystal lattice via coiled-coil interactions between their M4-ext helices. (e) Anomalous-difference electron-density for heavy atom derivatives of K163W Oraicryst in the I41 space group. One channel of the asymmetric unit is depicted as ribbons. The map for the platinum (Pt) derivative (magenta mesh, calculated from 25 to 8.0 Å, and contoured at 4.5 σ) was calculated from data collected from a crystal soaked in PIP (Table 2), using anomalous differences as amplitudes and phases from (a). The analogous map for the mercury (Hg) derivative (green mesh, calculated from 25 to 8.0 Å, and contoured at 5 σ) was calculated from data collected from a crystal soaked in PCMB (Table 2). Each channel in the asymmetric unit has anomalous-difference density at these sites (24 sites for each derivative, Table 2). Cys215 and Met321 residues, to which the heavy atoms presumably bind, are depicted as sticks.
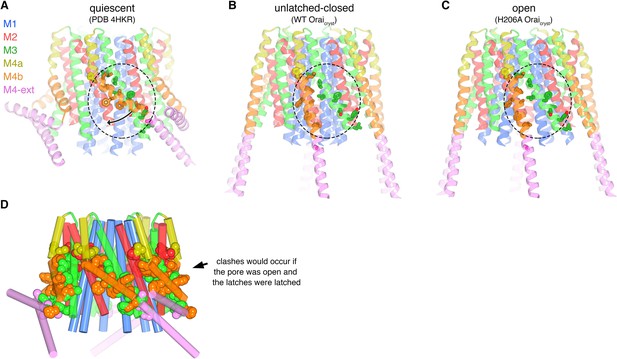
Unlatching is necessary for pore opening.
(a) Quiescent conformation (PDB ID: 4HKR) highlighting interactions between M4b and M3. The channel is shown in cartoon representation. On one of the Orai subunits, amino acids in the interface between M4b (orange) and M3 (green), which is highlighted by a dashed oval, are shown as sticks and colored accordingly. This interface exists for all six subunits and is stabilized by the pairing of M4-ext helices (pink). An arrow denotes the movement of M4b between (a) and (b). (b) Conformation of WT Oraicryst showing the released latches and closed pore. The amino acids that had been in the interface between M4b and M3 in the quiescent conformation are drawn as sticks, with the dashed region showing the same region as in (a). (c) Open conformation (H206A Oraicryst), depicted as in (b). (d) Unlatching is necessary for pore opening. A hybrid atomic model of the channel was generated using the conformations of the M1, M2 and M3 helices from the open conformation and the conformations of M4a/b and M4-ext from the quiescent conformation. In this model (shown in cartoon representation), molecular clashes exist between M4b and M3. Amino acids involved in the clashes are depicted as space-filling spheres (colored according to channel region: red, M2; green, M3; yellow, M4a; orange, M4b; pink, M4-ext). These steric hindrances would prevent opening of the pore while the latches are fastened. With unlatching, the repositioning of the M4b helix would allow the outward motion of M1-M3 that opens the pore.
Hypothetical dimensions of the open pore.
(a) The closed pore from the 3.35 Å resolution structure of the quiescent conformation of Orai (PDB 4HKR) (Hou et al., 2012). The gray surface represents the radial distance to the nearest van der Waals contact. Ca2+ is represented as a green sphere. Red spheres represent the iron complex and anion binding sites in the closed pore. (b) Hypothetical dimensions of the open pore, which is depicted as in (a), based on the atomic coordinates of the H206A Oraicryst structure. There is a large amount of uncertainty in the pore dimensions and in the conformations of side chains due to the limited resolution of the diffraction data. A scale bar is shown for reference. Two hypothetical Ca2+ ions in the selectivity filter are depicted as green spheres.
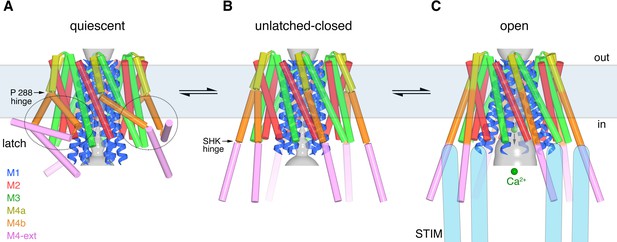
Proposed sequence of channel activation.
(a) Quiescent conformation of Orai prior to binding of STIM (from PDB ID 4HKR, cylinders and ribbons). The pore (gray surface) is closed and the latches are fastened (two latches are indicated with dashed ovals). The M4 helices are bent at Pro288, delineating them into M4a and M4b. The M4b portions (orange) interact with the M3 helices (green), in six-fold fashion, and prevent the pore from opening by constraining the cytosolic region of the M3 helices. The interaction between M4b and M3 and the bend at Pro288 are stabilized by three sets of paired M4-ext helices. (b) An unlatched-closed conformation: structure of WT Oraicryst in which the pore is closed but the latches are released. Conformations of M1-M4a are indistinguishable from (a) (Figure 8D). When unlatched, mobile M4-ext regions are hypothesized to be available to interact with cytosolic regions of STIM that would become exposed as a result of depletion of Ca2+ from the ER. Spontaneous unlatching would not necessarily require STIM binding and does not necessarily open the pore. (c) Open conformation. The structure of H206A Oraicryst is shown (cylinders and ribbons), with approximate dimensions of the pore shown as a gray surface. Following store depletion, we hypothesize that STIM (blue shapes) engages with cytosolic regions of Orai and stabilizes the pore in an open conformation. Unlatching is required to allow the widening of the pore and the influx of Ca2+ (green spheres). Arrows between conformations denote equilibria and the horizontal rectangle indicates approximate boundaries of the plasma membrane. The depiction of the cytosolic region of STIM is conceptual and is not meant to imply stoichiometry or conformation.
Video showing hypothetical opening sequence as illustrated in Figure 12.
https://doi.org/10.7554/eLife.36758.023Tables
H206A Oraicryst data collection, phasing and refinement statistics.
Data collection statistics are from HKL3000 (Otwinowski and Minor, 1997) or XDS (I- experiment) (Kabsch, 2010). Rsym = Σ | Ii- < Ii > | / Σ Ii, where < Ii > is the average intensity of symmetry-equivalent reflections. CC1/2, CCwork and CCfree are defined in (Karplus and Diederichs, 2012). Phasing power = RMS (|F|/ε), where |F| is the heavy-atom structure factor amplitude and ε is the residual lack of closure error. Rcullis is the mean residual lack of closure error divided by the dispersive or anomalous difference. Rwork = Σ | Fobs – Fcalc | / Σ | Fobs |, where Fobs and Fcalc are the observed and calculated structure factors, respectively. Rfree is calculated using a subset (~10%) of reflection data chosen randomly and omitted throughout refinement. Figure of merit is indicated after density modification and phase extension starting from 9.0 Å in DM. R.m.s.d: root mean square deviations from ideal geometry. Numbers in parentheses indicate the highest resolution shells and their statistics.
| H206A Oraicryst | ||||
|---|---|---|---|---|
| Native | Ba2+ | Gd3+ | I- | |
| Space group | I41 | I41 | I41 | I41 |
| Datasets source | APS 24ID-C | APS 24ID-C | APS 23ID-D | APS 23ID-D |
| Wavelength (Å) | 1.1000 | 1.7000 | 1.7000 | 1.7085 |
| Cell dimensions: | ||||
| a, b, c (Å) | 262.3, 262.3, 220.4 | 265.0, 265.0, 219.9 | 255.4, 255.4, 216.0 | 266.4, 266.4, 221.5 |
| α= β= γ (°) | 90 | 90 | 90 | 90 |
| Resolution (Å) | 50–6.70 (6.82–6.70) | 50–7.40 (7.53–7.40) | 50–7.90 (8.04–7.90) | 50–7.6 (7.68–7.60) |
| No. of crystals | 1 | 1 | 1 | 2 |
| Rsym (%) | 8.3 (>100) | 22.5 (>100) | 12.1 (>100) | 14.0 (>100) |
| Rpim (%) | 1.1 (57.0) | 3.4 (46.7) | 2.4 (42.8) | 1.1 (62.0) |
| CC1/2 (in outer shell) | 0.214 | 0.173 | 0.158 | 0.493 |
| I/σI | 92.4 (1.1) | 47.0 (1.4) | 37.5 (2.0) | 26.8 (0.2) |
| Completeness (%) | 100.0 (100.0) | 100.0 (100.0) | 100.0 (100.0) | 99.9 (99.7) |
| Redundancy | 55.8 (58.7) | 46.5 (51.8) | 25.1 (26.0) | 165.1 (172.5) |
| Figure of Merit (DM) | 0.729 [20–6.7 Å] | |||
| Refinement | PDB ID: 6BBF | |||
| Resolution (Å) | 20–6.7 | |||
| No. of reflections | 12844 | |||
| Rwork (%) | 30.6 | |||
| Rfree (%) | 33.9 | |||
| CCwork/CCfree (in outer shell) | 0.285/0.220 | |||
| No. atoms | 27120 | |||
| Ramachandran (%) | ||||
| Favored | 97.22 | |||
| Outliers | 1.04 | |||
| R.m.s.d: | ||||
| bond lengths (Å) | 0.005 | |||
| bond angles (°) | 1.15 | |||
Data collection, phasing and refinement statistics for WT and K163W Oraicryst.
Data collection statistics are from HKL3000 (Otwinowski and Minor, 1997). Rsym = Σ | Ii- < Ii > | / Σ Ii, where < Ii > is the average intensity of symmetry-equivalent reflections. CC1/2, CCwork and CCfree are defined in (Karplus and Diederichs, 2012). Phasing power = RMS (|F|/ε), where |F| is the heavy-atom structure factor amplitude and ε is the residual lack of closure error. Rcullis is the mean residual lack of closure error divided by the dispersive or anomalous difference. Rwork = Σ | Fobs – Fcalc | / Σ | Fobs |, where Fobs and Fcalc are the observed and calculated structure factors, respectively. Rfree is calculated using a subset (~10%) of reflection data chosen randomly and omitted throughout refinement. Figure of merit is indicated after density modification and phase extension starting from 8.0 Å in DM. R.m.s.d: root mean square deviations from ideal geometry. Numbers in parentheses indicate the highest resolution shells and their statistics.
| K163W Oraicryst | K163W Oraicryst | WT Oraicryst | |||
|---|---|---|---|---|---|
| Native | Native | Derivative 1 PCMB | Derivative 2 PIP | Native | |
| Space group | P42212 | I41 | I41 | I41 | I41 |
| Datasets source | NSLS X25 | NSLS X25 | NSLS X29 | NSLS X29 | NSLS X25 |
| Wavelength (Å) | 1.1000 | 1.1000 | 1.0074 | 1.0712 | 1.738 |
| Cell dimensions: | |||||
| a, b, c (Å) | 118.7, 118.7, 122.4 | 247.5, 247.5, 210.2 | 246.1, 246.1, 210.0 | 250.6, 250.6, 211.8 | 250.4, 250.4, 210.4 |
| α= β= γ (°) | 90 | 90 | 90 | 90 | 90 |
| Resolution (Å) | 60–4.35 (4.42–4.35) | 50–6.10 (6.20–6.10) | 50–6.10 (6.20–6.10) | 50–6.90 (7.02–6.90) | 50–6.9 (7.02–6.90) |
| No. of crystals | 1 | 1 | 1 | 1 | 1 |
| Rsym (%) | 6.0 (>100) | 5.9 (>100) | 5.7 (>100) | 12.0 (>100) | 9.4 (>100) |
| Rpim (%) | 1.7 (55.7) | 1.3 (>100) | 1.9 (>100) | 3.1 (>100) | 3.0 (>100) |
| CC1/2 (in outer shell) | 0.265 | 0.376 | 0.378 | 0.170 | 0.123 |
| I/σI | 61.1 (1.0) | 69.0 (1.0) | 49.3 (0.7) | 37.0 (.07) | 39.1 (0.5) |
| Completeness (%) | 100.0 (100.0) | 100.0 (100.0) | 99.9 (100) | 99.9 (100) | 99.7 (99.8) |
| Redundancy | 16.4 (16.9) | 22.9 (23.9) | 21.5 (20.5) | 16.3 (16.7) | 11.0 (11.6) |
| MIRAS Phasing | |||||
| No. of sites | 24 | 24 | |||
| Phasing power (iso/ano) | 0.523/0.559 | 0.405/0.686 | |||
| Rcullis (iso/ano) | 0.764/0.945 | 0.952/0.919 | |||
| Figure of Merit (DM) | 0.627 [20–4.35 Å] | 0.629 [20–6.1 Å] | 0.777 [20–6.9 Å] | ||
| Refinement | PDB ID: 6BBI | PDB ID: 6BBH | PDB ID: 6BBG | ||
| Resolution (Å) | 20–4.35 | 20–6.1 | 20–6.9 | ||
| No. of reflections | 6035 | 14682 | 10287 | ||
| Rwork (%) | 30.6 | 31.4 | 33.4 | ||
| Rfree (%) | 32.9 | 34.0 | 35.4 | ||
| CCwork/CCfree (in outer shell) | 0.487/0.441 | 0.346/0.333 | 0.332/0.414 | ||
| No. atoms | 3338 | 27360 | 27240 | ||
| Ramachandran (%) | |||||
| Favored | 95.5 | 97.2 | 96.6 | ||
| Outliers | 0.24 | 1.01 | 0.93 | ||
| R.m.s.d: | |||||
| Bond lengths (Å) | 0.006 | 0.005 | 0.005 | ||
| Bond angles (°) | 1.15 | 1.06 | 1.07 | ||
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.36758.024





