CYK-4 functions independently of its centralspindlin partner ZEN-4 to cellularize oocytes in germline syncytia
Figures
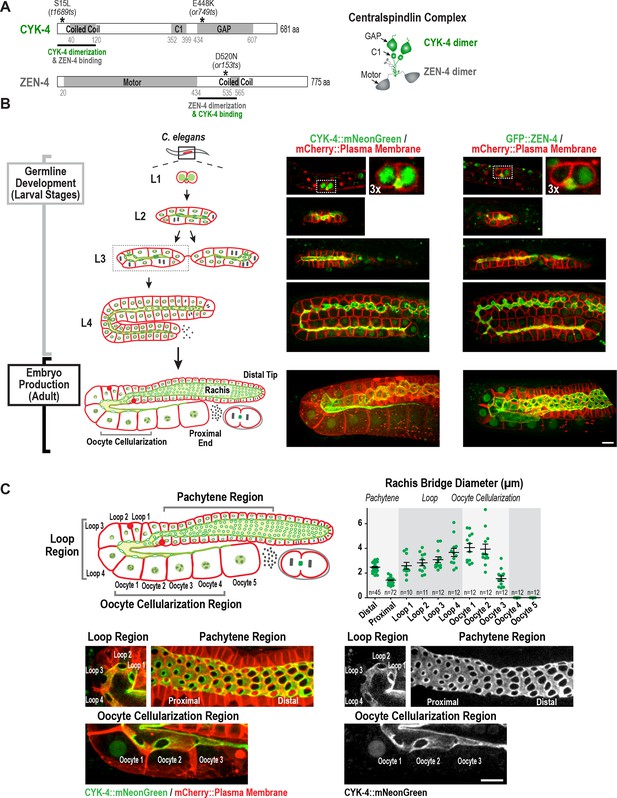
Both centralspindlin subunits localize to intercellular bridges throughout C. elegans germline development.
(A) (Left) Schematics highlight the domain structure of the two molecular components, CYK-4 and ZEN-4, of the heterotetrameric centralspindlin complex, and the location of the temperature sensitive mutations used in this study. (Right) Schematic showing that centralspindlin is formed by the association of a dimer of CYK-4 with a dimer of ZEN-4. (B) (Left) Schematics illustrate the development of the syncytial germline. See text for details. (Right) Fluorescence confocal images of germlines in worms at the indicated stages expressing an mCherry-tagged plasma membrane marker and either CYK-4::mNeonGreen or in situ-tagged GFP::ZEN-4. Images for L4 and adult stage germlines are maximum intensity projections. (C) Rachis bridges increase in diameter during oocyte loading prior to their closure during cellularization. (Upper left) Schematic shows the location of the germline regions in the images and the nomenclature for labeling the compartments in the Loop (Loop 1–4) and Oocyte Cellularization (Oocyte 1–5) Regions. As indicated, Oocyte 1 was the first compartment after the turn. (Upper right) Graph plotting the diameters of the rachis bridges (mean ± SEM) in the indicated regions of the germline measured in the CYK-4::mNeonGreen images. n = number of cells analyzed at the indicated position/region. (Lower panels) Maximum intensity projections of confocal images of the pachytene, loop, and oocyte cellularization regions of adult germlines acquired in the strain expressing an mCherry-tagged plasma membrane marker and CYK-4::mNeonGreen. Merged images are shown alongside single color images showing the CYK-4::mNeonGreen signal. Scale bars are 10 µm.
-
Figure 1—source data 1
Rachis bridges increase in diameter during oocyte loading prior to their closure during oocyte cellularization.
- https://doi.org/10.7554/eLife.36919.005
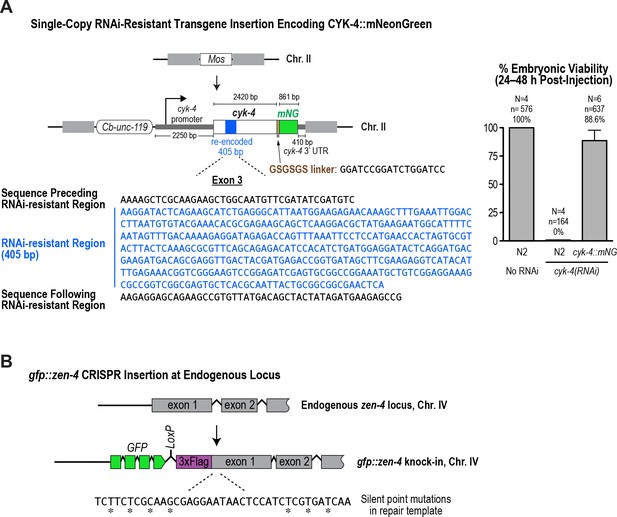
Generation of a functional single-copy transgene encoding CYK-4::mNeonGreen and in situ-tagged GFP::ZEN-4.
(A) (Left) Schematic of the RNAi-resistant cyk-4::mNeonGreen transgene integrated into a specific site (Mos transposon insertion) on Chromosome II. (Right) Graph plotting embryonic viability (mean ± SD) following depletion of endogenous CYK-4 by RNAi. N = number of worms, n = number of embryos. (B) Schematic showing the location where sequences encoding GFP were inserted prior to the first exon of the gene encoding ZEN-4.
-
Figure 1—figure supplement 1—source data 1
The RNAi-resistant transgene encoding CYK-4::mNeonGreen rescues depletion of endogenous CYK-4.
- https://doi.org/10.7554/eLife.36919.006
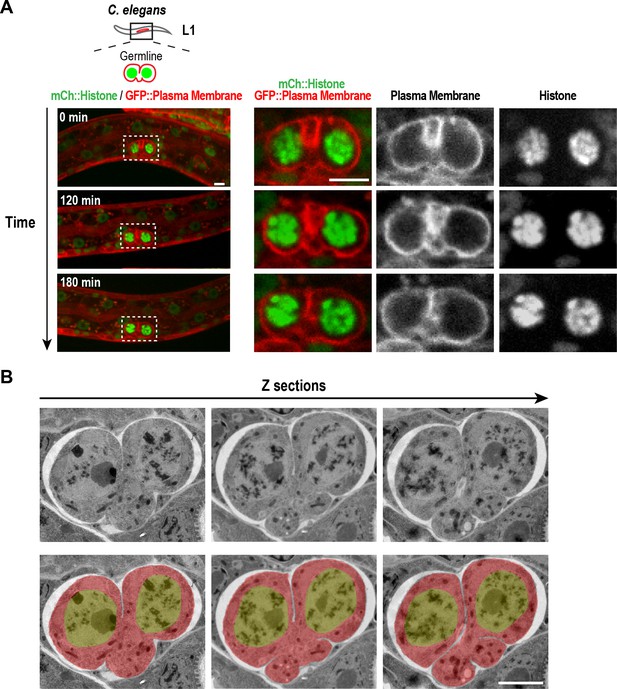
Structure of the nascent syncytial germline and rachis in the L1 larva.
(A) Timelapse images collected using the worm trap described in Figure 4A over a 3-hr period of the germline in an L1 stage worm. At this stage, the intercellular bridge connecting the two nuclear compartments extends out on one side. Scale bars are 5 µm. (B) Selected images from a stack of serial 100 nm sections of an L1 germline (to view the entire image series see Video 1) that were collected and imaged by transmission electron microscopy after fixation by high-pressure freezing and freeze-substitution. Images are shown without (top) and with (bottom) superposition of a pseudocolor model in which the regions within the boundary of the two-compartment germline syncytium (red) and germline nuclei (green) are highlighted. The two nuclei-containing compartments sit side-by-side and open into a small cytoplasm containing bridge (nascent rachis) that extends out to one side. Images above and below the central planes reveal that the intercellular bridge/rachis has a lobed structure. Scale bar is 2 µm.
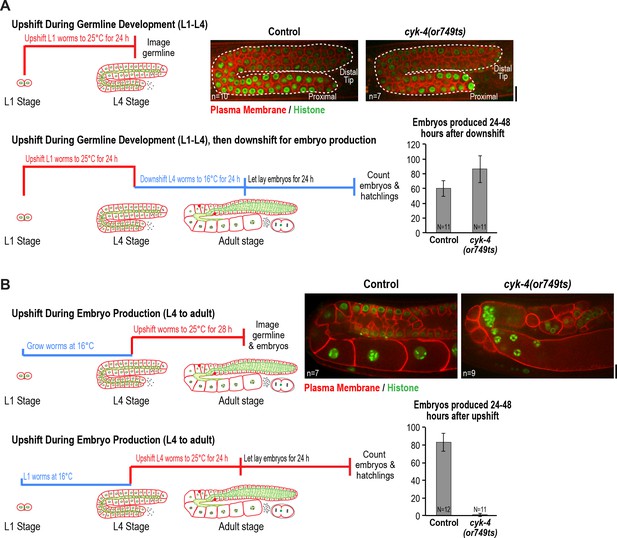
CYK-4 is required for oocyte production by the adult germline.
(A) The CYK-4 C-terminus is not required for compartment proliferation during germline development. (Upper left) Larvae were upshifted to the non-permissive temperature (25°C) at the L1 stage and the germlines were examined at the L4 stage as indicated in the schematic. (Upper right) Representative single plane confocal images of germlines in L4 stage worms (control or cyk-4(or749ts)) expressing a GFP-tagged plasma membrane probe (shown in red) and mCherry::histone H2B (shown in green) after the upshift protocol. Dashed lines mark the germline boundaries. (Lower left) Schematic outline of the upshift protocol used to assess embryo production. (Lower right) Graph plots the number of embryos laid by the worms between 24 and 48 hr after downshift. (B) The CYK-4 C-terminus is required for oocyte production in the adult. (Upper left) Larvae were upshifted to the non-permissive temperature (25°C) at the L4 stage and the germlines were examined 28 hr later as indicated in the schematic. (Upper right) Representative single plane confocal images of germlines in adult stage worms (control or cyk-4(or749ts)) expressing a GFP-tagged plasma membrane probe (shown in red) and mCherry::histone H2B (shown in green) after the upshift. (Lower left) Schematic outline of the upshift protocol used to assess embryo production. (Lower right) Graph plots the number of embryos produced between 24 and 48 hr after upshift. n in images in A and B = number of imaged germlines. N in graphs = number of assayed worms. Scale bars are 10 µm.
-
Figure 2—source data 1
The CYK-4 C-terminal region is not required for germline compartment proliferation.
- https://doi.org/10.7554/eLife.36919.009
-
Figure 2—source data 2
The CYK-4 C-terminal region is required for oocyte production.
- https://doi.org/10.7554/eLife.36919.010
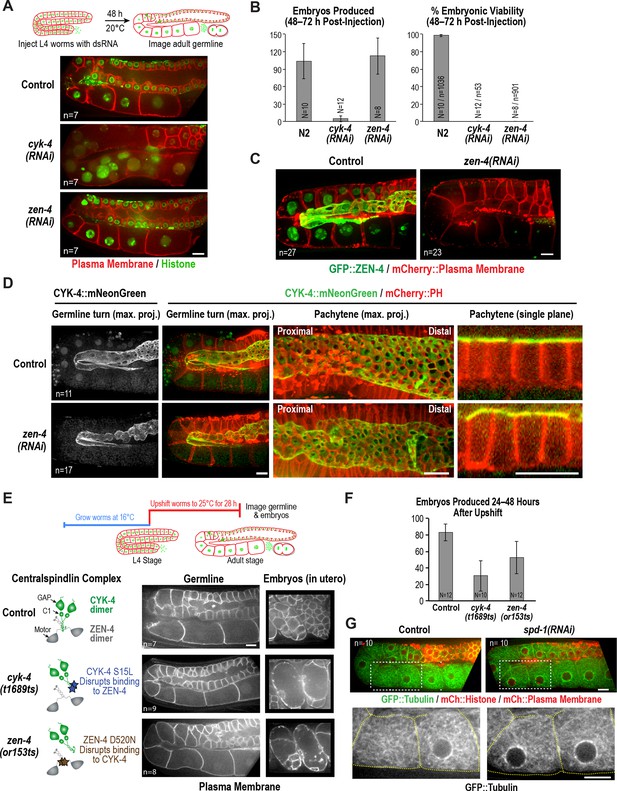
ZEN-4 is not essential for embryo production by the adult syncytial germline.
(A) (Top) Schematic outline of the imaging experiment following dsRNA injection. (Bottom) Single plane confocal images of adult germlines expressing a GFP-tagged plasma membrane probe (shown in red) and mCherry::histone H2B (shown in green) following depletion of CYK-4 or ZEN-4 by RNAi. n = number of worms imaged. (B) Graphs plotting the number of embryos laid by the worms during the indicated time intervals (left) or embryonic viability (right) (mean ± SD) for the indicated conditions. N = number of worms, n = number of embryos. (C) Maximum intensity projections of the adult germline in control and zen-4(RNAi) worms expressing in situ-tagged GFP::ZEN-4 and an mCherry-tagged plasma membrane probe. n = number of worms imaged. (D) (left three columns) Maximum intensity projection images of adult germlines in control (top) and zen-4(RNAi) (bottom) worms expressing CYK-4::mNeonGreen (green) and a mCherry-tagged plasma membrane probe (red). (right column) Single plane images of a portion of the pachytene region. n = number of imaged worms. (E) (Top) Schematic outline of the experiment imaging adult worms following temperature upshift at the L4 stage. (Bottom) Single plane confocal images showing the GFP-tagged plasma membrane probe in adult germlines and embryos in the uterus of worms subjected to the upshift protocol. n = number of worms imaged. (F) Graph plotting the number of embryos produced (between 24 and 48 hr after temperature upshift at the L4 stage. Embryos in the uterus were counted in addition to embryos laid because centralspindlin mutants, especially cyk-4(t1689ts), are somewhat egg laying defective under these conditions. (G) Images of adult germlines in control (left) and spd-1(RNAi) (right) worms expressing GFP::β-tubulin, mCherry::histone and an mCherry-tagged plasma membrane probe. Insets (bottom panels) show the GFP::β-tubulin signal in the boxed regions (oocyte cellularization region) with yellow dashed lines outlining the cell boundaries). n = number of imaged worms. As illustrated in (A), after dsRNA injection, worms were incubated for 48 hr at 20°C before imaging in (C), (D) and (G). Scale bars are 10 µm.
-
Figure 3—source data 1
ZEN-4 is required for embryo viability but not production.
- https://doi.org/10.7554/eLife.36919.013
-
Figure 3—source data 2
The CYK-4—ZEN-4 interaction is not required for embryo production.
- https://doi.org/10.7554/eLife.36919.014
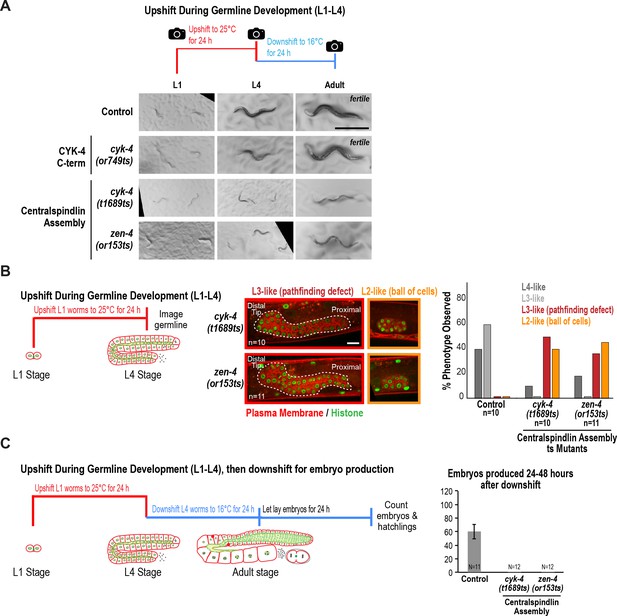
The interaction between the CYK-4 and ZEN-4 dimers is globally required for worm growth and development between the L1 and L4 stages.
(A) Low-resolution images of worms grown as indicated in the schematic. Briefly, L1 worms were picked from asynchronous populations grown at 16°C, and pictures were snapped immediately before upshifting to 25°C for 24 hr. Worms were imaged again after 24 hr at the non-permissive temperature. At this timepoint, control and CYK-4 C-terminal mutant cyk-4(or749ts) worms have developed to L4 stage, whereas the mutants that disrupt centralspindlin assembly (cyk-4(t1689ts) and zen-4(or153ts)) appeared markedly smaller and sick; the larger cyk-4(t1689ts) and zen-4(or153ts) worms were picked for subsequent steps. Worms were downshifted to 16°C and pictures were taken again after 24 hr. Adult control and cyk-4(or749ts) worms appeared fertile, whereas cyk-4(t1689ts) and zen-4(or153ts) worms exhibited defects including protruding or exploded vulvas. (B) (Left) Schematic illustrates the experimental procedure. (Center) Representative single plane images for the indicated conditions showing germlines expressing a GFP-tagged plasma membrane probe (shown in red) and mCherry-tagged histone (shown in green) after upshift of worms between the L1 and L4 stages. Control image is shown in Figure 2B. Dashed line indicates boundary of germline. n = number of worms. (Right) Graph showing the percentage of the indicated germline phenotypes observed after upshifting them at L1 stage to 25°C for 24 hr. (C) (Left) Schematic illustrates the experimental procedure. (Right) Graph shows the number of embryos produced between 24 and 48 hr after downshift (mean ± SD) by worms upshifted between the L1 and L4 stages. Embryos in the uterus were counted in addition to embryos laid because centralspindlin mutants, especially cyk-4(t1689ts), are somewhat egg laying defective under these conditions. Control data is reproduced from Figure 2A for comparison. N = number of worms. Scale bars in (A) and (B) are 500 and 10 µm.
-
Figure 3—figure supplement 1—source data 1
The CYK-4—ZEN-4 interaction is required for larval growth and germline pathfinding.
- https://doi.org/10.7554/eLife.36919.015
-
Figure 3—figure supplement 1—source data 2
Aberrant germlines in L1-L4 upshifted centralspindlin assembly mutants fail to produce embryos following downshift.
- https://doi.org/10.7554/eLife.36919.016
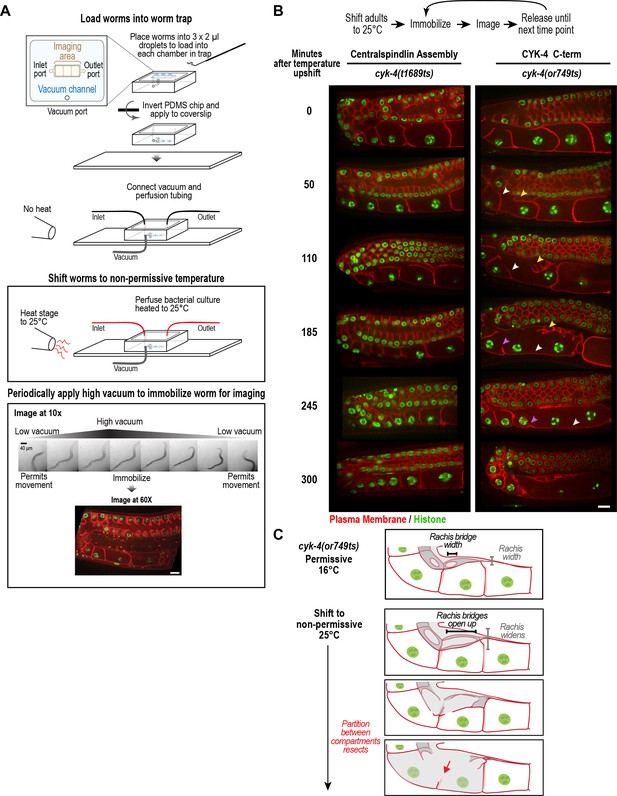
CYK-4 is required for oocyte cellularization.
Individual CYK-4 mutant worms were longitudinally monitored using a custom vacuum-actuated microfluidic device (the ‘worm trap’). (A) Schematics summarize the experimental procedure for mounting worms in the trap (top), temperature shift (middle), and periodic immobilization for imaging (bottom). Shown are a series of images demonstrating gradual immobilization and release of a single worm at low magnification (10x), and a single plane high-resolution (60x) image of immobilized worm. (B) (top) Individual young adult worms carrying the cyk-4(t1689ts) mutation that blocks the interaction between CYK-4 and ZEN-4 dimers (n = 7; Centralspindlin Assembly) or the cyk-4(or749ts) mutation that compromises the CYK-4 C-terminus (n = 9; CYK-4 C-term) expressing a GFP-tagged plasma membrane probe (shown in red) and an mCherry fusion with histone H2B (shown in green) were tracked for 5 hr at restrictive temperature in the worm trap. Worms were immobilized periodically for imaging, as indicated in the schematic in (A). Single plane images from the time course are shown. (C) Schematics illustrate how the phenotype in cyk-4(or749ts) worms arises. Following upshift, the partition between the last two uncellularized oocytes resects due to the combined effects of the rachis widening and the rachis bridges opening up (for example, see white arrowheads in the sequence in (B)). Resection of the next and subsequent partitions in a similar fashion (purple arrowheads in the sequence in (B)) leads to a hollow multinucleated ‘tubulated’ proximal germline. Scale bars are 10 µm.
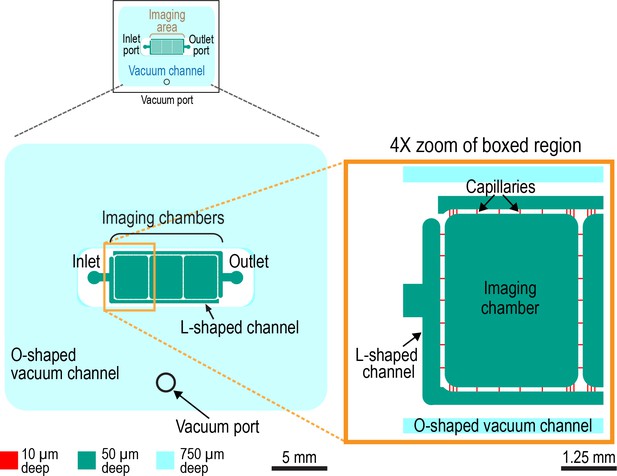
Design of the worm trap microfluidic chip.
Schematic of microfluidic device (left) and 4X zoom of left-most imaging chamber (right). The worm trap microfluidic device is assembled out of a PDMS chip with microchannels engraved on its surface and a 35 × 50 mm #1.5 microscope coverslip, which seals the microchannels (not shown). The microchannels of the device are of three different depths, 10, 50, and 750 µm (shown in red, green, and cyan, respectively). The 10 and 50 µm deep microchannels form a liquid-filled network with one inlet and one outlet as indicated that is surrounded by a separate O-shaped 750 µm deep channel (cyan). This last channel serves as a vacuum cup. When the chip is sealed with a coverslip, and vacuum is applied to this 750 µm deep channel (through a dedicated vacuum port) the chip and coverslip are pulled together. The application of vacuum also leads to partial collapse of all channels of the device, including the liquid-filled microchannels, making it possible to dynamically reduce the microchannel depth in a controlled way by adjusting the level of vacuum. The main functional elements of the liquid-filled microchannel network are the three identical 3.7 × 3 mm imaging chambers, one of which is highlighted in zoom panel, which are 50 µm deep and have rounded corners. The 50 µm depth (as well as somewhat reduced depths of 40–45 µm, when the chambers partially collapse under a low vacuum of −5 kPa) was empirically found to be sufficient for larval and young adult worms to move freely and feed. The imaging chambers are connected to each other and to the device inlet and outlet through capillary microchannels with cross-sections of 10 × 10 µm. The small cross-section of these capillary microchannels makes it impossible for worms to enter them. Hence, worms cannot escape from the imaging chambers. The inlet and outlet are connected to the capillary microchannels through 50 µm deep, 300 µm wide L-shaped microchannels. Relatively low fluidic resistance of these last microchannels (as compared with the 10 × 10 µm capillaries) facilitates even distribution of flow between the capillaries and even perfusion of the imaging chambers with bacterial suspension, which introduced via the inlet.
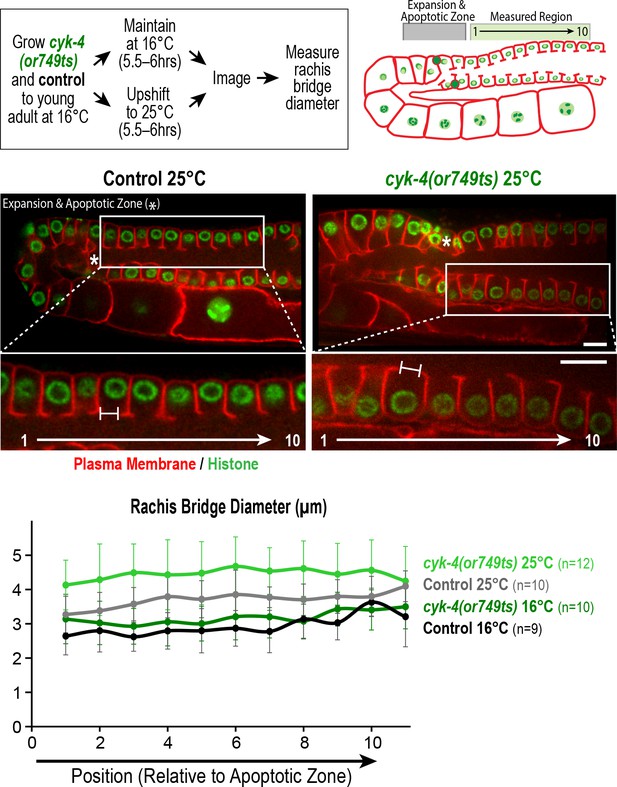
Measurement of the rachis bridges in the CYK-4 C-terminal mutant cyk-4(or749ts).
(Top left) Schematic shows the temperature shift procedure for the experiment. (Top right and middle panels) Schematic and single plane confocal images of adult germlines in control and cyk-4(or749ts) worms expressing a GFP-tagged plasma membrane marker (shown in red) and mCherry::histone (shown in green). Insets show the position of the cells in the pachytene region, right next to the expansion and apoptotic zone as indicated by an asterisk, that were used for measurement of the rachis bridges. (Bottom) Graph plotting the diameters of the rachis bridges in the indicated region of the germline measured in the CYK-4::mNeonGreen images. n = number of cells analyzed at the indicated position/region. Scale bars are 10 µm. Error bars are the SD.
-
Figure 4—figure supplement 2—source data 1
Rachis bridges in the pachytene region are wider in upshifted cyk-4(or749ts) mutant worms than in controls.
- https://doi.org/10.7554/eLife.36919.020
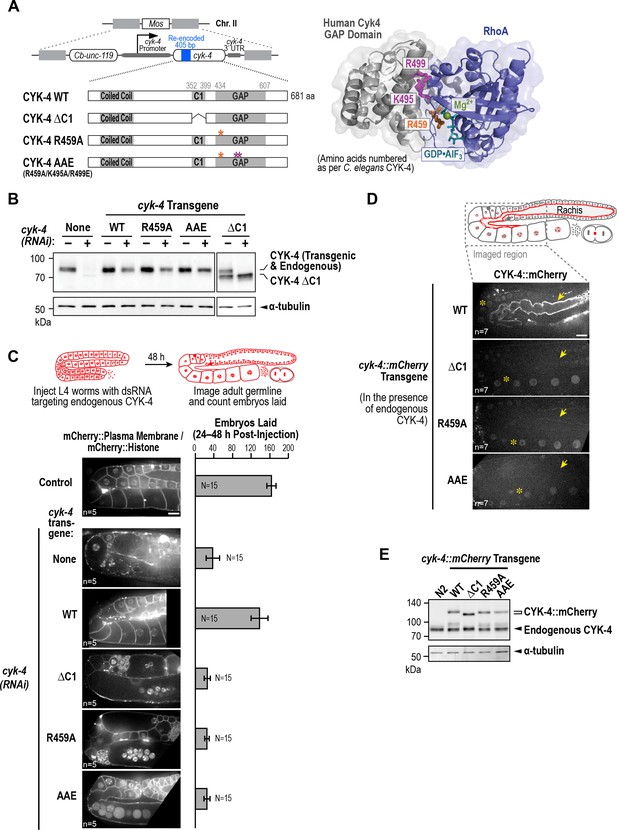
CYK-4 requires both its C1 domain and the Rho GTPase interaction interface of its GAP to target to the rachis surface and bridges and promote oocyte cellularization.
(A) (Left) Schematic illustrates the set of single copy untagged RNAi-resistant cyk-4 transgenes inserted into a specific location on Chr. II generated to analyze the role of the C1 and GAP domains. (Right) Structure of the complex of human Cyk4 GAP domain (grey) and RhoA (blue) (reproduced based on PBD ID 5C2K). Key residues in the binding pocket (numbered as per C. elegans CYK-4) are highlighted in orange (R459, catalytic arginine finger) and magenta (K495 and R499, important for GTPase binding). (B) Immunoblots of extracts prepared from worms lacking a transgene (None; N2 strain) or with the transgenes outlined in (A) in the presence (+) or absence (-) of RNAi to deplete endogenous CYK-4. Blots were probed with antibodies to CYK-4 (top) and α-tubulin as a loading control (bottom). With the exception of the ΔC1 mutant, which runs at a lower molecular weight, the transgene-encoded proteins ran at the same molecular weight as endogenous CYK-4. In the absence of a cyk-4 transgene, the CYK-4 band disappears, confirming the effectiveness of our RNAi. The protein running at the level of endogenous CYK-4 after RNAi in the WT, R459A, AAE samples is the transgenic protein. (C) (Left) Single central plane images of the germline in adult worms with the indicated transgenes that were also expressing mCherry::histone and an mCherry plasma membrane maker after depletion of endogenous CYK-4 by RNAi. n = number of worms. (Right) Graph plotting the number of embryos laid 24–48 hr post-injection (mean ± SD) for strains expressing the indicated cyk-4 transgenes (without histone or plasma membrane markers). N = number of worms. (D) Single central plane images of the germline in adult worms expressing the indicated CYK-4::mCherry fusions. Asterisks highlight the localization of each of the fusions to nuclei and arrows point to the rachis surface in each germline. n = number of imaged worms. (E) Immunoblot of extracts prepared from worms expressing the mCherry-tagged RNAi-resistant cyk-4 transgenes probed with antibodies to CYK-4 (top) and α-tubulin (bottom) as a loading control. Scale bars are 10 µm.
-
Figure 5—source data 1
Both the C1 domain and the GTPase binding interface of the GAP domain are required for the germline function of CYK-4.
- https://doi.org/10.7554/eLife.36919.023
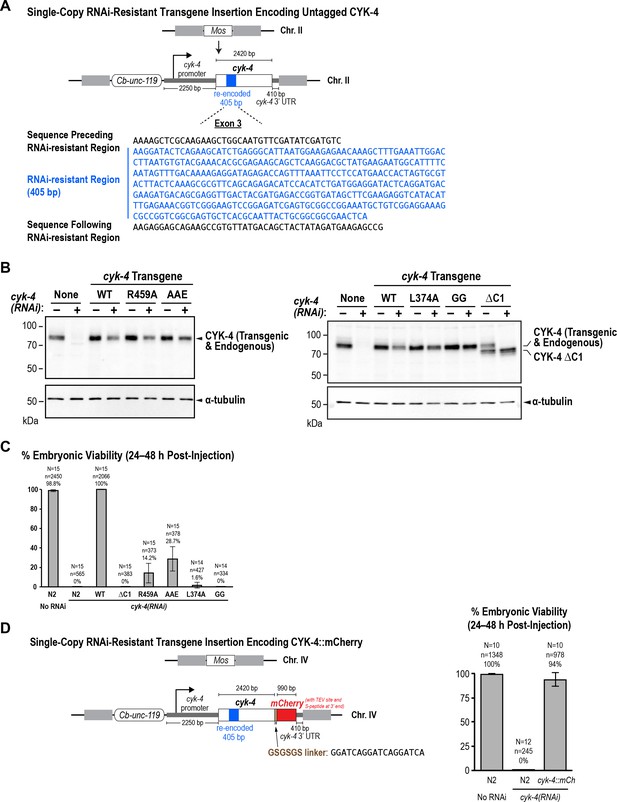
The WT, GTPase binding interface mutant, and ΔC1 mutant CYK-4 proteins encoded by the single copy untagged transgenes are expressed at levels comparable to endogenous CYK-4.
(A) Schematic of the RNAi-resistant untagged cyk-4 transgene integrated into a specific site (Mos transposon insertion) on Chromosome II. (B) Full versions of the blots in Figure 5B. L374A and GG are point mutants in the C1 domain that are not used in this study. (C) Graph plotting embryonic viability (mean ± SD) following depletion of endogenous CYK-4 by RNAi. N = number of worms, n = number of embryos. (C) (Left) Schematic of the RNAi-resistant cyk-4::mCherry transgene integrated into a specific site (Mos transposon insertion) on Chromosome IV. (Right) Graph plotting embryonic viability (mean ± SD) following depletion of endogenous CYK-4 by RNAi. N = number of worms, n = number of embryos.
-
Figure 5—figure supplement 1—source data 1
Both the C1 and the GTPase binding interface of the GAP domain are required for embryonic viability.
- https://doi.org/10.7554/eLife.36919.024
-
Figure 5—figure supplement 1—source data 2
The RNAi-resistant transgene encoding CYK-4::mCherry rescues depletion of endogenous CYK-4.
- https://doi.org/10.7554/eLife.36919.025
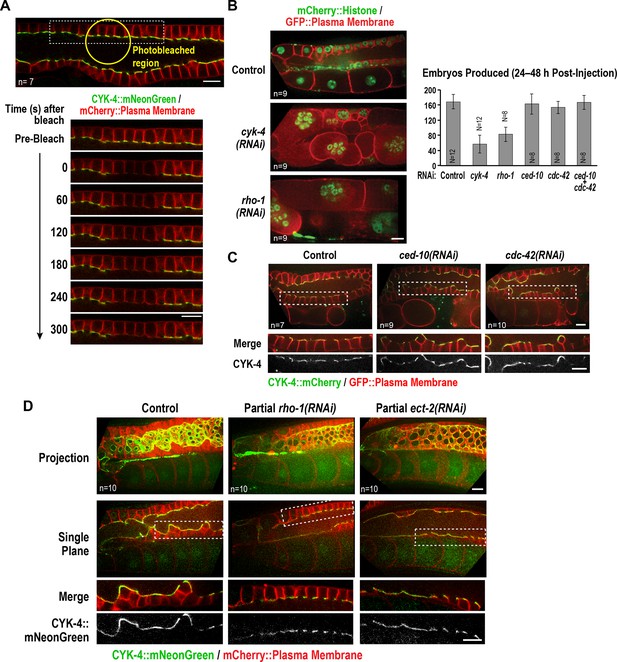
The Rho GTPase binding interface of the CYK-4 GAP domain may recruit CYK-4 to the rachis surface/bridges by binding RhoARHO-1.
(A) Central plane confocal images acquired in adult worms expressing CYK-4::mNeonGreen (green) and an mCherry-tagged plasma membrane probe (red) before and after photobleaching of the CYK-4::mNeonGreen signal in a section of the rachis in the pachytene region. (B) (Left) Single central plane images of germlines in adult worms expressing mCherry::H2B (shown in green) and a GFP-tagged plasma membrane probe (shown in red) following CYK-4 or RhoARHO-1 depletion. n = number of worms. (Right) Plot of number of embryos laid by the worms 24–48 hr after dsRNA injection (mean ± SD) for the indicated targets in the N2 background. N = number of worms. (C) Single central plane images of germlines in adult worms expressing CYK-4::mCherry (shown in green) and a GFP-tagged plasma membrane probe (shown in red) following depletion of RacCED-10 or Cdc42CDC-42 (48 hr post-injection). (D) Single central plane images of germlines in adult worms expressing CYK-4::mNeonGreen (green) and an mCherry-tagged plasma membrane probe (red) following partial depletion of RhoARHO-1 or ECT-2 (20 hr post-injection). Insets (bottom) show the boxed regions magnified 1.5x. n = number of worms.
-
Figure 6—source data 1
Depletion of RhoA or CYK-4 leads to a comparable reduction in embryo production.
- https://doi.org/10.7554/eLife.36919.030
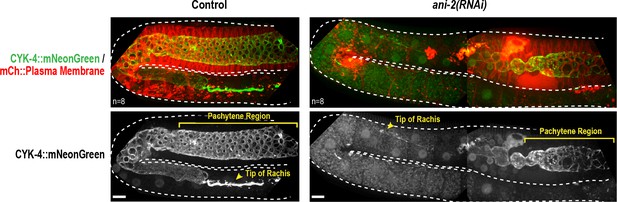
The anillin homolog, ANI-2, is not required to recruit CYK-4 recruitment to the rachis surface/bridges.
Maximum intensity projections of germlines in adult control and ani-2(RNAi) (48 hr after introduction of dsRNA by soaking) animals expressing CYK-4::mNeonGreen and an mCherry-tagged plasma membrane probe. Dotted region outlines the boundary of the germline. ANI-2 depletion leads to germline defects that increase the number of unfertilized oocytes within the germline and push the rachis tip and pachytene region backwards within the body of the worm. Thus, a larger region of ani-2(RNAi) worms was imaged and the images were stitched together to allow comparison of the ability of CYK-4 to localize to the rachis surface. n = number of worms. Scale bar is 10 µm.
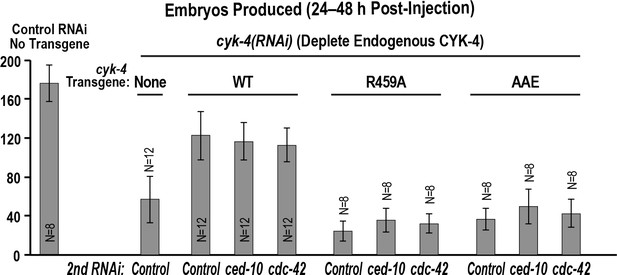
Depletion of RacCED-10 or CDC-42 cannot rescue the effects of mutants in the Rho GTPase binding interface on the germline.
Graph shows the number of embryos laid by the worms (mean ± SD) 24–48 hr post-injection for the indicated strains and RNAi conditions. N = number of worms.
-
Figure 6—figure supplement 2—source data 2
Depletion of Racor CDC-42 does not rescue the germline defects of the CYK-4 Rho GTPase binding interface mutants.
- https://doi.org/10.7554/eLife.36919.029
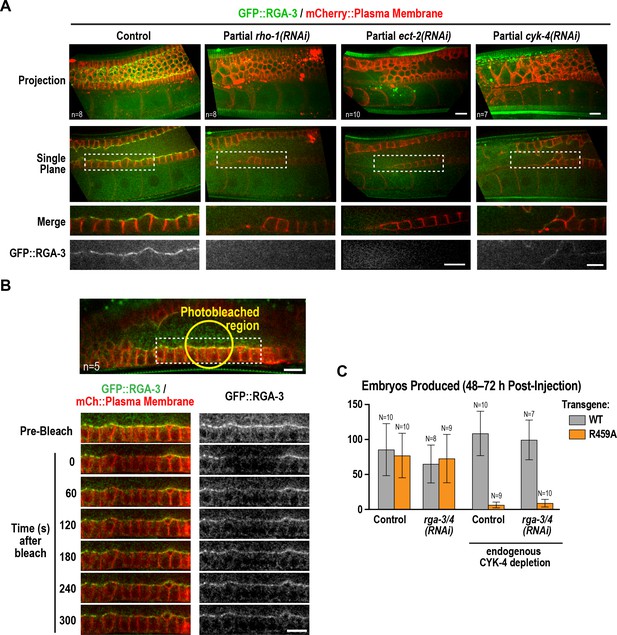
Active RhoARHO-1 localization to the rachis surface/bridges depends on ECT-2 and CYK-4.
(A) Maximum intensity projections (top) and single central plane images (middle) of the germline in adult worms expressing GFP::RGA-3 (green) and an mCherry-tagged plasma membrane probe (red) following partial depletion of RhoARHO-1, ECT-2, or CYK-4 (20 hr post-injection). Insets (bottom) show the boxed regions magnified 1.5x. n = number of worms. (B) Central plane confocal images acquired in adult worms expressing GFP::RGA-3 (green) and an mCherry-tagged plasma membrane probe (red) before and after photobleaching of the GFP::RGA-3 signal in a section of the rachis in the pachytene region. (C) Graph plotting the number of embryos laid (mean ± SD) by worms expressing wildtype or R459A CYK-4::mCherry from RNAi-resistant transgenes following depletion of endogenous CYK-4 and/or RGA-3/4 as indicated. N = number of worms. Scale bars are 10 µm.
-
Figure 7—source data 1
RGA-3/4 depletion cannot rescue the effect of the R459A mutation on embryo production.
- https://doi.org/10.7554/eLife.36919.032
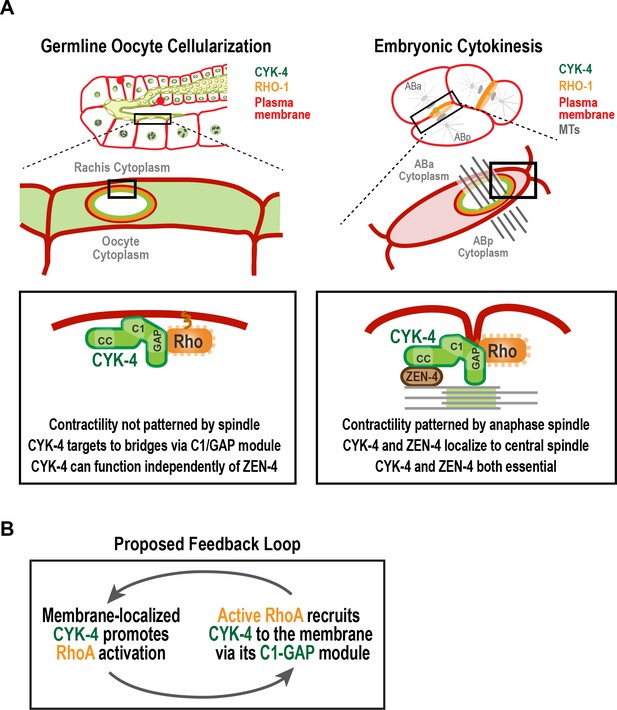
A C-terminal C1 domain-GAP module targets CYK-4 to the rachis surface/bridges to enable oocyte celluarization.
(A) Schematics compare the closure of compartment bridges during oocyte cellularization in the germline (left) to cytokinesis (shown here in a two-cell stage embryo, right). (Left) During bridge closure in the germline, the CYK-4 C1 domain and the Rho GTPase binding interface collaborate to recruit CYK-4 to the rachis surface/bridges. In the germline, bridges are not bisected by a central spindle-like microtubule bundles and the kinesin-6, ZEN-4, although present, is not essential for CYK-4 targeting or cellularization. (Right) During cytokinesis, cortical contractility is patterned by the anaphase spindle. Both ZEN-4 and CYK-4 are required to assemble the central spindle and contractile ring. Targeting to the central spindle and other ZEN-4/microtubule-dependent mechanisms may help deliver CYK-4 to the cell surface. We speculate that the C-terminal module-based mechanism that operates in the germline may also contribute to targeting CYK-4 to the plasma membrane during cytokinesis. (B) Our data suggest that CYK-4 associated with the rachis surface promotes RhoA activation. If active RhoARHO-1 generated by membrane-associated CYK-4 enables the recruitment of additional CYK-4 via interaction with its C1-GAP module, this would result in a positive feedback that promotes RhoA activation and CYK-4 recruitment.
Videos
Structure of the syncytial germline at the two-compartment stage in an L1 larva.
Video shows images of a stack of serial 100 nm serial sections of an L1 germline that were collected and imaged by transmission electron microscopy. Images are shown without and then with superposition of a pseudocolor model in which the regions within the boundary of the two-compartment germline syncytium (red) and germline nuclei (green) are highlighted.
Tables
C. elegans strains used in this study.
https://doi.org/10.7554/eLife.36919.034| Strain # | Genotype | Figure |
|---|---|---|
| N2 | wild type (ancestral) | 1S1, 1S2B, 2B, 3B, 5B, 5C, 5E, 5S1, 6B, 6S2 |
| OD95 | unc-119(ed3) III; ltIs37 [pAA64; Ppie-1::mCherry::his-58; unc-119 (+)] IV; ltIs38 [pAA1; Ppie-1::GFP::PH(PLC1delta1); unc-119(+)] III | 1S2, 2A, 2B, 3A, 3E, 3F, 3S1, 4S2, 6B |
| OD239 | cyk-4(or749ts) III; ltIs38 [pAA1; Ppie-1::GFP::PH(PLC1delta1) unc-119 (+)] III; ltIs37 [pAA64; Ppie-1::mCherry::H2B his-58; unc-119 (+)] IV | 2A, 2B, 3S1, 4B, 4S2 |
| OD241 | cyk-4(t1689ts) III; ltIs38 [pAA1; Ppie-1::GFP::PH(PLC1delta1) unc-119 (+)] III; ltIs37 [pAA64; Ppie-1::mCherry::H2B his-58; unc-119 (+)] IV | 3E, 3F, 3S1, 4B |
| OD1176 | unc-119(ed3) III; ItSi346 [pKL3; Pcyk-4::CYK-4reencoded::mCherry; cb-unc-119(+)] IV | 5D, 5E, 5S1D, 6C, 7C |
| OD1178 | unc-119(ed3) III; ItSi348 [pKL4; Pcyk-4::CYK-4reencoded(R459A)::mCherry; cb-unc-119(+)] IV | 5D, 5E, 7C |
| OD1211 | ItSi346 [pKL3; Pcyk-4::CYK-4reencoded::mCherry; cb-unc-119(+)] IV; ltIs38 [pAA1; Ppie-1::GFP::PH(PLC1delta1); unc-119 (+)] III | 6C |
| OD1364 | unc-119(ed3) III; ItSi432[pKL33; Pcyk-4::CYK-4reencoded(∆C1)::mCherry; cb-unc-119(+)] IV | 5D, 5E |
| OD1970 | ltSi835 [pKL62; Pcyk-4::CYK-4reencoded; cb-unc-119(+)]II; unc-119(ed3) III | 5B, 5S1B, 5S1C, 6S2 |
| OD1972 | ltSi837 [pKL64; Pcyk-4::CYK-4reencoded(R459A); cb-unc-119(+)] II; unc-119(ed3) III | 5B, 5S1B, 5S1C, 6S2 |
| OD1974 | ltSi839 [pKL65; Pcyk-4::CYK-4reencoded(R459A/K495A/R499E); cb-unc-119(+)] II; unc-119(ed3) III | 5B, 5S1B, 5S1C, 6S2 |
| OD1978 | ltSi843 [pKL67; Pcyk-4::CYK-4reencoded(∆C1); cb-unc-119(+)] II; unc-119(ed3) III | 5B, 5S1B, 5S1C |
| OD2064 | ltSi849[pKL120; Pmex-5::mCherry-PH::tbb-2 3'UTR; cb-unc-119(+)]I; ItSi641[pKL89; Pcyk-4::CYK-4reencoded::GFP::MEI-1 (1–224); cb-unc-119(+)]I; unc-119(ed3)III; ltIs37[pAA64; pie-1/mCherry::H2B his-58; unc-119(+)] IV | 5C |
| OD2083 | ltSi849 [pKL120; Pmex-5::mCherry::PH(PLC1delta1)::tbb-2 3'UTR; cb-unc-119(+)] I; ItSi641 [pKL89; Pcyk-4::CYK-4reencoded::GFP::MEI-1 (1–224); cb-unc-119(+)] I; ltSi835 [pKL62; Pcyk-4::CYK-4reencoded; cb-unc-119(+)]II; unc-119(ed3) III; ltIs37 [pAA64; Ppie-1::mCherry::H2B his-58; unc-119(+)] IV | 5C |
| OD2084 | ltSi849 [pKL120; Pmex-5::mCherry:: PH(PLC1delta1)::tbb-2 3'UTR; cb-unc-119(+)] I; ItSi641 [pKL89; Pcyk-4::CYK-4reencoded::GFP::MEI-1 (1–224); cb-unc-119(+)] I; ltSi837 [pKL64; Pcyk-4::CYK-4reencoded(R459A); cb-unc-119(+)] II; unc-119(ed3) III; ltIs37 [pAA64; Ppie-1::mCherry::H2B his-58; unc-119(+)] IV | 5C |
| OD2085 | ltSi849 [pKL120; Pmex-5::mCherry::PH(PLC1delta1)::tbb-2 3'UTR; cb-unc-119(+)] I; ItSi641 [pKL89; Pcyk-4::CYK-4reencoded::GFP::MEI-1 (1–224); cb-unc-119(+)] I; ltSi839 [pKL65; Pcyk-4::CYK-4reencoded(R459A/K495A/R499E); cb-unc-119(+)] II; unc-119(ed3) III; ltIs37 [pAA64; Ppie-1::mCherry::H2B his-58; unc-119(+)] IV | 5C |
| OD2087 | ltSi849 [pKL120; Pmex-5::mCherry::PH(PLC1delta1)::tbb-2 3'UTR; cb-unc-119(+)]I; ItSi641 [pKL89; Pcyk-4::CYK-4reencoded::GFP::MEI-1 (1–224); cb-unc-119(+)] I; ltSi843 [pKL67; Pcyk-4::CYK-4reencoded(∆C1); cb-unc-119(+)] II; unc-119(ed3) III; ltIs37 [pAA64; pie-1::mCherry::H2B his-58; unc-119(+)] IV | 5C |
| OD2127 | ltSi220 [pOD1249/pSW077; Pmex-5::GFP-tbb-2-operon-linker-mCherry-his-11; cb-unc-119(+)] I; ltSi849 [pKL120; Pmex-5::mCh-PH::tbb-2 3'UTR; cb-unc-119(+)] I | 3G |
| OD2286 | unc-119(ed3) III; ItSi867 [pKL142; Pcyk-4::CYK-4reencoded(R459A/K495A/R499E)::mCherry; cb-unc-119(+)] IV | 5D, 5E |
| OD3639 | ltSi849 [pKL120; Pmex-5::mCherry::PH(PLC1delta1)::tbb-2 3'UTR; cb-unc-119(+)] I; ltSi17 [pOD928/EZ-36; Prga-3::GFP::RGA-3; cb-unc-119(+)] II; unc-119(ed3) III | 7A, 7B |
| OD3640 | ltSi849 [pKL120; Pmex-5::mCherry::PH(PLC1delta1)::tbb-2 3'UTR; cb-unc-119(+)] I; unc-119(ed3) III(?); zen-4(lt30[GFP::loxP::zen-4]) IV | 1B, 3C |
| OD3686 | ltSi849 [pKL120; Pmex-5::mCherry::PH(PLC1delta1)::tbb-2 3'UTR; cb-unc-119(+)] I; ltSi1124[pKL177/pSG092; Pcyk-4::CYK-4reencoded::mNeonGreen::cyk-4 3'-UTR; cb- unc-119(+)] II; unc-119(ed3) III | 1B, 1C, 3D, 6A, 6D, 6S1 |
| JCC754 | unc-119(ed3) III?; ltIs38 [pAA1; Ppie-1::GFP::PH(PLC1delta1); unc-119 (+)] III; zen-4(or153ts)IV; ltIs37 [pAA64; Ppie-1::mCherry::his-58; unc-119 (+)] IV | 3E, 3F, 3S1 |
Oligos used for dsRNA production.
https://doi.org/10.7554/eLife.36919.035| Gene | Oligonucleotide 1 (5’→3’) | Oligonucleotide 2 (5→3’) | Template | Concentration (mg/ml) |
|---|---|---|---|---|
| cyk-4 (K08E3.6) | CGTAATACGACTCACTATAGGTGTCA AAGACACTCAGAAAC | CGTAATACGACTCACTATAGGCCTC TTCGAATTGGCAGCAGC | N2 cDNA | 2.0 |
| zen-4 (M03D4.1) | AATTAACCCTCACTAAAGGAATTGGT TATGGCTCCGAGA | TAATACGACTCACTATAGGATTGGA GCTGTTGGATGAGC | N2 cDNA | 1.3 |
| ect-2 (T19E10.1) | TAATACGACTCACTATAGGTCTCCGA TAAATCTGTGGGG | AATTAACCCTCACTAAAGGCAGCAG TTTGCGAAAATGAA | N2 genomic DNA | 2.0 |
| spd-1 (Y34D9A.4) | TAATACGACTCACTATAGGTCGTTGA CGCGTACTCAACT | AATTAACCCTCACTAAAGGGAATTC GAAATCCGACTCCA | N2 cDNA | 1.8 |
| rga-3/4 (K09H11.3/Y75B7AL.4) | TAATACGACTCACTATAGGCCTTCCT GAGCACGACTTTC | AATTAACCCTCACTAAAGGAGCTTT CGCGACCTTAAACA | N2 genomic DNA | 2.6 |
| rho-1 (Y51H4A.3) | AATTAACCCTCACTAAAGGATCGTC TGCGTCCACTCTCT | TAATACGACTCACTATAGGCTCGGC TGAAATTTCCAAAA | N2 genomic DNA | 1.9 |
| ced-10 (C09G12.8) | AATTAACCCTCACTAAAGGATCGCC TCATCGA AAACTTG | TAATACGACTCACTAT AGGTCAAAT GTGTCGT CGTTGGT | N2 cDNA | 2.0 |
| cdc-42 (R07G3.1) | AATTAACCCTCACTAAAGGGTTTGG CATTTTTCAGGGAA | TAATACGACTCACTATAGGACGTGT GCGTGCACATTTAT | N2 genomic DNA | 2.0 |
| hyls-1 (C05C8.9) | AATTAACCCTCACTAAAGGTGGCA AATTTTACCACTGAAA | TAATACGACTCACTATAGGTGATATC TTGTGACCGGATCA | N2 cDNA | 2.0 |
| gfp | AATTAACCCTCACTAAAGGCCAA CACTTGTCACTACTTTCTGTTATGG | TAATACGACTCACTATAGGGTATAGT TCATCCATGCCATGTGTAATCCC | Plasmid | 2.0 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.36919.036





