Depleting Trim28 in adult mice is well tolerated and reduces levels of α-synuclein and tau
Figures
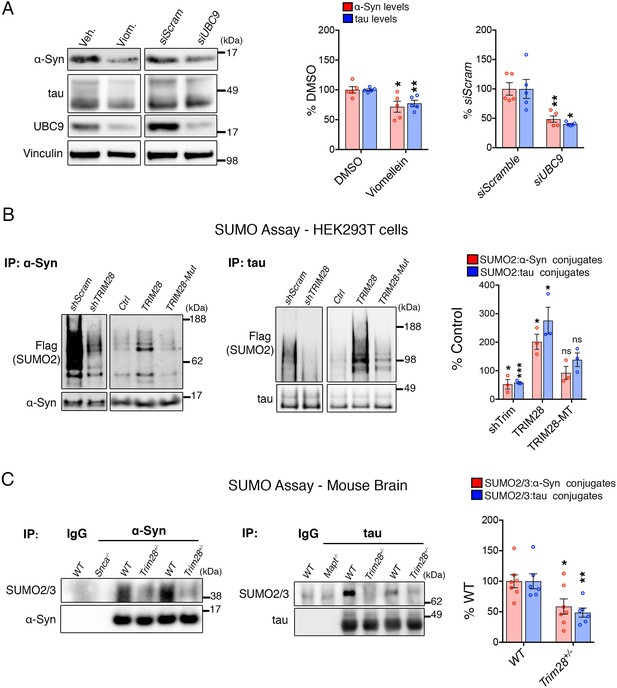
Trim28 mediates the SUMOylation of α-Syn and tau.
(A) Blocking SUMOylation – by either pharmacological inhibition using viomellein or siRNA-mediated suppression of the sole SUMO E2 ligase, UBC9 – decreases α-Syn and tau levels by western blot. (B) SUMO assay in human cells reveals that TRIM28 mediates the formation of SUMO2 adducts on α-Syn and tau. This effect is lost upon mutation of the RING domain of TRIM28 (TRIM28-Mut). (C) In vivo SUMO assay from denatured mouse brain lysates of WT and Trim28+/- mice. Snca-/- and Mapt-/- mice and IP: IgG serve as negative controls. *, **, *** and ns denote p<0.05, p<0.01, p<0.001 and p>0.05, respectively.
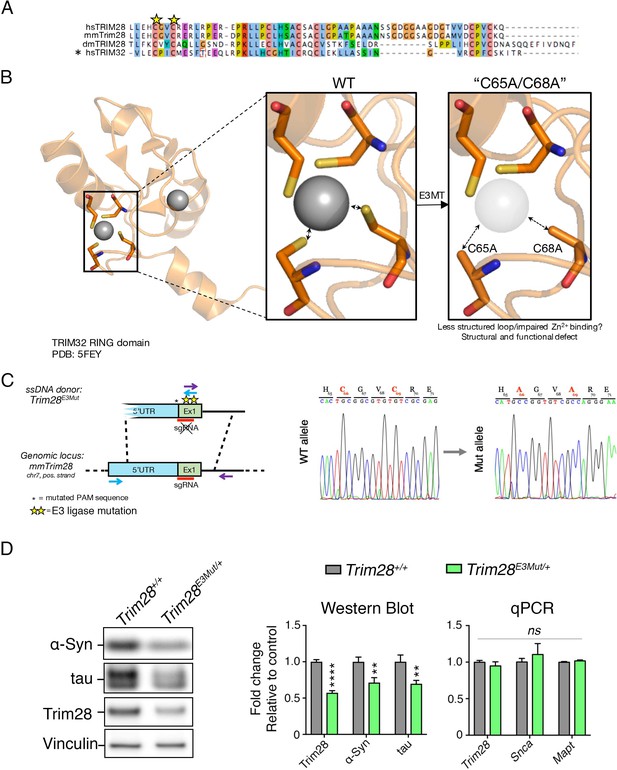
Ablating endogenous Trim28 catalytic activity dramatically reduces its stability, concomitantly decreasing α-Syn and tau levels.
(A), Structural rationale for targeting the RING domain of Trim28. Alignment of human TRIM28 (hsTRIM28) with mouse (mmTrim28) and Drosophila melanogaster (dmTRIM28) TRIM28 in addition to human TRIM32 (hsTRIM32). * denotes the sequence upon which modeling was conducted. Modeling of RING domain disruption in PyMOL using the TRIM32 RING domain (PDB: 5FEY). (B), Approach to mutate endogenous Trim28 catalytic activity and Sanger sequencing confirmation of mutation insertion. (C), Western blot and qPCR analysis of Trim28, Snca and Mapt transcripts in Trim28 E3 mutant heterozygous mice (Trim28E3MT/+) compared to littermate controls. In (C), n = 4–9 mice per genotype. Error bars denote s.e.m. **, **** and ns denote p<0.01, p<0.0001 and p>0.05, respectively.
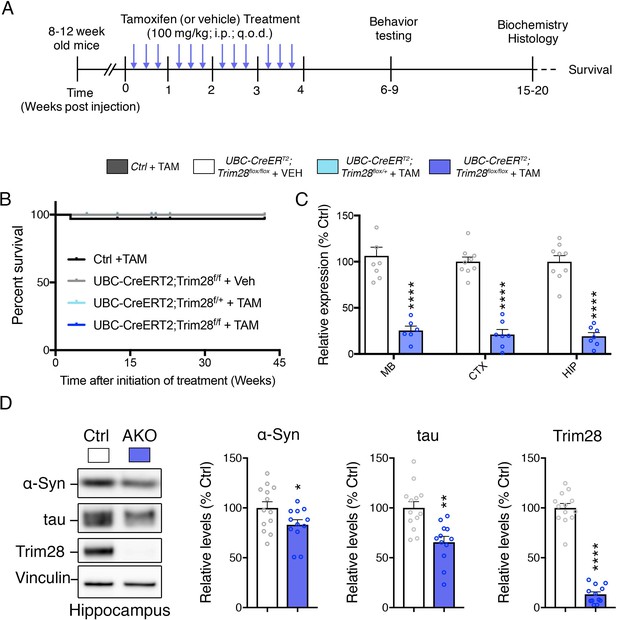
Trim28 adult knockout mice are viable and demonstrate reduced α-Syn and tau levels.
(A) Experimental approach to delete Trim28 from the adult body. (B) Kaplan-Meier survival curve of Adult knockout mice (UBC-CREERT2; Trim28flox/flox + TAM vs littermate controls). No significant differences in survival are observed. (C) qPCR analysis for Trim28 expression in midbrain (MB), cortex (CTX) and hippocampus (HIP) of Trim28 adult knockout mice and control littermates. (D) Western blot analysis of α-Syn, tau and Trim28 levels in hippocampi from Trim28 adult knockout mice and control littermates. In (B), n = 14–33 per group. In (C and D), n = 12–13 per group.
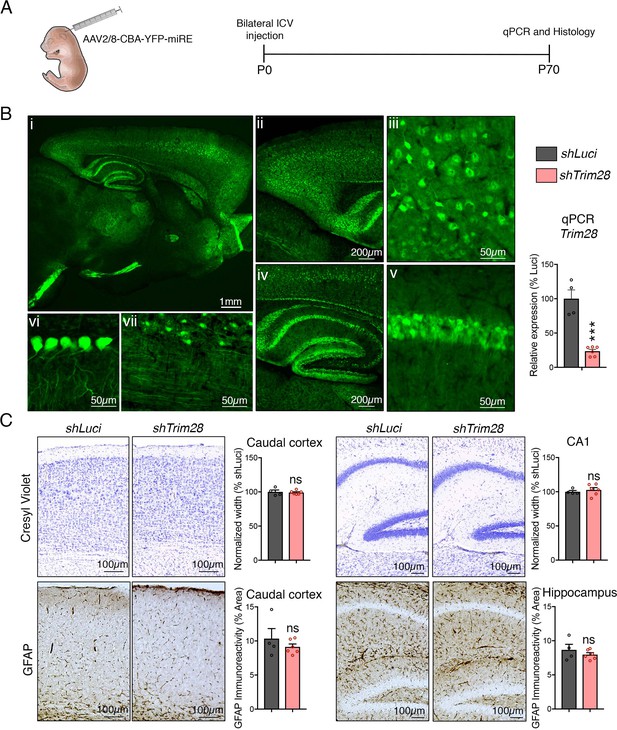
Perinatal suppression of Trim28 in the brain is safe and decreases α-Syn and tau levels.
(A) Approach to deplete Trim28 in the postnatal brain via RNAi. (B) Left panel demonstrates widespread expression of AAV across the brain. Epifluorescence of YFP expression in a representative 10-week-old mouse demonstrating widespread expression of the AAV throughout various brain regions. Brain regions depicted: (i) whole forebrain; (ii) caudal cortex; (iii) caudal cortex (zoom); (iv) hippocampus; (v) hippocampal CA1 region (zoom); (vi) Purkinje cells; and (vii) brainstem). Right panel denotes evidence of Trim28 depletion by qPCR from mice harboring AAV-encoded shRNAs against Trim28 (shTrim28) compared to control, shRNAs against Luciferase (shLuci). (C) Histological examination at the level of the cortex (left panel) and hippocampus (right panel) of mice expressing shTrim28 or shLuci. Cortical and CA1 thickness are measured via cresyl violet staining and astrogliosis is measured using GFAP staining. In (B) and (C), n = 4–6. *** and ns denote p<0.001 and p>0.05, respectively.
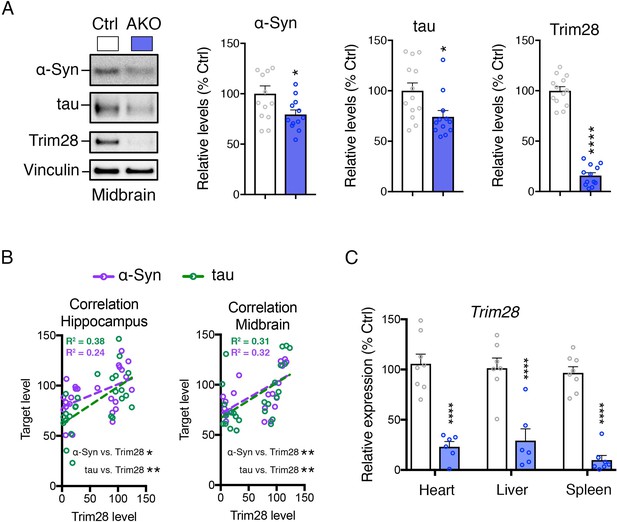
α-Syn and tau levels are reduced in multiple brain regions from Trim28 adult knockout mice.
(A) Western blot analysis of α-Syn, tau and Trim28 levels in the midbrain from adult knockout mice and littermate controls. (B) Correlation analysis between normalized Trim28 levels and α-Syn or tau levels in hippocampal or midbrain extracts from Trim28 adult knockout mice and littermate controls (obtained from Western blot analysis in Figure 2D and Figure 2—figure supplement 2A). R2 values are presented for each linear regression. (C) qPCR analysis of Trim28 expression from peripheral organs in adult knockout mice and littermate controls. In (A and B), n = 12–13 per group. In (C), n = 7–8 per group. *, ** and **** denote p<0.05, p<0.01 and p<0.001, respectively.
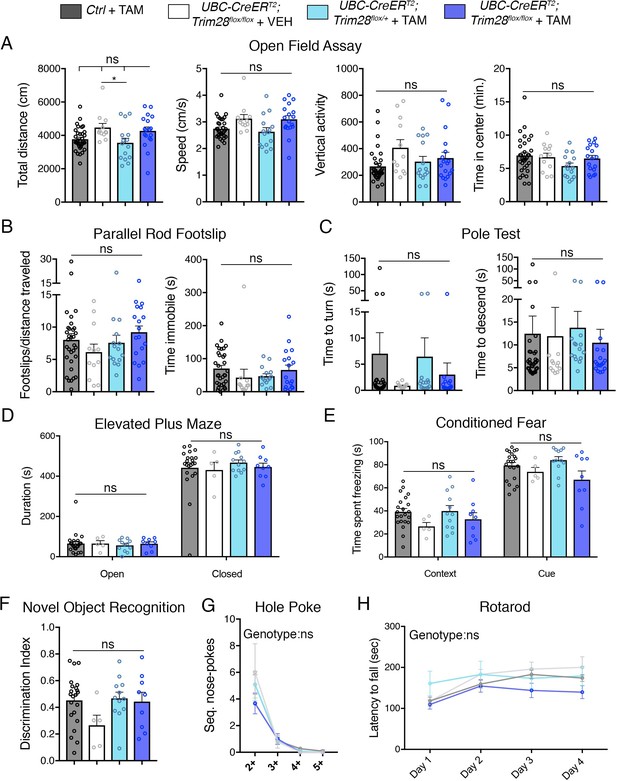
Adult depletion of Trim28 does not cause behavioral abnormalities.
Adult knockout mice and littermate controls were subjected to: (A) Open field assay where total distance, speed, vertical activity and time in center were measured over a period of 30 min. (B) Parallel rod footslip analysis where number of footslips and time spent immobile were measured on a grid over a period of 10 min. (C) Pole test where the time to turn and descend were measured to a mouse on top (facing upward) of a 18’ pole. (D) Elevated plus maze measured the time spent in open vs. closed arms during a period of 10 min. (E) Pavlovian conditioned fear analysis in both context and cued settings (day 2). (F) Novel object recognition assay showing the discrimination index for identifying the novel vs. familiar object. (G) Hole poke analysis of repetitive behavior measuring the number of sequential nose pokes. (H) Rotarod analysis measuring the motor coordination and learning of mice over a period of four days. For each test, n = 5–33; ns denotes p>0.05.
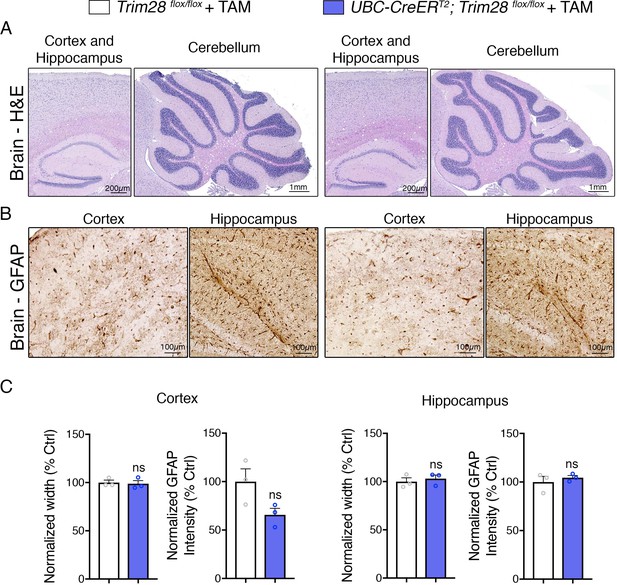
Adult depletion of Trim28 does not cause pathological abnormalities in the adult brain.
Representative photomicrographs of the cortex, hippocampus and cerebellum stained with (A) H and E and (B) GFAP. (C) Quantification of cortical and hippocampal width as well as normalized GFAP intensity. For each test, n = 3; ns denotes p>0.05.
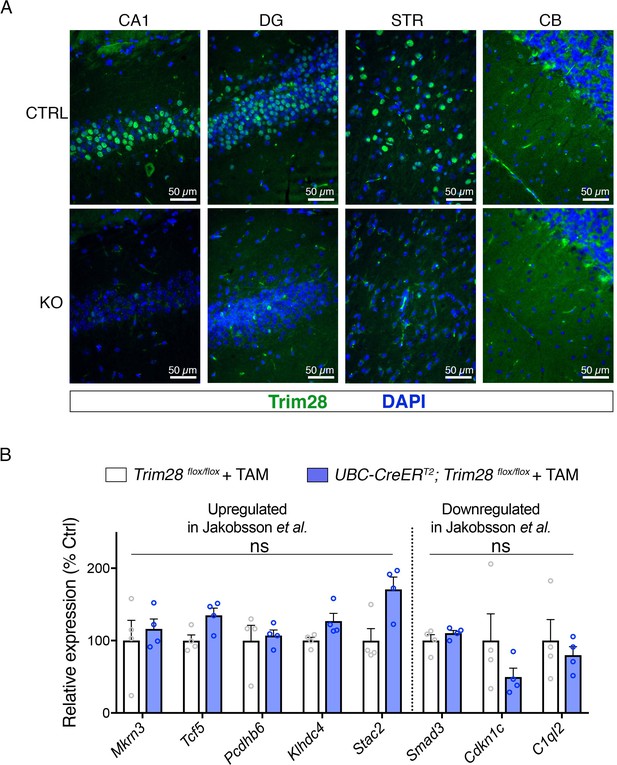
Trim28 is expressed in the adult brain and can be effectively excised from adult mice.
(A) Representative photomicrographs of Trim28 staining in the CA1 and dentate gyrus (DG) of the hippocampus, striatum (STR) and cerebellum (CB) in Trim28 adult knockouts (KO) compared to littermate controls (CTRL). (B) qPCR on genes previously reported to be disrupted upon Trim28 loss in the juvenile hippocampus.
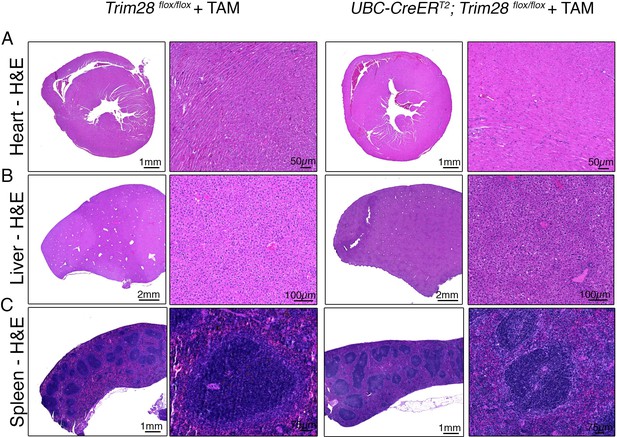
Adult depletion of Trim28 does not cause peripheral pathological abnormalities.
Representative photomicrographs of (A) Heart, (B) Liver, and (C) Spleen from Trim28 adult knockouts compared to littermate controls stained for hematoxylin and eosin. Each photomicrograph is representative of three independent animals for both genotypes.
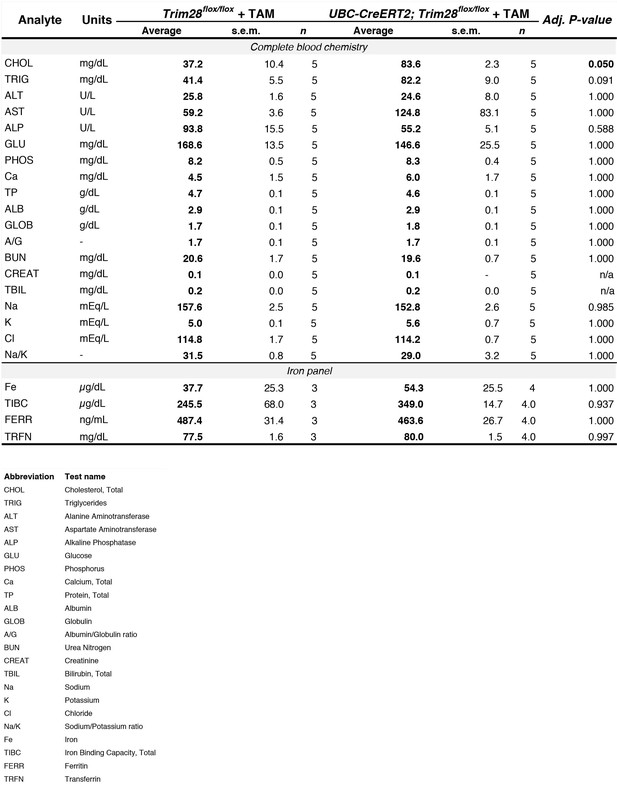
Loss of Trim28 in adult mice does not disrupt global blood chemistry or iron homeostasis.
Complete blood chemistry of Trim28 adult knockout mice compared to control littermates. Adjusted P-values from multiple t-tests (Holm-Sidak corrected) for each value are presented on the right. Legend for each analyte is presented below.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (M. musculus) | Trim28E3MT (C66A, C69A, R72G) | This study | Pure C57Bl/6J background | |
| Strain, strain background (M. musculus) | Trim28flox B6.129S2(SJL)- Trim28tm1.1Ipc/J | Jackson laboratory | Stock #018552 | Pure C57Bl/6J background |
| Strain, strain background (M. musculus) | UBC-CreERT2 B6.Cg-Ndor1 Tg(UBC-cre/ERT2)1Ejb/1J | Jackson laboratory | Stock #007001 | Pure C57Bl/6J background |
| Strain, strain background (M. musculus) | FVB/NCrl | Charles River | Code #207 | Pure C57Bl/6J background |
| Strain, strain background (M. musculus) | Trim28+/- | Rousseaux et al. (2016); this study | Crossing Jax stock #018552 to #006054 | |
| Strain, strain background (M. musculus) | Snca-/- B6;129 × 1-Sncatm1 Rosl/J | Jackson laboratory | Stock #003692 | |
| Strain, strain background (M. musculus) | Mapt-/- B6.129 × 1-Mapttm1 Hnd/J | Jackson laboratory | Stock #007251 | |
| Cell line (H. sapiens) | 293T | ATCC | CRL-3216 | |
| Cell line (H. sapiens) | 293T-shScram | This study; shScram from Rousseaux et al. (2016). | 293 T cells infected with retrovirus (pMSCV) harboring shScramble. Selected with 1 µg/mL of puromycin for at least 1 week before commencing experimentation | |
| Cell line (H. sapiens) | 293T-shTRIM28 | This study; shTRIM28 from Rousseaux et al. (2016). | 293 T cells infected with retrovirus (pMSCV) harboring shTRIM28. Selected with 1 µg/mL of puromycin for at least 1 week before commencing experimentation | |
| Transfected construct (H. sapiens) | Flag-SUMO2 | This study | ||
| Transfected construct (H. sapiens) | pKH3-HA-TRIM28 | Addgene | #45569 | |
| Transfected construct (H. sapiens) | pKH3-HA-TRIM28-C65A/ C68A | Rousseaux et al. (2016); Addgene | #92199 | |
| Transfected construct (H. sapiens) | pKH3 | Addgene | #12555 | |
| Transfected construct (M. musculus) | AAV8-YFP-shScramble | This study | accgcctgaagtctctgattaa | |
| Transfected construct (M. musculus) | AAV8-YFP-shTrim28 | This study | ttgttgaactgtttgaacatgc | |
| Antibody | alpha-synuclein (C-20), Rabbit polyclonal | Santa Cruz Biotechnology | sc-7011-R | This antibody has been discontinued. |
| Antibody | alpha-synuclein (Clone 42), Mouse monoclonal | BD Biosciences | 610786 | |
| Antibody | Tau, Rabbit polyclonal | Dako | A0024 | |
| Antibody | Tau (Tau-5), Mouse monoclonal | Abcam | ab80579 | |
| Antibody | Trim28 (20C1), Mouse monoclonal | Abcam | ab22553 | |
| Antibody | SUMO2/3, Rabbit polyclonal | Abcam | ab3742 | |
| Antibody | Flag (M2), Mouse monoclonal | Sigma Aldrich | F1804 | |
| Antibody | UBC9, Goat polyclonal | Novus Biologicals | NB300-812 | |
| Antibody | Vinculin (hVIN-1), Mouse monoclonal | Sigma Aldrich | V9131 | |
| Antibody | GFAP (G-A-5), Mouse monoclonal | Sigma Aldrich | G3893 | |
| Sequence-based reagent (M. musculus), qPCR | Mkrn3-f | ccatggagaaatatgcgaca | ||
| Sequence-based reagent (M. musculus), qPCR | Mkrn3-r | ctgagctgcatcccaagg | ||
| Sequence-based reagent (M. musculus), qPCR | Tcf5-f | tgatgcaatccggatcaa | ||
| Sequence-based reagent (M. musculus), qPCR | Tcf5-r | cacgtgtgttgcgtcagtc | ||
| Sequence-based reagent (M. musculus), qPCR | Pcdhb6-f | gccactagaagggctcgaat | ||
| Sequence-based reagent (M. musculus), qPCR | Pcdhb6-r | tgtctccacatctagctgcaa | ||
| Sequence-based reagent (M. musculus), qPCR | Klhdc4-f | cctggacaaaagttgacatcc | ||
| Sequence-based reagent (M. musculus), qPCR | Klhdc4-r | caaactccccaccgaagac | ||
| Sequence-based reagent (M. musculus), qPCR | Stac2-f | tgtctactagaaatcggtagccaag | ||
| Sequence-based reagent (M. musculus), qPCR | Stac2-r | agcgtcttgttctccacctg | ||
| Sequence-based reagent (M. musculus), qPCR | Smad3-f | ctcttggagcacatcctggt | ||
| Sequence-based reagent (M. musculus), qPCR | Smad3-r | gcccagctggaaatatgc | ||
| Sequence-based reagent (M. musculus), qPCR | Cdkn1c-f | caggacgagaatcaagagca | ||
| Sequence-based reagent (M. musculus), qPCR | Cdkn1c-r | gcttggcgaagaagtcgt | ||
| Sequence-based reagent (M. musculus), qPCR | C1ql2-f | tcacgtaccacattctcatgc | ||
| Sequence-based reagent (M. musculus), qPCR | C1ql2-r | tgttgctggcgtagtcgta | ||
| Sequence-based reagent (M. musculus), qPCR | Snca-f | gaagacagtggagggagctg | ||
| Sequence-based reagent (M. musculus), qPCR | Snca-r | caggcatgtcttccaggatt | ||
| Sequence-based reagent (M. musculus), qPCR | Mapt-f | gagaatgccaaagccaagac | ||
| Sequence-based reagent (M. musculus), qPCR | Mapt-r | gtgagtccaccatgtcgatg | ||
| Sequence-based reagent (M. musculus), qPCR | Trim28-f | gctgctgccctgtctacatt | ||
| Sequence-based reagent (M. musculus), qPCR | Trim28-r | cacactggacaatccaccat | ||
| Sequence-based reagent (M. musculus), qPCR | S16-f | aggagcgatttgctggtgtgg | ||
| Sequence-based reagent (M. musculus), qPCR | S16-r | gctaccagggcctttgagatg | ||
| Sequence-based reagent (H. sapiens), siRNA | siScramble | ThermoFisher Scientific | AM4611 | |
| Sequence-based reagent (H. sapiens), siRNA | siUBC9 | ThermoFisher Scientific | AM16708-120322 | |
| Chemical compound, drug | Viomeillin | BioViotica | BVT-0359-C500 | |
| Chemical compound, drug | N-ethylmaleimide (NEM) | Sigma Aldrich | E3876-5G | |
| Chemical compound, drug | Tamoxifen | Sigma Aldrich | T5648-5G |
Additional files
-
Supplemental file 1
Detailed statistical analysis for all data throughout the manuscript.
- https://doi.org/10.7554/eLife.36768.012
-
Transparent reporting form
- https://doi.org/10.7554/eLife.36768.013





