Structure of the human lipid-gated cation channel TRPC3
Figures
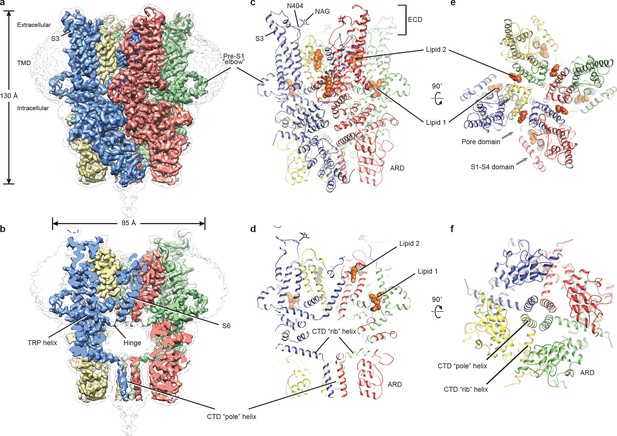
Architecture of human TRPC3.
(a) Three-dimensional reconstruction viewed parallel to the membrane. The transparent envelope denotes the unsharpened reconstruction. (b) Slice view of the reconstruction showing the interior of the channel. (c–f) Atomic model of TRPC3 viewed parallel to the membrane (c–d), from the extracellular side (e), and from the intracellular side (f). Each subunit is colored differently.
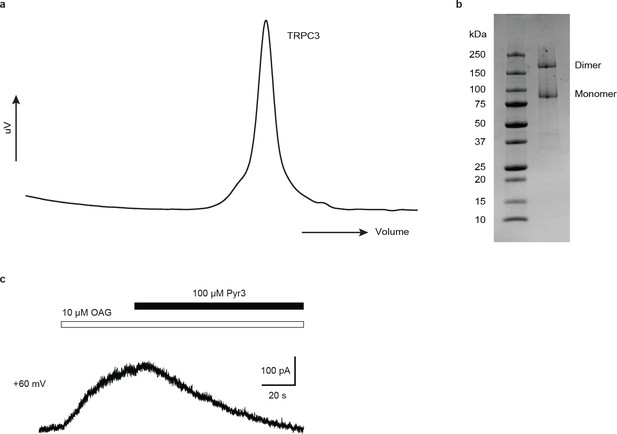
Preparation, electrophysiological characterization of human full-length TRPC3.
(a) Size-exclusion chromatography profile of TRPC3. (b) SDS gel of purified TRPC3. (c) Under whole-cell voltage clamp configuration, HEK-293 cells infected by virus encoding WT TRPC3 receptor gene show robust current when exposing to 10 µM OAG, and this current can be completely blocked by 10 µM OAG +100 µM Pyr3 (n = 3 cells).
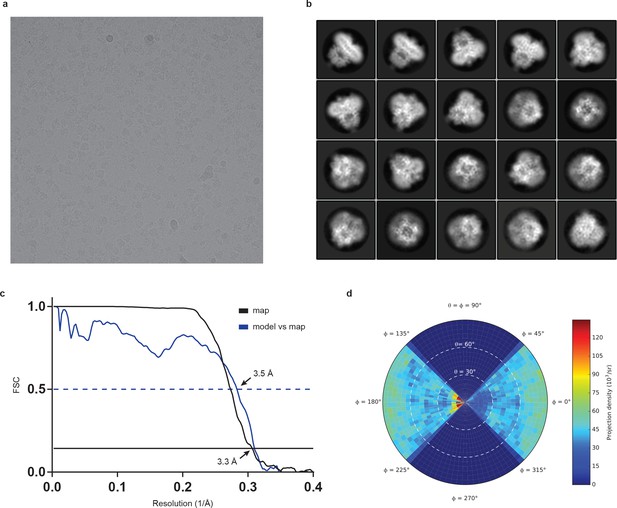
Cryo-EM analysis of human full-length TRPC3.
(a) Representative electron micrograph. (b) Selected two-dimensional class averages of the electron micrographs. (c) The gold-standard Fourier shell correlation curve for the EM maps is shown in black and the FSC curve between the atomic model and the final EM map is shown in blue. (d) Angular distribution of particles used for refinement.
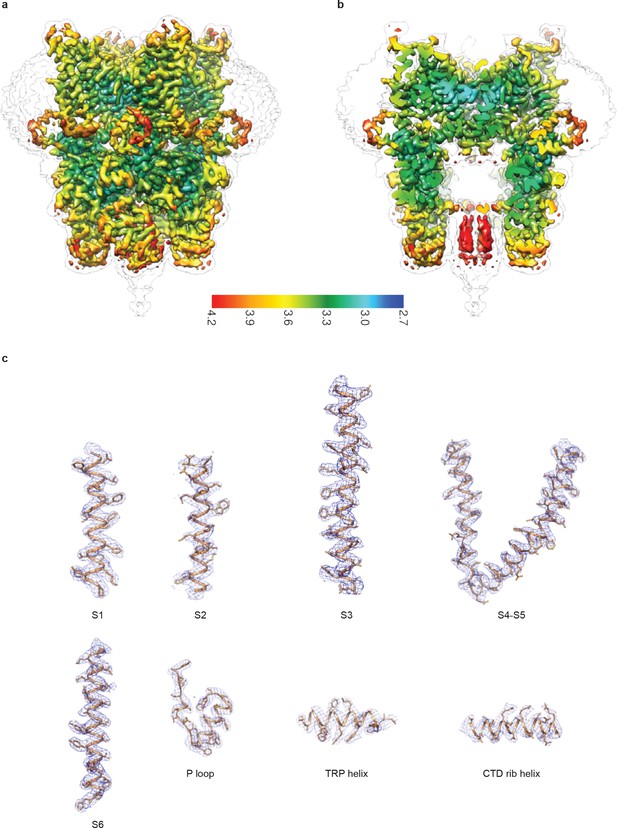
Cryo-EM map of human full-length TRPC3.
(a-b) Local resolution estimation. The map is colored according to local resolution estimation. (c) Representative densities. Density maps are shown in blue meshes, and the atomic models are shown in cartoon representation with side chains as sticks.

Secondary structure arrangement of human TRPC3 and sequence alignment of TRPC family channels.
The TRPC2 is from Mus musculus, whereas all the other proteins are from human. The sequences were aligned using the Clustal Omega program on the Uniprot website and coloured using BLOSUM62 score by conservation. The secondary structural elements are color-coded to match Figure 2a.
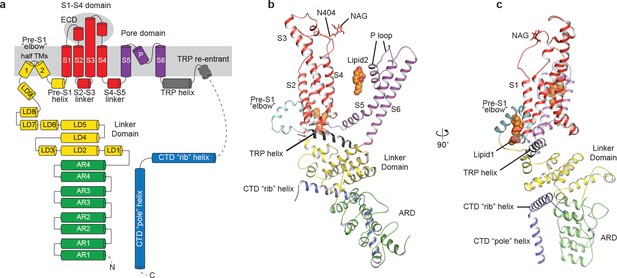
Structure of a single subunit.
(a) The schematic representation of TRPC3 domain organization. Dashed lines indicate the regions that have not been modeled. (b–c) Cartoon representation of one subunit color-coded to match panel a.
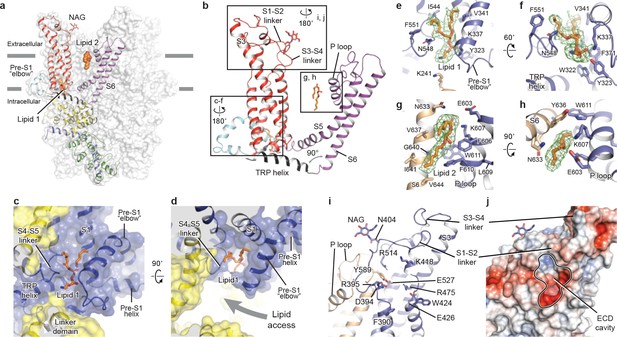
Transmembrane domain, extracellular domain, and lipid-binding sites.
(a) Domain organization. The channel is shown in surface representation, with one subunit shown in cartoon representation. The colors match those in Figure 2a. (b) Details of the transmembrane domain and extracellular domain. (c–d) Pre-S1 elbow and binding site of lipid 1. The lipid molecule is buried inside the pocket formed by pre-S1 elbow, S1, and the S4-S5 linker. Two adjacent subunits (blue and yellow) are shown in both cartoon and surface representations. The lipid molecule is shown as sticks. (e–f) Residues that interact with lipid 1 are shown in sticks, and protein is shown in cartoon representation. Lipid density is shown in mesh. (g–h) Lipid 2 binds between S6 and the P loop of adjacent subunits, which are in light blue and wheat. (i) Structure of the ECD. Key residues forming the cavity is shown in sticks. Adjacent subunits are in light blue and wheat. (j) Surface representation of the ECD, colored according to the electrostatic surface potential. The color gradient is from −5 to 5 kT/e (red to blue).
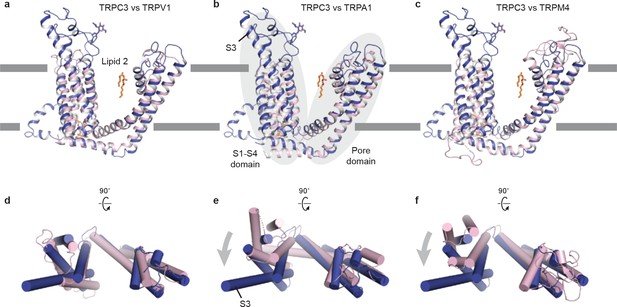
Comparison of the TMD of TRPC3 with TRPV1.
(a) TRPA1 (b) and TRPM4 (c). Structures are aligned using main chain atoms of the pore domain. Only the TMD of one subunit is shown in cartoon representation, viewed in parallel to the membrane. TRPC3 is in blue; TRPV1, TRPA1 and TRPM4 are in pink. (d-e) TMD viewed from extracellular side. The relative organization of the S1-S4 domain with the pore domain in TRPC3 is similar to that in TRPV1, but the S1-S4 domain in TRPC3 exhibits a clockwise rotation relative to TRPA1 or TRPM4.
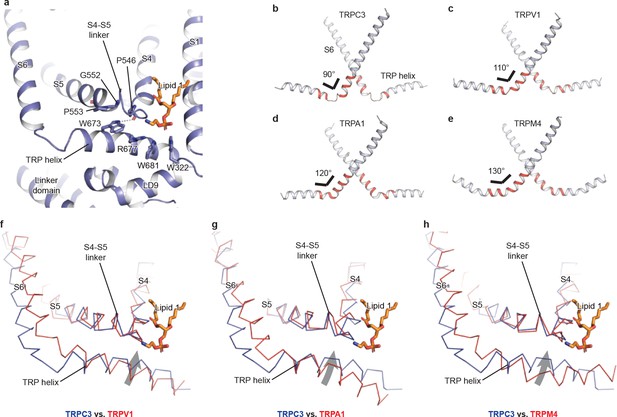
The TRP domain.
(a) Cartoon representation of the TRP helix, pre-S1 elbow, TMD, and linker domain, showing their interaction. Lipid 1 is shown in sticks. W673 on the TRP helix stacks with P553 and G552 forming a hydrogen bond with the backbone oxygen (dashed line) of P546 on the S4-S5 linker. The side chain of R677 is in close contact with the head group of lipid 1. (b–e) The pore lining helix S6 and the TRP helix in TRPC3 (b), TRPV1 (c), TRPA1 (d), and TRPM4 (e). The angle between the S6 and TRP helices are indicated; only two subunits are shown for clarity. The hinge connecting the S6 and TRP helix is highlighted in red. (f–h) Comparison of TRPC3 with TRPV1, TRPA1, and TRPM4, respectively, focusing on the S4, S5 and TRP helix. Structures are superimposed using backbone atoms in S4 and S5. TRPC3 is in blue, whereas TRPV1, TRPA1 and TRPM4 are in red. Proteins are shown in ribbon representation, and lipid 1 in TRPC3 is shown in sticks. Arrows indicate the relative movement of the TRP helix in TRPC3 compared to TRPV1, TRPA1 or TRPM4.
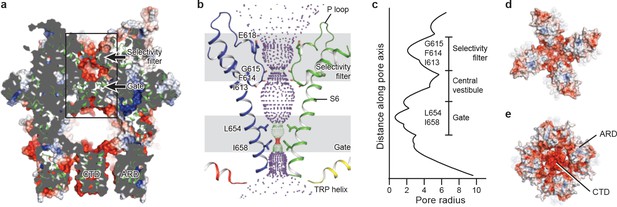
The ion-conducting pore.
(a, d, e) Surface representation of TRPC3, viewed (a) parallel to the membrane, (d) from the extracellular side, and (e) from the intracellular side. The surface is colored according to electrostatic surface potential; the color gradient is from −5 to 5 kT/e (red to blue). The protein is also shown in cartoon representation in (a). (b) The shape and size of the ion-conducting pore (boxed area in panel a). The P loop and S6 of two subunits and the TRP helix of the other two subunits are shown as cartoons, and the side chains of restriction residues are shown as sticks. Purple, green, and red spheres define radii of >2.3, 1.2–2.3, and <1.2 Å, respectively. (c) Plot of pore radius as a function of distance along the pore axis in Angstroms.
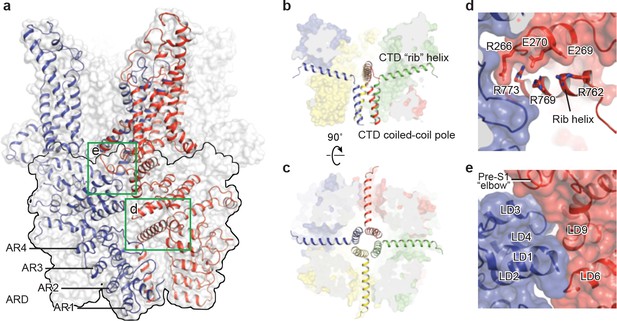
The intracellular domain.
(a) Surface representation of TRPC3 with two adjacent subunits shown in cartoon representation. The intracellular domain is highlighted in the black frame. Two interfaces highlighted in green frames are enlarged in (d–e). (b–c) Cartoon representation of the CTD coiled-coil pole and the rib helix. The intracellular domain is shown in surface representation, viewed in parallel to the membrane (b) and from the intracellular side (c). (d) Inter-subunit interface formed by the CTD rib helix with adjacent ARD and LD. Protein is shown in cartoon and surface representations. Two adjacent subunits are in blue and red. Charged residues forming hydrogen bond or polar interaction with each other are shown as sticks. (e) Interface between adjacent LDs and pre-S1. Alpha helices involved in the inter-subunit interaction are indicated.
Tables
Statistics of EM data processing and model refinement.
https://doi.org/10.7554/eLife.36852.013| Data collection/processing | |
|---|---|
| Microscope | Titan Krios (FEI) |
| Voltage (kV) | 300 |
| Defocus range (µM) | 1.0–2.5 |
| Exposure time (s) | 8 |
| Dose rate (e-/Å2/s) | 6.76 |
| Number of frames | 40 |
| Pixel size (Å) | 1.074 |
| Particles refined | 143855 |
| Resolution (Å) | 3.3 |
| FSC threshold | 0.143 |
| Resolution range (Å) | 412.4–3.3 |
| Model statistics | |
| Number of atoms | 20988 |
| Protein | 20744 |
| Ligand | 244 |
| r.m.s. deviations | |
| Bond length (Å) | 0.005 |
| Bond angle (°) | 1.008 |
| Ramachandran plot | |
| Favored (%) | 94.09 |
| Allowed (%) | 5.77 |
| Disallowed (%) | 0.14 |
| Rotamer outlier (%) | 0.85 |
| Clashscore | 3.0 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.36852.014





