Genetic predisposition to uterine leiomyoma is determined by loci for genitourinary development and genome stability
Figures
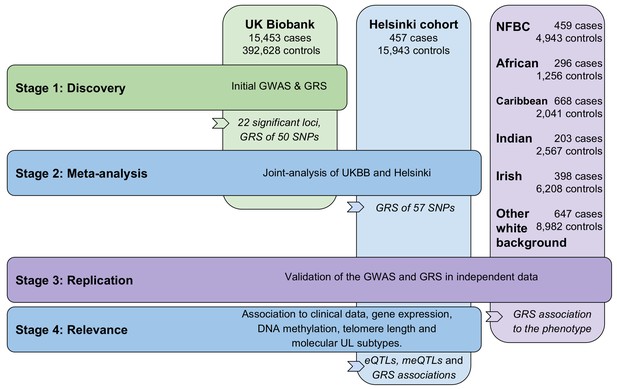
Outline of the study stages and genotyping cohorts.
GRS, genomic risk score. NFBC, Northern Finland Birth Cohort.
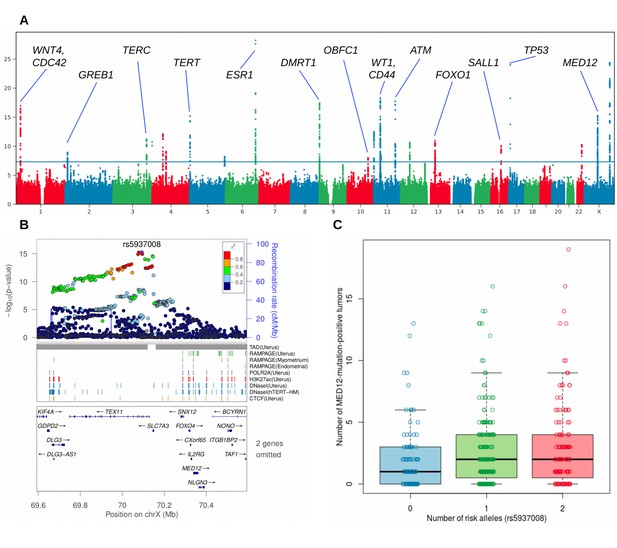
Overview of the uterine leiomyoma risk loci and the effect of increased number of MED12-mutated lesions per rs5937008 risk allele.
(A), Manhattan plot of the UK Biobank GWAS on 15,453 UL cases and 392,628 controls. On Y-axis, logarithm transformed association values, and on X-axis, autosomes and the X chromosome. The blue horizontal line denotes genome-wide significance (p=5 × 10−8). Gene symbols shown for reference. (B), MED12 region in more detail. ENCODE tracks (details in Supplementary Methods) are shown for reference. (C), The risk allele near MED12 (rs5937008) is observed with a significant increase in number of MED12-mutation-positive tumors (p=0.009; negative binomial regression; RR = 1.23 per risk allele; n = 457 Helsinki cohort patients).
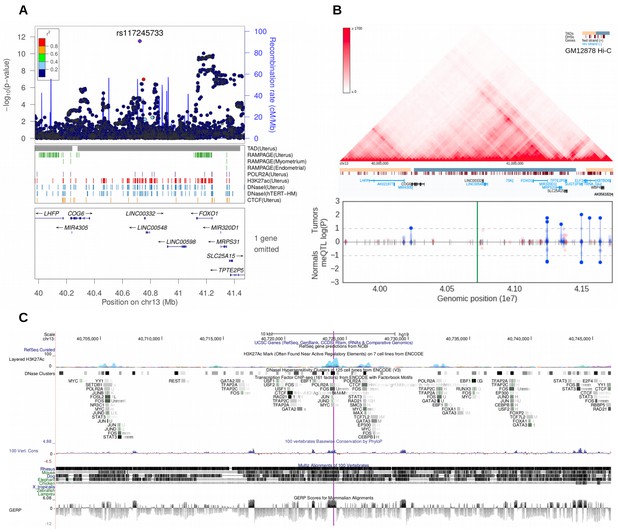
Meta-analysis of UL risk revealed rs117245733 at a gene poor region of 13q14.11.
(A) meta-analysis P-values and the genomic context at the locus. Gene symbols and ENCODE tracks (details in Supplementary Methods) are shown for reference; coordinates follow hg19. (B) Hi-C, TADs and CpG methylation around the locus with a 1 Mb flank. The needle plot shows the meQTL associations (dashed lines at 10% FDR; green line denotes the SNP; gray ticks denote all CpGs tested; blue needle for positive coefficient, red for negative coefficient) for tumors (above x-axis; nAA = 53, nAB = 3) and normals (below x-axis; nAA = 33, nAB = 2). (C) UCSC genome browser tracks related to conservation and regulation at the locus.
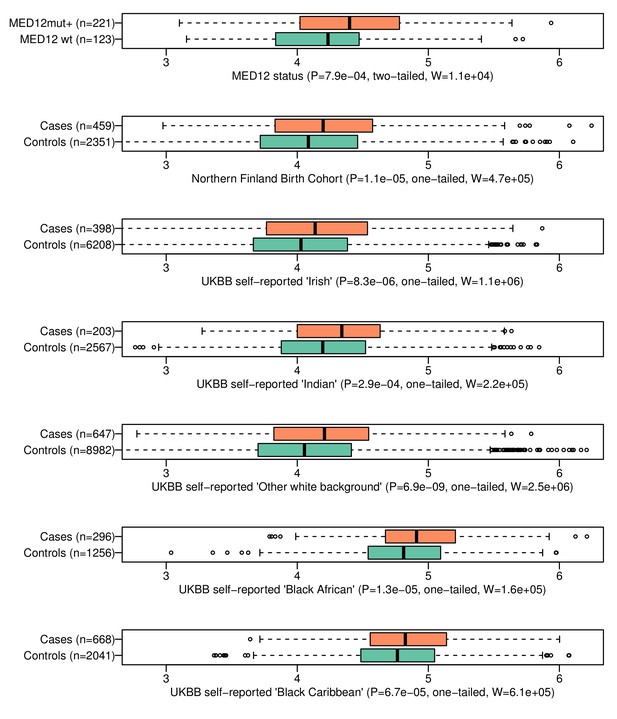
The genomic risk score is elevated in patients with MED12-mutated lesions and in respect to the UL phenotype in the six follow-up cohorts.
On top, GRS association to MED12 mutation status. The rest show GRS association to the UL phenotype in six independent replication cohorts. Associations (P) and test statistics (W) are from Wilcoxon rank-sum tests. Only females were included as the control samples. The X-axes show the GRS distributions for each phenotype.
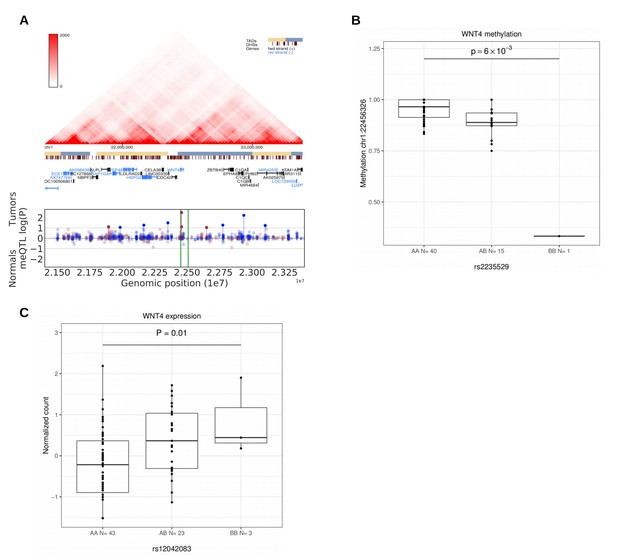
Methylation and expression differences in WNT4.
(A), Hi-C, TADs and CpG methylation around the locus with an 1 Mb flank. The needle plot shows the meQTL associations (dashed lines at 10% FDR; green lines denote the two SNPs, rs2235529 and rs2092315; gray ticks denote all CpGs tested; blue needle for positive coefficient, red for negative coefficient) for tumors (above x-axis; nAA = 40, nAB = 15, nBB = 1 for rs2235529 and nAA = 32, nAB = 23 for rs2092315) and normals (below x-axis; nAA = 23, nAB = 12 and nAA = 17, nAB = 17, nBB = 1). (B), Methylation differences in tumors (n = 56) at CpG chr1:22456326 by SNP rs2235529. (C), WNT4 expression differences in tumors (n = 41) stratified by the rs12042083 genotype. B is the risk allele, and the P-value is corrected for local multiple testing (permutation based test).
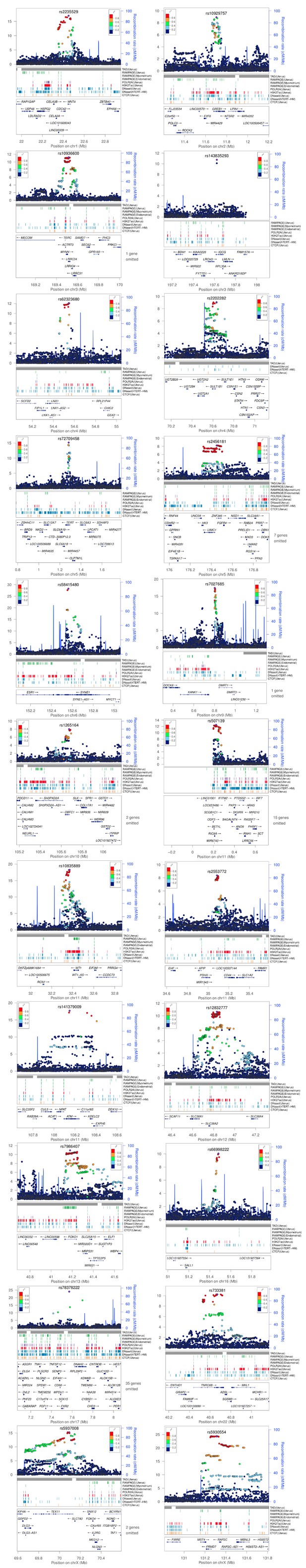
Structure of UL predisposition loci.
The lead SNPs from stage one and their genomic context. The LD estimates (r2) were taken from UK10k ALSPAC. Associations are based on the UKBB cohort. Gene symbols and ENCODE tracks (details in Supplementary Methods) are shown for reference; coordinates follow hg19. See main text Table 1 for more information. In total 22 figures, ordered by genomic position.
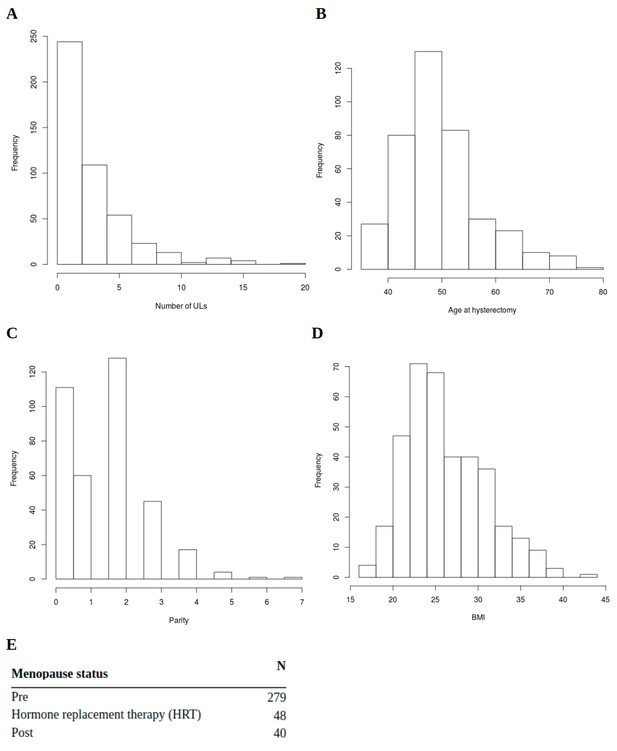
Clinical background information for the Helsinki cohort patients.
(A,) the number of ULs per patient (n = 457). (B), patient’s age at hysterectomy (n = 392). (C), parity (n = 367). (D), body mass index (BMI; n = 366). (E), menopause status (n = 367).
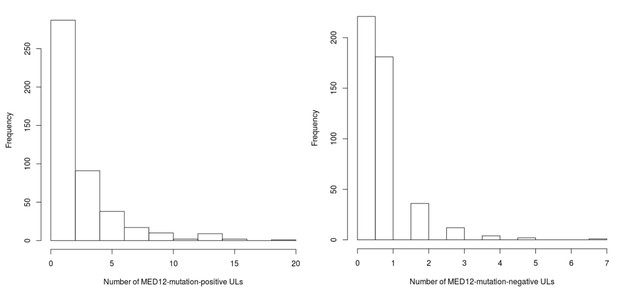
MED12 mutation status distributions for the Helsinki cohort patients.
On left, mutation-positive tumors per patient (n = 457), and on right, mutation-negative tumors per patient (n = 457).
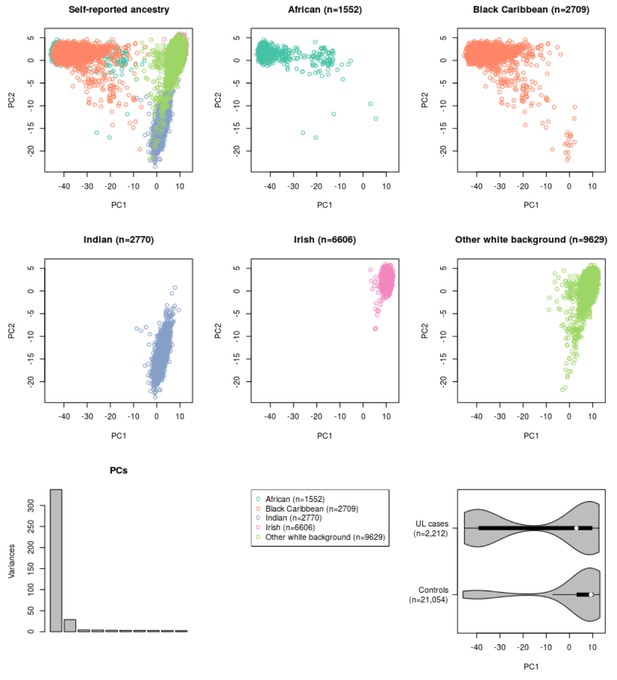
Genetic separation between the self-reported ancestries in UK Biobank.
Principal components (PC) analysis of ancestry informative markers. In total 1396 autosomal, ancestry informative SNPs were used and the resulting first two PCs are shown: top-left plot shows all 23,266 samples colored according to their self-reported ancestry. The subsequent plots show the same data divided by the self-reported ancestry. Bottom-left plot displays the variance explained by the first ten PCs. Bottom-right violin plot displays the distribution of the first principal component for cases (n = 2,212) and controls (n = 21,054). The phenotype is more prevalent in individuals with increased African ancestry (p<2.2 × 10−16; Wilcoxon rank sum W = 3 × 107).
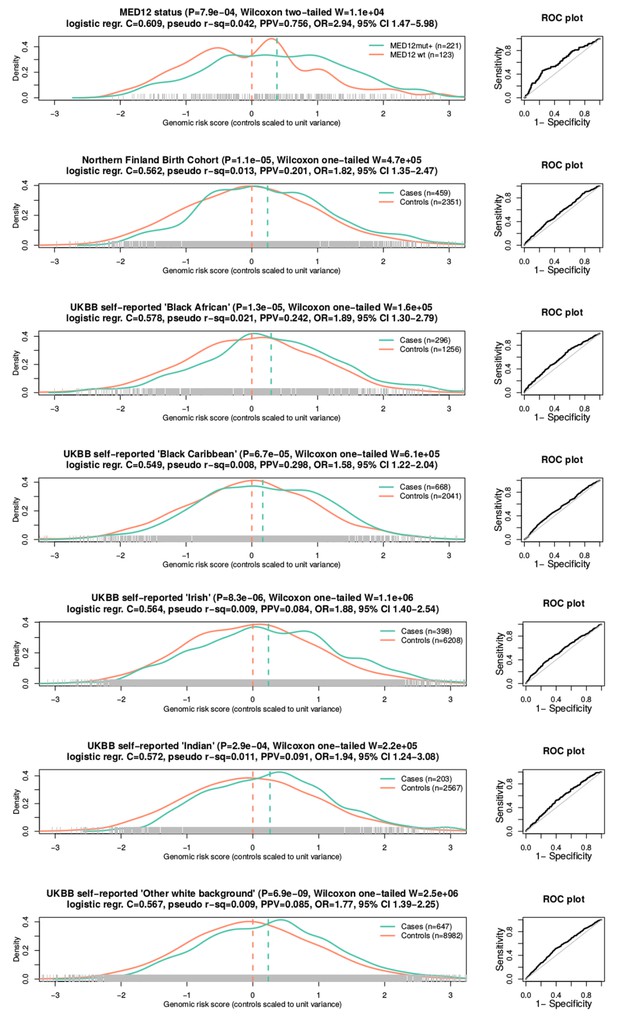
Association between GRS and phenotype.
One somatic phenotype (MED12 mutation status) and six independent case-control replication cohorts. On left, density plots (bandwidth = 0.2) for each phenotype. X-axis gives GRS values scaled to unit variance with respect to the controls. Dashed lines denote the mean GRS for cases and controls. Associations (P) and test statistics (W) are from Wilcoxon rank-sum tests. C-scores, Nagelkerke’s R2 (pseudo R2) and Receiver operating characteristic (ROC; on right) are from a logistic regression model. Positive predictive values (PPV) were computed from the highest GRS quartile, and odds ratios (OR and 95% CI) from the top and bottom GRS quartiles.
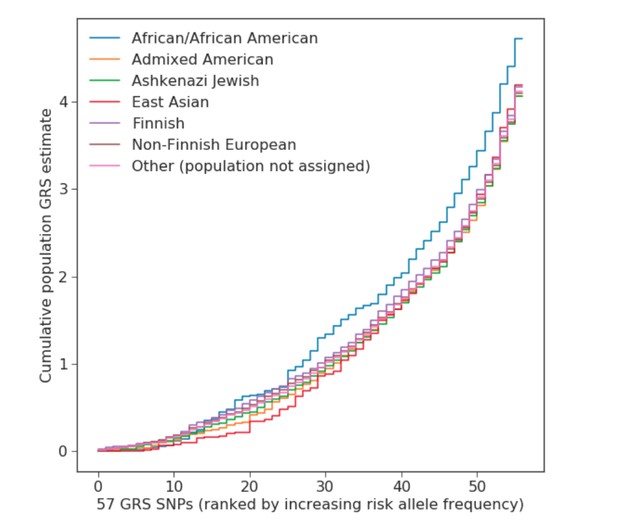
Genomic risk scores in gnomAD populations.
Population-specific genomic risk scores (GRS) as seen based on gnomAD allele frequencies. Y-axis shows the cumulative effect of each of the 57 GRS SNPs; the X-axis is ordered by risk allele frequency. The population names displayed here follow the naming convention of the gnomAD database.
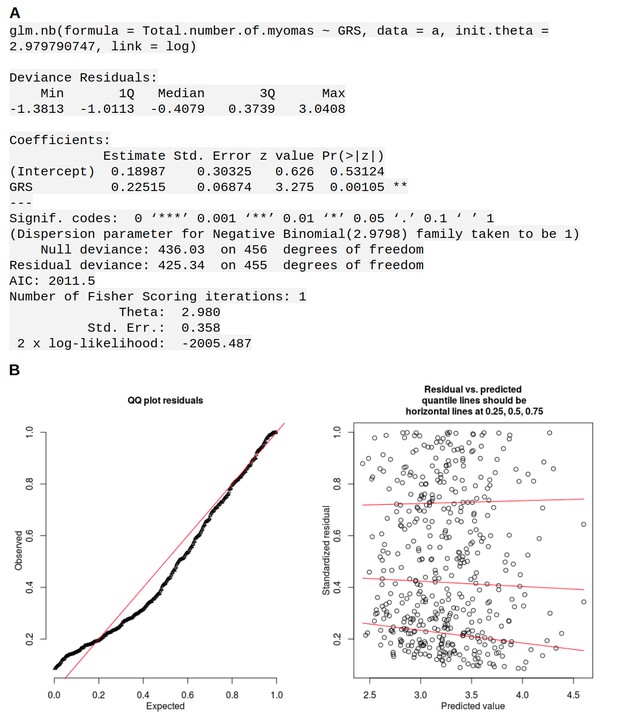
Association between GRS and number of ULs per patient.
The numbers are based on the Helsinki cohort data of 457 patients with all distinct tumors of ≥1 cm diameter harvested at hysterectomy; details on sample collection are given in Supplementary Methods. A, summary of the model. B, diagnostic plots for the model.
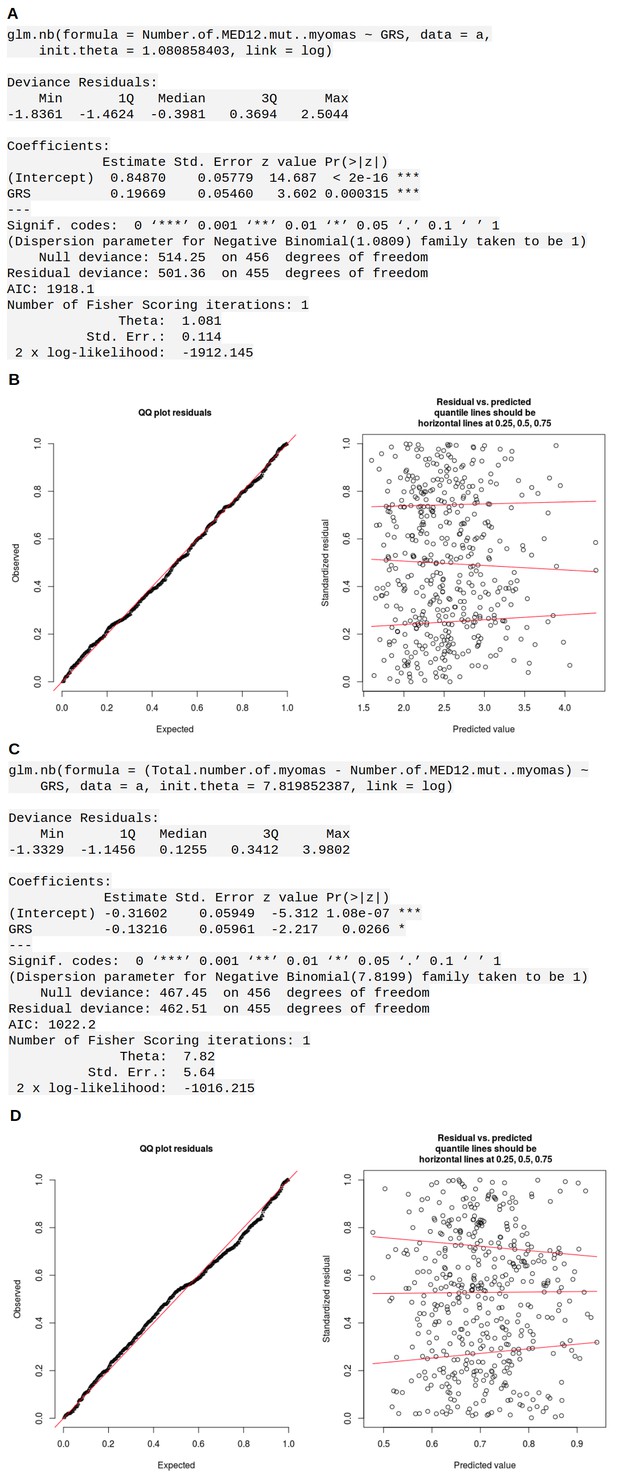
Association between GRS and MED12 mutations.
The numbers are based on the Helsinki cohort data (457 patients). (A) summary of the negative binomial model for MED12-mutation-positive tumor counts. (B) diagnostic plots for the negative binomial model. (C, D) similar model for the MED12-mutation-negative tumor counts.
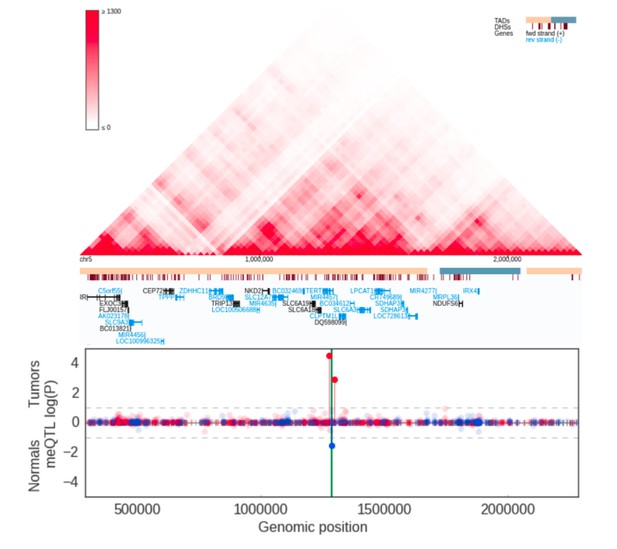
meQTLs at TERT region.
Hi-C, TADs and CpG methylation around rs2736100 with an 1Mbp flank. The needle plot shows the meQTL associations (dashed lines at 10% FDR; green line denotes the two SNPs; gray ticks denote all CpGs tested; blue needle for positive coefficient, red for negative coefficient) for tumors (above x-axis; nAA = 8, nAB = 32, nBB = 16) and normals (below x-axis; nAA = 5, nAB = 22, nBB = 7). The only significant meQTLs were seen around TERT.
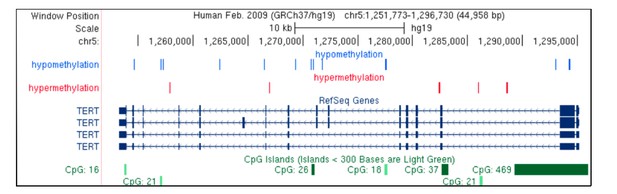
Overview of the CpG methylation around TERT in tumor tissue.
There are several CpGs in TERT whose methylation level is associated with rs2736100 genotype (nominal p<0.05). Some of them are also detected in normal tissue (Appendix 1—Table 9). Hypomethylation refers to decreased methylation in BB vs. AA genotype and hypermethylation the opposite.
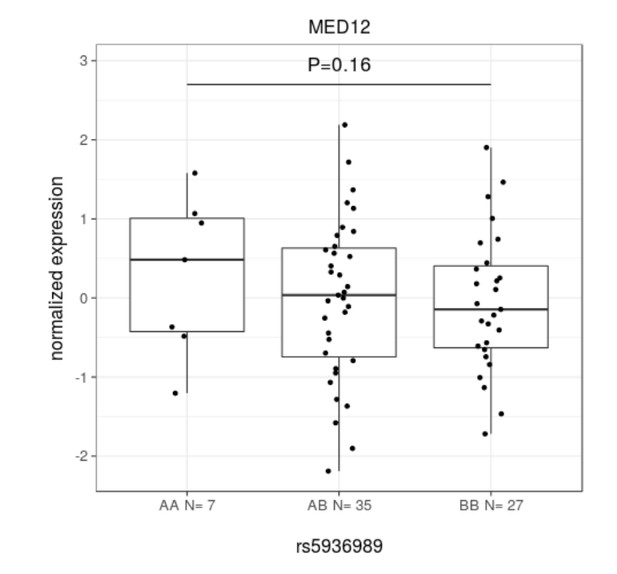
MED12 expression.
Linear model. Here, A is the risk allele. Beta 0.247 ± 0.177.
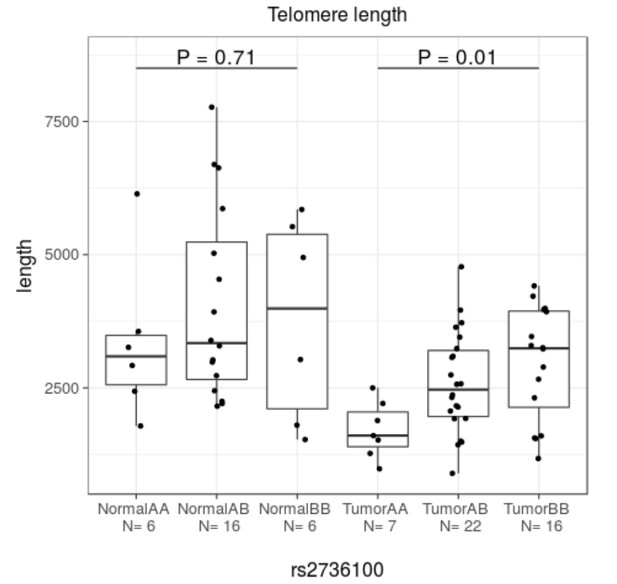
Telomere lengths in tumors and normals.
Here A denotes the risk allele (see Appendix 1—Table 1 for the allele information). In tumors, shorter telomere length is significantly associated with the risk allele at 5p15.33 (rs2736100) (Kruskal-Wallis test).
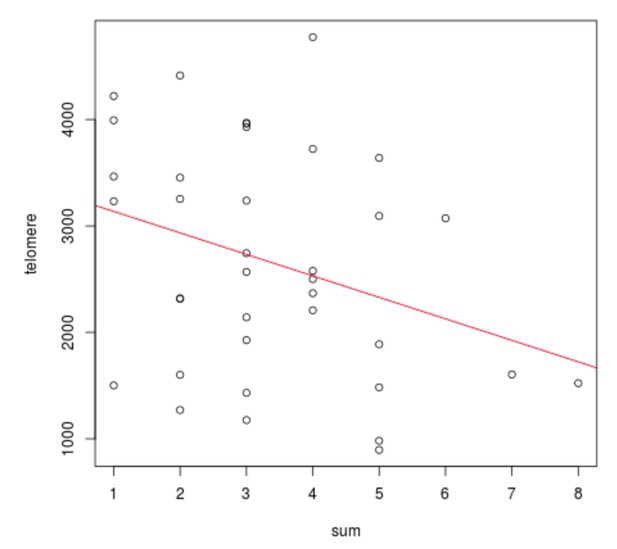
The association between risk allele count and telomere length On X-axis, total number of risk alleles at TERT (rs72709458, rs2736100, rs2853676), TERC (rs10936600) and OBFC1 (rs1265164).
On Y-axis, estimated telomere length. Linear regression model p=0.055; 95% CI −408.511 – 4.704 per one risk allele.
Tables
Predisposition loci for uterine leiomyoma.
https://doi.org/10.7554/eLife.37110.005| Chr | Position | rs-code | A | B | B freq | OR | P | Likely disease gene |
|---|---|---|---|---|---|---|---|---|
| 6 | 152,562,271 | rs58415480 | C | G | 0.155 | 1.18 | 6.0E-29 | ESR1 |
| X | 131,312,089 | rs5930554 | T | C | 0.311 | 1.14 | 4.3E-25 | ? |
| 17 | 7,571,752 | rs78378222 # | T | G | 0.013 | 1.53 | 9.7E-25 | TP53 |
| 11 | 32,370,380 | rs10835889 | G | A | 0.159 | 1.14 | 5.5E-19 | WT1 |
| 11 | 108,149,207 | rs141379009 | T | G | 0.027 | 1.32 | 2.0E-18 | ATM |
| 9 | 802,228 | rs7027685 | A | T | 0.402 | 1.11 | 3.8E-18 | DMRT1 |
| 1 | 22,450,487 | rs2235529 # | C | T | 0.157 | 1.14 | 1.1E-17 | WNT4/CDC42 |
| X | 70,093,038 | rs5937008 | C | T | 0.520 | 0.91 | 5.6E-16 | MED12 |
| 5 | 1,283,755 | rs72709458 # | C | T | 0.206 | 1.12 | 6.9E-16 | TERT |
| 11 | 225,196 | rs507139 * | G | A | 0.074 | 0.84 | 3.2E-13 | ? |
| 4 | 54,546,192 | rs62323680 | G | A | 0.067 | 1.16 | 8.3E-13 | ? |
| 3 | 169,514,585 | rs10936600 # | A | T | 0.244 | 0.91 | 6.4E-12 | TERC |
| 13 | 41,179,798 | rs7986407 | A | G | 0.310 | 1.09 | 1.2E-11 | FOXO1 |
| 3 | 197,623,337 | rs143835293 | A | G | 0.002 | 1.75 | 1.8E-11 | ? |
| 12 | 46,831,129 | rs12832777 | T | C | 0.701 | 1.09 | 2.3E-11 | ? |
| 22 | 40,669,648 | rs733381 * | A | G | 0.213 | 1.10 | 5.7E-11 | ? |
| 16 | 51,481,596 | rs66998222 | G | A | 0.201 | 0.91 | 8.9E-11 | SALL1 |
| 4 | 70,634,441 | rs2202282 | C | T | 0.497 | 1.07 | 8.7E-10 | ? |
| 2 | 11,702,661 | rs10929757 | A | C | 0.579 | 1.08 | 1.2E-09 | GREB1 |
| 11 | 35,085,453 | rs2553772 | T | G | 0.538 | 1.07 | 4.4E-09 | CD44 |
| 5 | 176,450,837 | rs2456181 | C | G | 0.484 | 1.07 | 6.3E-09 | ? |
| 10 | 105,674,854 | rs1265164 | A | G | 0.869 | 0.91 | 1.0E-08 | OBFC1 |
-
The numbers for B allele frequency (B Freq), odds-ratio (OR, where B is the effect allele) and association (P) are based on the UKBB cohort (15,453 UL cases). Gene symbols are shown for reference. The genomic coordinates follow hg19 and dbSNP build 147. All genome-wide significant (p<5 × 10−8) loci and their highest-association SNP are shown.
* Previously implicated predisposition to ULs.
-
# Previous associations to endometriosis, lung adenocarcinoma, glioma or telomere length; see literature in Appendix 1—table 11
Summary of the UKBB cohort individuals.
The UKBB cohort was split into six disjoint, self-reported ancestries. For each ancestry, we summarized the background information for the phenotype (number of UL cases and female controls), proportion of cases (%), age at first assessment visit (mean and SD), number of live births (mean and SD), body mass index (BMI; mean and SD), proportion of hysterectomy cases (%) and age at hysterectomy (mean and SD).
| Age at first assessment visit | Number of live births | BMI | Age at hysterectomy | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Self-reported ancestry | Phenotype | N | Cases (%) | Mean | SD | Mean | SD | Mean | SD | Ever had hyst. (%) | Mean | SD |
| White British | cases | 15453 | 7.0 | 56.9 | 7.5 | 1.7 | 1.2 | 27.7 | 5.2 | 31.9 | 46.1 | 9.0 |
| controls | 205157 | 56.7 | 7.9 | 1.8 | 1.2 | 27.0 | 5.1 | 7.0 | 42.2 | 10.3 | ||
| Black African | cases | 296 | 19.1 | 49.6 | 6.8 | 2.0 | 1.7 | 31.2 | 5.6 | 22.1 | 45.1 | 8.8 |
| controls | 1256 | 51.8 | 8.1 | 2.8 | 1.8 | 31.3 | 5.7 | 6.5 | 36.4 | 15.4 | ||
| Black | cases | 668 | 24.7 | 50.6 | 6.8 | 1.5 | 1.4 | 30.0 | 6.3 | 24.0 | 41.2 | 12.1 |
| Caribbean | controls | 2041 | 53.0 | 8.2 | 2.2 | 1.7 | 29.8 | 5.9 | 9.7 | 37.2 | 14.8 | |
| White Irish | cases | 398 | 6.0 | 56.5 | 7.6 | 1.8 | 1.4 | 27.8 | 5.2 | 32.6 | 46.6 | 7.2 |
| controls | 6208 | 56.4 | 8.1 | 1.9 | 1.4 | 26.9 | 5.0 | 7.5 | 42.9 | 10.8 | ||
| Asian Indian | cases | 203 | 7.3 | 53.8 | 7.9 | 2.0 | 1.1 | 27.6 | 4.2 | 33.5 | 44.1 | 11.9 |
| controls | 2567 | 53.7 | 8.0 | 2.0 | 1.2 | 27.1 | 4.8 | 6.6 | 39.8 | 13.9 | ||
| Other white | cases | 647 | 6.7 | 54.8 | 7.9 | 1.3 | 1.2 | 26.7 | 5.3 | 23.6 | 46.5 | 8.6 |
| background | controls | 8982 | 54.6 | 8.3 | 1.6 | 1.2 | 26.4 | 5.2 | 5.4 | 42.1 | 11.0 | |
Discovery stage GWAS for the UKBB cohort.
The information for B allele frequency (B Freq), odds-ratio (OR) and association (Beta; standard error of Beta; χ2 and P) were collected from the UKBB cohort. All genome-wide significant, LD-independent (r2 ≤0.3 pruned in order of UKBB association) SNPs that passed imputation QC are shown. SNPs with r2 = NA are the lead-SNPs of each distinct locus. The A and B alleles are on GRCh37 forward strand. OR was computed from SNP dosages and B as the effect allele. Rows are sorted by genomic position.
| rs-code | Chr | Position | A | B | r2 (reference SNP) | B freq | OR | Beta | SE | χ2 | P |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs2235529 | 1 | 22450487 | C | T | NA | 0.157 | 1.14 | −0.005 | 5.81E-04 | 73.373 | 1.1E-17 |
| rs2092315 | 1 | 22507684 | C | T | 0.14 (rs2235529) | 0.248 | 1.07 | −0.003 | 4.90E-04 | 30.465 | 3.4E-08 |
| rs10929757 | 2 | 11702661 | A | C | NA | 0.579 | 1.08 | −0.003 | 4.32E-04 | 36.947 | 1.2E-09 |
| rs11674184 | 2 | 11721535 | T | G | 0.30 (rs10929757) | 0.386 | 0.94 | 0.002 | 4.36E-04 | 30.082 | 4.1E-08 |
| rs10936600 | 3 | 169514585 | A | T | NA | 0.244 | 0.91 | 0.003 | 4.91E-04 | 47.191 | 6.4E-12 |
| rs143835293 | 3 | 197623337 | A | G | NA | 0.002 | 1.75 | −0.043 | 6.33E-03 | 45.143 | 1.8E-11 |
| rs62323680 | 4 | 54546192 | G | A | NA | 0.067 | 1.16 | −0.006 | 8.49E-04 | 51.212 | 8.3E-13 |
| rs2202282 | 4 | 70634441 | C | T | NA | 0.497 | 1.07 | −0.003 | 4.22E-04 | 37.593 | 8.7E-10 |
| rs72709458 | 5 | 1283755 | C | T | NA | 0.206 | 1.12 | −0.004 | 5.28E-04 | 65.165 | 6.9E-16 |
| rs2736100 | 5 | 1286516 | C | A | 0.23 (rs72709458) | 0.498 | 0.91 | 0.003 | 4.23E-04 | 60.879 | 6.1E-15 |
| rs2853676 | 5 | 1288547 | T | C | 0.23 (rs2736100) | 0.731 | 0.91 | 0.003 | 4.77E-04 | 49.761 | 1.7E-12 |
| rs2456181 | 5 | 176450837 | C | G | NA | 0.484 | 1.07 | −0.002 | 4.24E-04 | 33.743 | 6.3E-09 |
| rs4870084 | 6 | 152543949 | C | T | 0.29 (rs6904757) | 0.189 | 0.92 | 0.003 | 5.44E-04 | 32.941 | 9.5E-09 |
| rs6928363 | 6 | 152546094 | G | A | 0.17 (rs58415480) | 0.485 | 0.92 | 0.003 | 4.24E-04 | 46.849 | 7.7E-12 |
| rs58415480 | 6 | 152562271 | C | G | NA | 0.155 | 1.18 | −0.007 | 5.89E-04 | 124.672 | 6.0E-29 |
| rs75510204 | 6 | 152592680 | T | G | 0.07 (rs58415480) | 0.012 | 1.29 | −0.012 | 2.07E-03 | 32.664 | 1.1E-08 |
| rs6904757 | 6 | 152593102 | A | G | 0.08 (rs6928363) | 0.363 | 0.93 | 0.003 | 4.42E-04 | 41.035 | 1.5E-10 |
| rs144444583 | 6 | 152684585 | T | C | 0.30 (rs58415480) | 0.128 | 1.12 | −0.004 | 6.37E-04 | 45.286 | 1.7E-11 |
| rs138821078 | 9 | 674217 | C | G | 0.13 (rs10975820) | 0.021 | 1.23 | −0.008 | 1.50E-03 | 31.819 | 1.7E-08 |
| rs10975820 | 9 | 684160 | G | A | 0.00 (rs7027685) | 0.142 | 1.12 | −0.004 | 6.08E-04 | 48.602 | 3.1E-12 |
| rs7027685 | 9 | 802228 | A | T | NA | 0.402 | 1.11 | −0.004 | 4.33E-04 | 75.424 | 3.8E-18 |
| rs114680331 | 9 | 815682 | T | C | 0.12 (rs7027685) | 0.100 | 1.11 | −0.004 | 7.21E-04 | 35.577 | 2.5E-09 |
| rs4742448 | 9 | 826585 | C | G | 0.28 (rs7027685) | 0.458 | 1.07 | −0.003 | 4.33E-04 | 33.714 | 6.4E-09 |
| rs2277163 | 9 | 827224 | A | G | 0.07 (rs7027685) | 0.947 | 0.87 | 0.005 | 9.51E-04 | 33.211 | 8.3E-09 |
| rs1265164 | 10 | 105674854 | A | G | NA | 0.869 | 0.91 | 0.004 | 6.27E-04 | 32.772 | 1.0E-08 |
| rs11246003 | 11 | 213723 | T | G | 0.00 (rs507139) | 0.044 | 0.85 | 0.006 | 1.03E-03 | 32.337 | 1.3E-08 |
| rs507139 | 11 | 225196 | G | A | NA | 0.074 | 0.84 | 0.006 | 8.11E-04 | 53.082 | 3.2E-13 |
| rs2207548 | 11 | 32368744 | C | A | 0.24 (rs10835889) | 0.423 | 1.09 | −0.003 | 4.30E-04 | 62.360 | 2.9E-15 |
| rs10835889 | 11 | 32370380 | G | A | NA | 0.159 | 1.14 | −0.005 | 5.81E-04 | 79.234 | 5.5E-19 |
| rs7120483 | 11 | 32406983 | G | C | 0.30 (rs10835889) | 0.120 | 1.12 | −0.004 | 6.51E-04 | 44.481 | 2.6E-11 |
| rs11031783 | 11 | 32459923 | C | A | 0.28 (rs10835889) | 0.204 | 1.09 | −0.003 | 5.25E-04 | 36.687 | 1.4E-09 |
| rs2553772 | 11 | 35085453 | T | G | NA | 0.538 | 1.07 | −0.002 | 4.24E-04 | 34.458 | 4.4E-09 |
| rs59021565 | 11 | 107999907 | C | G | 0.27 (rs141379009) | 0.091 | 1.11 | −0.004 | 7.38E-04 | 30.882 | 2.7E-08 |
| rs141379009 | 11 | 108149207 | T | G | NA | 0.027 | 1.32 | −0.011 | 1.30E-03 | 76.719 | 2.0E-18 |
| rs4988023 | 11 | 108168995 | A | C | 0.00 (rs141379009) | 0.144 | 0.89 | 0.004 | 6.01E-04 | 43.699 | 3.8E-11 |
| rs12223381 | 11 | 108354102 | C | T | 0.12 (rs59021565) | 0.406 | 1.07 | −0.002 | 4.31E-04 | 30.234 | 3.8E-08 |
| rs9669403 | 12 | 46798900 | G | A | 0.28 (rs12832777) | 0.402 | 1.07 | −0.003 | 4.35E-04 | 36.998 | 1.2E-09 |
| rs12832777 | 12 | 46831129 | T | C | NA | 0.701 | 1.09 | −0.003 | 4.61E-04 | 44.734 | 2.3E-11 |
| rs117245733 | 13 | 40723944 | G | A | 0.00 (rs7986407) | 0.016 | 1.26 | −0.010 | 1.74E-03 | 34.519 | 4.2E-09 |
| rs7986407 | 13 | 41179798 | A | G | NA | 0.310 | 1.09 | −0.003 | 4.56E-04 | 45.943 | 1.2E-11 |
| rs66998222 | 16 | 51481596 | G | A | NA | 0.201 | 0.91 | 0.003 | 5.28E-04 | 42.053 | 8.9E-11 |
| rs78378222 | 17 | 7571752 | T | G | NA | 0.013 | 1.53 | −0.020 | 1.93E-03 | 105.457 | 9.7E-25 |
| rs733381 | 22 | 40669648 | A | G | NA | 0.213 | 1.10 | −0.003 | 5.16E-04 | 42.919 | 5.7E-11 |
| rs5936989 | X | 70022420 | T | A | 0.27 (rs5937008) | 0.782 | 0.92 | 0.005 | 9.30E-04 | 32.606 | 1.1E-08 |
| rs5937008 | X | 70093038 | C | T | NA | 0.520 | 0.91 | 0.006 | 7.68E-04 | 65.570 | 5.6E-16 |
| rs7059898 | X | 70149078 | C | A | 0.27 (rs5937008) | 0.359 | 1.07 | −0.005 | 8.11E-04 | 31.756 | 1.7E-08 |
| rs7888560 | X | 131171122 | A | G | 0.19 (rs5930554) | 0.228 | 1.08 | −0.005 | 9.16E-04 | 31.830 | 1.7E-08 |
| rs5930554 | X | 131312089 | T | C | NA | 0.311 | 1.14 | −0.009 | 8.29E-04 | 107.076 | 4.3E-25 |
| rs5933158 | X | 131578034 | A | G | 0.21 (rs5930554) | 0.586 | 1.08 | −0.005 | 7.98E-04 | 38.833 | 4.6E-10 |
| rs5975338 | X | 131626317 | A | G | 0.28 (rs5930554) | 0.129 | 1.10 | −0.006 | 1.14E-03 | 32.201 | 1.4E-08 |
Meta-analysis of the UKBB and Helsinki cohorts.
All the genome-wide significant, LD-independent (r2 ≤0.3) associations from the meta-analysis stage. Seven SNPs were LD-independent when compared to the discovery stage SNPs, and rs117245733 is shown for reference. The numbers for regression coefficients (Beta), standard error of beta (SE) and association (P) were collected from the UKBB and Helsinki summary statistics and their fixed effect model meta-analysis. The bolded SNP was the only one to reach a suggestive association (p<10−5) in both cohorts.
| UKBB cohort | Helsinki cohort | Meta-analysis (fixed eff.) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs-code | Chr | Position | A | B | r2 (reference SNP) | Beta | SE | P | Beta | SE | P | Beta | SE | P |
| rs17631680 | 2 | 67090367 | T | C | NA | 0.004 | 0.0007 | 2.1E-07 | 0.005 | 0.0029 | 6.2E-02 | 0.004 | 0.0007 | 4.29E-08 |
| rs1735537 | 3 | 128122820 | T | C | NA | −0.003 | 0.0005 | 1.2E-07 | −0.004 | 0.0021 | 8.7E-02 | −0.003 | 0.0005 | 3.01E-08 |
| rs67751869 | 4 | 54568834 | T | C | 0.14 (rs62323680) | −0.005 | 0.0009 | 9.9E-08 | −0.011 | 0.0039 | 3.7E-03 | −0.005 | 0.0009 | 5.05E-09 |
| rs6901631 | 6 | 152567047 | T | C | 0.26 (rs6904757) | 0.003 | 0.0006 | 5.4E-08 | 0.005 | 0.0031 | 1.2E-01 | 0.003 | 0.0006 | 1.76E-08 |
| rs11790408 | 9 | 876418 | G | T | 0.10 (rs4742448) | 0.002 | 0.0004 | 6.5E-08 | 0.002 | 0.0019 | 3.9E-01 | 0.002 | 0.0004 | 4.89E-08 |
| rs117245733 | 13 | 40723944 | G | A | 0.00 (rs7986407) | −0.010 | 0.0017 | 4.2E-09 | −0.024 | 0.0053 | 8.1E-06 | −0.012 | 0.0017 | 3.18E-12 |
| rs10415391 | 19 | 22652436 | C | T | NA | −0.004 | 0.0007 | 2.8E-07 | −0.006 | 0.0028 | 2.0E-02 | −0.004 | 0.0007 | 2.87E-08 |
| rs62132801 | 19 | 49267882 | A | T | NA | 0.004 | 0.0007 | 3.3E-07 | 0.006 | 0.0029 | 2.8E-02 | 0.004 | 0.0007 | 4.20E-08 |
Genomic risk score.
Summary of the GRS and its weights based on the discovery and meta-analysis stages. Dosage-based odds-ratios (OR) and log-odds were collected from the UKBB summary statistics. The A and B alleles are on GRCh37 forward strand, and the B allele is the effect allele. In total 50 SNPs from stage 1 and seven SNPs from stage 2.
| rs-code | Chr | Position | A | B | OR | Log-odds | Stage * |
|---|---|---|---|---|---|---|---|
| rs2235529 | 1 | 22450487 | C | T | 1.14 | 0.132 | Stage 1 |
| rs2092315 | 1 | 22507684 | C | T | 1.07 | 0.072 | Stage 1 |
| rs10929757 | 2 | 11702661 | A | C | 1.08 | 0.073 | Stage 1 |
| rs11674184 | 2 | 11721535 | T | G | 0.94 | −0.067 | Stage 1 |
| rs10936600 | 3 | 169514585 | A | T | 0.91 | −0.095 | Stage 1 |
| rs143835293 | 3 | 197623337 | A | G | 1.75 | 0.558 | Stage 1 |
| rs62323680 | 4 | 54546192 | G | A | 1.16 | 0.153 | Stage 1 |
| rs2202282 | 4 | 70634441 | C | T | 1.07 | 0.071 | Stage 1 |
| rs72709458 | 5 | 1283755 | C | T | 1.12 | 0.111 | Stage 1 |
| rs2736100 | 5 | 1286516 | C | A | 0.91 | −0.090 | Stage 1 |
| rs2853676 | 5 | 1288547 | T | C | 0.91 | −0.089 | Stage 1 |
| rs2456181 | 5 | 176450837 | C | G | 1.07 | 0.066 | Stage 1 |
| rs4870084 | 6 | 152543949 | C | T | 0.92 | −0.087 | Stage 1 |
| rs6928363 | 6 | 152546094 | G | A | 0.92 | −0.078 | Stage 1 |
| rs58415480 | 6 | 152562271 | C | G | 1.18 | 0.167 | Stage 1 |
| rs75510204 | 6 | 152592680 | T | G | 1.29 | 0.257 | Stage 1 |
| rs6904757 | 6 | 152593102 | A | G | 0.93 | −0.078 | Stage 1 |
| rs144444583 | 6 | 152684585 | T | C | 1.12 | 0.111 | Stage 1 |
| rs138821078 | 9 | 674217 | C | G | 1.23 | 0.205 | Stage 1 |
| rs10975820 | 9 | 684160 | G | A | 1.12 | 0.112 | Stage 1 |
| rs7027685 | 9 | 802228 | A | T | 1.11 | 0.103 | Stage 1 |
| rs114680331 | 9 | 815682 | T | C | 1.11 | 0.108 | Stage 1 |
| rs4742448 | 9 | 826585 | C | G | 1.07 | 0.066 | Stage 1 |
| rs2277163 | 9 | 827224 | A | G | 0.87 | −0.140 | Stage 1 |
| rs1265164 | 10 | 105674854 | A | G | 0.91 | −0.097 | Stage 1 |
| rs11246003 | 11 | 213723 | T | G | 0.85 | −0.167 | Stage 1 |
| rs507139 | 11 | 225196 | G | A | 0.84 | −0.171 | Stage 1 |
| rs2207548 | 11 | 32368744 | C | A | 1.09 | 0.091 | Stage 1 |
| rs10835889 | 11 | 32370380 | G | A | 1.14 | 0.133 | Stage 1 |
| rs7120483 | 11 | 32406983 | G | C | 1.12 | 0.112 | Stage 1 |
| rs11031783 | 11 | 32459923 | C | A | 1.09 | 0.084 | Stage 1 |
| rs2553772 | 11 | 35085453 | T | G | 1.07 | 0.069 | Stage 1 |
| rs59021565 | 11 | 107999907 | C | G | 1.11 | 0.109 | Stage 1 |
| rs141379009 | 11 | 108149207 | T | G | 1.32 | 0.281 | Stage 1 |
| rs4988023 | 11 | 108168995 | A | C | 0.89 | −0.113 | Stage 1 |
| rs12223381 | 11 | 108354102 | C | T | 1.07 | 0.064 | Stage 1 |
| rs9669403 | 12 | 46798900 | G | A | 1.07 | 0.071 | Stage 1 |
| rs12832777 | 12 | 46831129 | T | C | 1.09 | 0.086 | Stage 1 |
| rs117245733 | 13 | 40723944 | G | A | 1.26 | 0.231 | Stage 1 |
| rs7986407 | 13 | 41179798 | A | G | 1.09 | 0.084 | Stage 1 |
| rs66998222 | 16 | 51481596 | G | A | 0.91 | −0.098 | Stage 1 |
| rs78378222 | 17 | 7571752 | T | G | 1.53 | 0.427 | Stage 1 |
| rs733381 | 22 | 40669648 | A | G | 1.10 | 0.091 | Stage 1 |
| rs5936989 | X | 70022420 | T | A | 0.92 | −0.081 | Stage 1 |
| rs5937008 | X | 70093038 | C | T | 0.91 | −0.094 | Stage 1 |
| rs7059898 | X | 70149078 | C | A | 1.07 | 0.068 | Stage 1 |
| rs7888560 | X | 131171122 | A | G | 1.08 | 0.077 | Stage 1 |
| rs5930554 | X | 131312089 | T | C | 1.14 | 0.129 | Stage 1 |
| rs5933158 | X | 131578034 | A | G | 1.08 | 0.074 | Stage 1 |
| rs5975338 | X | 131626317 | A | G | 1.10 | 0.094 | Stage 1 |
| rs17631680 | 2 | 67090367 | T | C | 0.90 | −0.102 | Stage 2 |
| rs1735537 | 3 | 128122820 | T | C | 1.07 | 0.071 | Stage 2 |
| rs67751869 | 4 | 54568834 | T | C | 1.13 | 0.121 | Stage 2 |
| rs6901631 | 6 | 152567047 | T | C | 0.91 | −0.097 | Stage 2 |
| rs11790408 | 9 | 876418 | G | T | 0.94 | −0.063 | Stage 2 |
| rs10415391 | 19 | 22652436 | C | T | 1.10 | 0.098 | Stage 2 |
| rs62132801 | 19 | 49267882 | A | T | 0.90 | −0.105 | Stage 2 |
-
* Discovered in stage 1 (UKBB GWAS) or in stage 2 (meta-analysis of UKBB and Helsinki)
Summary of association tests for SNPs.
Each of the 57 GRS SNPs was tested for an additive effect to age at hysterectomy, degree of somatic allelic imbalance (AI) and tumor counts. Somatic allele imbalance was defined as the mean of the length of somatic allelic imbalance over all tumors of a patient. Tests of MED12 mutation positive and negative tumors are denoted by MED12mut + and MED12mut-, respectively. The numbers for regression coefficient (Beta), standard error of beta (SE), test statistic (z) and association (P) were taken by fitting either a linear regression or negative binomial (NB) regression model (response ~predictor). The nominal P-values were adjusted for FDR (Q). The risk allele was used as the effect allele for Beta. In total 228 tests, all p<0.05 are shown.
| Predictor | Response | Model | Beta | SE | Statistic | P | Q |
|---|---|---|---|---|---|---|---|
| 12:46798900:G:A | MED12mut + count | NB | −0.22 | 0.08 | −2.88 | 0.004 | 0.51 |
| 9:674217:C:G | MED12mut + count | NB | −0.69 | 0.26 | −2.63 | 0.008 | 0.51 |
| X:70093038:C:T | MED12mut + count | NB | 0.21 | 0.08 | 2.62 | 0.009 | 0.51 |
| X:70022420:T:A | MED12mut + count | NB | 0.20 | 0.08 | 2.56 | 0.011 | 0.51 |
| 11:32459923:C:A | MED12mut + count | NB | 0.23 | 0.09 | 2.54 | 0.011 | 0.51 |
| 4:54568834:T:C | log(somatic AI basepairs) | Linear | −0.25 | 0.10 | −2.48 | 0.013 | 0.51 |
| 17:7571752:T:G | MED12mut- count | NB | −0.92 | 0.41 | −2.24 | 0.025 | 0.56 |
| 6:152546094:G:A | MED12mut- count | NB | −0.19 | 0.08 | −2.24 | 0.025 | 0.56 |
| 11:107999907:C:G | Age at hysterectomy | Linear | 3.00 | 1.34 | 2.24 | 0.026 | 0.56 |
| 13:40723944:G:A | MED12mut + count | NB | 0.36 | 0.17 | 2.17 | 0.030 | 0.56 |
| 22:40669648:A:G | MED12mut + count | NB | 0.19 | 0.09 | 2.15 | 0.031 | 0.56 |
| 5:1286516:C:A | MED12mut + count | NB | 0.17 | 0.08 | 2.15 | 0.031 | 0.56 |
| 22:40669648:A:G | Age at hysterectomy | Linear | 1.36 | 0.63 | 2.16 | 0.032 | 0.56 |
| 1:22450487:C:T | Age at hysterectomy | Linear | −1.48 | 0.72 | −2.05 | 0.041 | 0.65 |
| 16:51481596:G:A | log(somatic AI basepairs) | Linear | 0.17 | 0.08 | 2.02 | 0.044 | 0.65 |
| 5:1288547:T:C | Age at hysterectomy | Linear | 1.32 | 0.66 | 2.00 | 0.046 | 0.65 |
Summary of all the association tests for GRS.
All GRS related tests from the main text. The notation of MED12mut + and MED12mut- refer to the numbers of MED12-mutation-positive and -negative tumors, respectively. The tests include Wilcoxon rank-sum and models for linear and negative binomial (NB) regression (variable ~GRS). The P values were adjusted for FWER with the Holm-Bonferroni method (Q). Significant associations (Q < 0.05) are shown bolded. Note that population association tests include only the female controls.
| GRS * | Cohort | Variable | Test | N cases | N controls | Rate ratio | P | Q |
|---|---|---|---|---|---|---|---|---|
| Stage 1 | Helsinki | UL phenotype | Rank-sum (one-tailed) | 457 | 8899 | - | 8.3e-10 | 1.1e-08 |
| Stage 2 | NFBC | UL phenotype | Rank-sum (one-tailed) | 459 | 2351 | - | 1.1e-05 | 1.1e-04 |
| Stage 2 | Helsinki | Total number of ULs | NB | 457 | - | 1.25 | 0.00105 | 0.0032 |
| Stage 2 | Helsinki | Age at hysterectomy | Linear | 392 | - | 0.50 | 0.48 | 0.48 |
| Stage 2 | Helsinki | Number of MED12mut+ | NB | 457 | - | 1.43 | 3.2e-04 | 0.002 |
| Stage 2 | Helsinki | Number of MED12mut- | NB | 457 | - | 0.79 | 0.0266 | 0.053 |
| Stage 2 | Helsinki | One-or-more MED12mut+ | Rank-sum | 334 | 123 | - | 5.3e-04 | 0.0026 |
| Stage 2 | Helsinki | All MED12mut+ | Rank-sum | 221 | 123 | - | 7.9e-04 | 0.0032 |
| Stage 2 | African # | UL phenotype | Rank-sum (one-tailed) | 296 | 1256 | - | 1.3e-05 | 1.2e-04 |
| Stage 2 | Caribbean# | UL phenotype | Rank-sum (one-tailed) | 668 | 2041 | - | 6.7e-05 | 5.4e-04 |
| Stage 2 | Irish # | UL phenotype | Rank-sum (one-tailed) | 398 | 6208 | - | 8.3e-06 | 9.1e-05 |
| Stage 2 | Indian # | UL phenotype | Rank-sum (one-tailed) | 203 | 2567 | - | 2.9e-04 | 0.0020 |
| Stage 2 | Other white # | UL phenotype | Rank-sum (one-tailed) | 647 | 8982 | - | 6.9e-09 | 8.3e-08 |
-
*Based either on stage 1 (UKBB GWAS) or stage 2 (meta-analysis of UKBB and Helsinki)
# An independent subset of UKBB data (stratified based on self-reported UKBB annotation).
Population specific estimates for GRS.
Allele frequencies were collected from gnomAD (version 2.0.1). GRS weights were taken from UKBB summary statistics. The estimate for population specific GRS follows from weighting the allele frequencies with log-odds. The final values were scaled with a factor two to make them comparable with SNP dosage-based GRS. See Appendix-figure 6 for an explanation of the population acronyms.
| rs-code | Chr | Pos | A | B | AF_AFR | AF_AMR | AF_ASJ | AF_EAS | AF_FIN | AF_NFE | AF_OTH |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs58415480 | 6 | 152562271 | C | G | 0.228 | 0.066 | 0.090 | 0.000 | 0.237 | 0.161 | 0.158 |
| rs78378222 | 17 | 7571752 | T | G | 0.002 | 0.004 | 0.000 | 0.000 | 0.020 | 0.018 | 0.015 |
| rs10835889 | 11 | 32370380 | G | A | 0.384 | 0.091 | 0.202 | 0.052 | 0.069 | 0.150 | 0.124 |
| rs141379009 | 11 | 108149207 | T | G | 0.005 | 0.014 | 0.023 | 0.000 | 0.007 | 0.022 | 0.015 |
| rs7027685 | 9 | 802228 | A | T | 0.535 | 0.602 | 0.357 | 0.738 | 0.556 | 0.413 | 0.462 |
| rs2235529 | 1 | 22450487 | C | T | 0.029 | 0.313 | 0.046 | 0.484 | 0.164 | 0.158 | 0.171 |
| rs72709458 | 5 | 1283755 | C | T | 0.139 | 0.137 | 0.183 | 0.134 | 0.221 | 0.188 | 0.180 |
| rs2207548 | 11 | 32368744 | C | A | 0.553 | 0.228 | 0.393 | 0.179 | 0.321 | 0.411 | 0.378 |
| rs2736100 | 5 | 1286516 | C | A | 0.550 | 0.597 | 0.477 | 0.606 | 0.507 | 0.504 | 0.539 |
| rs507139 | 11 | 225196 | G | A | 0.028 | 0.058 | 0.079 | 0.007 | 0.094 | 0.072 | 0.084 |
| rs62323680 | 4 | 54546192 | G | A | 0.020 | 0.032 | 0.099 | 0.002 | 0.045 | 0.070 | 0.060 |
| rs2853676 | 5 | 1288547 | T | C | 0.765 | 0.801 | 0.613 | 0.856 | 0.783 | 0.739 | 0.763 |
| rs10975820 | 9 | 684160 | G | A | 0.669 | 0.397 | 0.212 | 0.586 | 0.257 | 0.171 | 0.255 |
| rs10936600 | 3 | 169514585 | A | T | 0.094 | 0.406 | 0.195 | 0.539 | 0.274 | 0.249 | 0.267 |
| rs6928363 | 6 | 152546094 | G | A | 0.692 | 0.693 | 0.541 | 0.851 | 0.575 | 0.508 | 0.568 |
| rs7986407 | 13 | 41179798 | A | G | 0.446 | 0.459 | 0.277 | 0.739 | 0.384 | 0.311 | 0.367 |
| rs144444583 | 6 | 152684585 | T | C | 0.050 | 0.045 | 0.044 | 0.006 | 0.230 | 0.139 | 0.155 |
| rs143835293 | 3 | 197623337 | A | G | 0.000 | 0.004 | 0.003 | 0.000 | 0.015 | 0.002 | 0.006 |
| rs12832777 | 12 | 46831129 | T | C | 0.918 | 0.740 | 0.762 | 0.902 | 0.651 | 0.675 | 0.669 |
| rs7120483 | 11 | 32406983 | G | C | 0.242 | 0.087 | 0.152 | 0.025 | 0.074 | 0.112 | 0.098 |
| rs4988023 | 11 | 108168995 | A | C | 0.022 | 0.044 | 0.142 | 0.006 | 0.236 | 0.146 | 0.149 |
| rs733381 | 22 | 40669648 | A | G | 0.095 | 0.471 | 0.169 | 0.247 | 0.258 | 0.216 | 0.239 |
| rs66998222 | 16 | 51481596 | G | A | 0.033 | 0.138 | 0.265 | 0.001 | 0.166 | 0.213 | 0.167 |
| rs6904757 | 6 | 152593102 | A | G | 0.443 | 0.579 | 0.401 | 0.625 | 0.379 | 0.392 | 0.401 |
| rs2202282 | 4 | 70634441 | C | T | 0.230 | 0.549 | 0.420 | 0.726 | 0.437 | 0.491 | 0.466 |
| rs10929757 | 2 | 11702661 | A | C | 0.116 | 0.505 | 0.556 | 0.517 | 0.674 | 0.591 | 0.556 |
| rs9669403 | 12 | 46798900 | G | A | 0.615 | 0.476 | 0.406 | 0.266 | 0.396 | 0.382 | 0.369 |
| rs11031783 | 11 | 32459923 | C | A | 0.262 | 0.364 | 0.288 | 0.748 | 0.197 | 0.204 | 0.241 |
| rs114680331 | 9 | 815682 | T | C | 0.065 | 0.178 | 0.067 | 0.128 | 0.190 | 0.115 | 0.133 |
| rs117245733 | 13 | 40723944 | G | A | 0.001 | 0.004 | 0.003 | 0.000 | 0.030 | 0.024 | 0.027 |
| rs2553772 | 11 | 35085453 | T | G | 0.342 | 0.656 | 0.600 | 0.769 | 0.524 | 0.544 | 0.569 |
| rs2456181 | 5 | 176450837 | C | G | 0.796 | 0.355 | 0.533 | 0.481 | 0.500 | 0.511 | 0.505 |
| rs4742448 | 9 | 826585 | C | G | 0.838 | 0.629 | 0.473 | 0.785 | 0.548 | 0.471 | 0.503 |
| rs2277163 | 9 | 827224 | A | G | 0.959 | 0.858 | 0.964 | 0.825 | 0.966 | 0.948 | 0.954 |
| rs4870084 | 6 | 152543949 | C | T | 0.100 | 0.421 | 0.175 | 0.188 | 0.234 | 0.211 | 0.238 |
| rs1265164 | 10 | 105674854 | A | G | 0.469 | 0.891 | 0.825 | 0.985 | 0.890 | 0.863 | 0.873 |
| rs75510204 | 6 | 152592680 | T | G | 0.002 | 0.002 | 0.000 | 0.000 | 0.015 | 0.011 | 0.011 |
| rs11246003 | 11 | 213723 | T | G | 0.060 | 0.047 | 0.033 | 0.149 | 0.022 | 0.035 | 0.038 |
| rs138821078 | 9 | 674217 | C | G | 0.002 | 0.004 | 0.000 | 0.000 | 0.017 | 0.025 | 0.014 |
| rs59021565 | 11 | 107999907 | C | G | 0.038 | 0.112 | 0.119 | 0.049 | 0.050 | 0.086 | 0.054 |
| rs2092315 | 1 | 22507684 | C | T | 0.132 | 0.323 | 0.242 | 0.504 | 0.200 | 0.243 | 0.210 |
| rs12223381 | 11 | 108354102 | C | T | 0.413 | 0.606 | 0.477 | 0.444 | 0.390 | 0.414 | 0.457 |
| rs11674184 | 2 | 11721535 | T | G | 0.259 | 0.476 | 0.334 | 0.414 | 0.403 | 0.384 | 0.389 |
| rs5930554 | X | 131312089 | T | C | 0.752 | 0.174 | 0.296 | 0.104 | 0.320 | 0.319 | 0.311 |
| rs5937008 | X | 70093038 | C | T | 0.145 | 0.383 | 0.414 | 0.563 | 0.486 | 0.520 | 0.485 |
| rs5933158 | X | 131578034 | A | G | 0.710 | 0.588 | 0.500 | 0.686 | 0.650 | 0.605 | 0.741 |
| rs5936989 | X | 70022420 | T | A | 0.742 | 0.888 | 0.767 | 0.994 | 0.677 | 0.778 | 0.810 |
| rs5975338 | X | 131626317 | A | G | 0.409 | 0.103 | 0.099 | 0.091 | 0.161 | 0.145 | 0.151 |
| rs7059898 | X | 70149078 | C | A | 0.143 | 0.167 | 0.355 | 0.001 | 0.340 | 0.342 | 0.276 |
| rs7888560 | X | 131171122 | A | G | 0.142 | 0.090 | 0.199 | 0.000 | 0.244 | 0.217 | 0.201 |
| rs67751869 | 4 | 54568834 | T | C | 0.085 | 0.030 | 0.099 | 0.022 | 0.053 | 0.062 | 0.067 |
| rs6901631 | 6 | 152567047 | T | C | 0.256 | 0.249 | 0.182 | 0.122 | 0.102 | 0.120 | 0.127 |
| rs10415391 | 19 | 22652436 | C | T | 0.202 | 0.074 | 0.110 | 0.225 | 0.130 | 0.107 | 0.144 |
| rs1735537 | 3 | 128122820 | T | C | 0.523 | 0.385 | 0.262 | 0.425 | 0.249 | 0.251 | 0.288 |
| rs62132801 | 19 | 49267882 | A | T | 0.013 | 0.070 | 0.053 | 0.009 | 0.112 | 0.103 | 0.092 |
| rs17631680 | 2 | 67090367 | T | C | 0.018 | 0.061 | 0.050 | 0.001 | 0.100 | 0.102 | 0.082 |
| rs11790408 | 9 | 876418 | G | T | 0.110 | 0.307 | 0.387 | 0.246 | 0.355 | 0.380 | 0.353 |
| AFR | AMR | ASJ | EAS | FIN | NFE | OTH | |||||
| GRS | 4.718 | 4.059 | 4.068 | 4.196 | 4.168 | 4.091 | 4.116 | ||||
eQTLs in tumor and normal tissue.
Here, B is the effect allele. All local permutation p<0.05 are shown. Full table of eQTLs is given in Supplementary file 4.
| Tissue | Gene | ID | N | Best SNP | Distance | Nominal P | Permutation P | FDR |
|---|---|---|---|---|---|---|---|---|
| T | RP11- 816J6.3 | ENSG00000269889.1 | 82 | rs67795055 | 42457 | 9.10E-05 | 4.03E-04 | 0.224 |
| T | RUFY3 | ENSG00000018189.8 | 8 | rs7660770 | −972945 | 7.84E-04 | 6.68E-04 | 0.370 |
| T | TRIP13 | ENSG00000071539.9 | 22 | rs2736099 | 394581 | 1.63E-04 | 2.27E-03 | 1 |
| N | RN7SL832P | ENSG00000243819.3 | 28 | rs7570979 | 886958 | 4.06E-04 | 2.75E-03 | 1 |
| N | LNX1 | ENSG00000072201.9 | 25 | rs62323674 | 212300 | 7.12E-04 | 3.22E-03 | 1 |
| N | RP11- 849F2.8 | ENSG00000269928.1 | 5 | rs143094271 | −665038 | 2.43E-03 | 4.00E-03 | 1 |
| T | MIR5006 | ENSG00000264190.1 | 158 | rs9549254 | −907660 | 6.10E-04 | 4.42E-03 | 1 |
| N | RP11- 423H2.3 | ENSG00000249684.1 | 50 | rs183686 | −883235 | 3.36E-03 | 5.55E-03 | 1 |
| N | SLC7A3 | ENSG00000165349.7 | 148 | rs4360450 | 965 | 6.75E-04 | 7.38E-03 | 1 |
| T | ALPL | ENSG00000162551.9 | 52 | . | 521370 | 8.48E-04 | 7.59E-03 | 1 |
| N | PRRG4 | ENSG00000135378.3 | 167 | rs11031737 | −478718 | 3.99E-04 | 8.59E-03 | 1 |
| N | RP11- 791G15.2 | ENSG00000272275.1 | 28 | rs6432220 | 802714 | 1.24E-03 | 8.81E-03 | 1 |
| T | ZDHHC11B | ENSG00000206077.6 | 22 | rs2736099 | 576864 | 9.74E-04 | 1.02E-02 | 1 |
| T | ACAP1 | ENSG00000072818.7 | 5 | rs78378222 | 331903 | 4.83E-03 | 1.04E-02 | 1 |
| N | HNRNPA1P48 | ENSG00000224578.3 | 20 | rs55881068 | −212266 | 4.97E-03 | 1.12E-02 | 1 |
| T | WNT4 | ENSG00000162552.10 | 52 | rs12042083 | 28933 | 1.30E-03 | 1.16E-02 | 1 |
| T | LRRIQ4 | ENSG00000188306.6 | 82 | rs13074500 | 25860 | 2.96E-03 | 1.20E-02 | 1 |
| T | TEX11 | ENSG00000120498.9 | 148 | rs11795627 | 208650 | 1.30E-03 | 1.23E-02 | 1 |
| T | AC011994.1 | ENSG00000265583.1 | 28 | rs11685032 | −73236 | 1.84E-03 | 1.32E-02 | 1 |
| T | FRMD7 | ENSG00000165694.5 | 215 | rs5933158 | 367012 | 8.40E-04 | 1.33E-02 | 1 |
| N | TRAPPC1 | ENSG00000170043.7 | 5 | rs138420351 | −133601 | 7.23E-03 | 1.44E-02 | 1 |
| T | RP11- 423H2.1 | ENSG00000170089.11 | 72 | rs353496 | −806511 | 9.37E-03 | 1.51E-02 | 1 |
| N | SDHAP3 | ENSG00000185986.10 | 22 | rs11742908 | −297655 | 1.42E-03 | 1.58E-02 | 1 |
| T | CDC42-IT1 | ENSG00000230068.2 | 52 | rs10917167 | 118400 | 2.10E-03 | 1.80E-02 | 1 |
| N | RP11- 362K14.6 | ENSG00000270096.1 | 82 | rs12638862 | −35245 | 4.49E-03 | 1.96E-02 | 1 |
| T | TNRC6B | ENSG00000100354.16 | 32 | rs12484951 | 262253 | 8.29E-03 | 1.97E-02 | 1 |
| N | DNAH2 | ENSG00000183914.10 | 5 | rs143094271 | −157571 | 9.73E-03 | 1.98E-02 | 1 |
| N | SALL1 | ENSG00000103449.7 | 20 | rs12933686 | 308913 | 9.53E-03 | 2.13E-02 | 1 |
| T | XXyac- YRM2039.2 | ENSG00000181404.13 | 71 | rs138821078 | 659705 | 1.69E-03 | 2.22E-02 | 1 |
| N | ITPRIP | ENSG00000148841.11 | 8 | rs9419958 | −395949 | 1.43E-02 | 2.39E-02 | 1 |
| T | RNASEK- C17orf49 | ENSG00000161939.14 | 5 | . | 501685 | 1.19E-02 | 2.43E-02 | 1 |
| T | UQCRFS1P1 | ENSG00000226085.2 | 32 | rs1807579 | 256926 | 1.06E-02 | 2.44E-02 | 1 |
| T | FOXO1 | ENSG00000150907.6 | 166 | rs9549260 | 124299 | 2.94E-03 | 2.61E-02 | 1 |
| T | NDUFS6 | ENSG00000145494.7 | 22 | rs7734992 | −521387 | 2.81E-03 | 2.82E-02 | 1 |
| T | RP11- 259O2.3 | ENSG00000249731.1 | 22 | rs7710703 | −680704 | 2.89E-03 | 2.84E-02 | 1 |
| T | CDC42 | ENSG00000070831.11 | 52 | rs41307810 | −25076 | 3.35E-03 | 2.91E-02 | 1 |
| T | RP3- 363L9.1 | ENSG00000227064.1 | 215 | rs3764771 | −830073 | 2.03E-03 | 2.97E-02 | 1 |
| T | RP11- 362K14.7 | ENSG00000270135.1 | 82 | rs67795055 | 18611 | 7.45E-03 | 3.07E-02 | 1 |
| N | AMIGO2 | ENSG00000139211.5 | 155 | rs9706162 | −606808 | 6.28E-03 | 3.08E-02 | 1 |
| T | LPIN1 | ENSG00000134324.7 | 28 | rs11685032 | −143427 | 5.06E-03 | 3.33E-02 | 1 |
| T | MBNL3 | ENSG00000076770.10 | 215 | rs1263155 | 190718 | 2.55E-03 | 3.37E-02 | 1 |
| N | TOX3 | ENSG00000103460.12 | 5 | rs12325192 | −983789 | 3.24E-02 | 3.38E-02 | 1 |
| N | DANCR | ENSG00000226950.2 | 23 | rs62325482 | 885823 | 8.24E-03 | 3.54E-02 | 1 |
| T | DMRT1 | ENSG00000137090.7 | 71 | rs12004436 | −158261 | 3.09E-03 | 3.64E-02 | 1 |
| N | NT5C2 | ENSG00000076685.14 | 8 | rs1265164 | 828913 | 2.05E-02 | 3.75E-02 | 1 |
| T | TMEM256- PLSCR3 | ENSG00000187838.12 | 5 | rs138420351 | 407016 | 1.77E-02 | 3.81E-02 | 1 |
| T | RP11- 401P9.4 | ENSG00000261685.2 | 20 | rs12933686 | 799079 | 1.64E-02 | 3.84E-02 | 1 |
| N | CTC1 | ENSG00000178971.9 | 5 | rs143094271 | −667090 | 1.89E-02 | 4.03E-02 | 1 |
| N | SULT1B1 | ENSG00000173597.4 | 19 | rs13133166 | 13318 | 1.49E-02 | 4.04E-02 | 1 |
| N | TNK1 | ENSG00000174292.8 | 5 | rs143094271 | 179248 | 2.24E-02 | 4.21E-02 | 1 |
| T | SOX15 | ENSG00000129194.3 | 5 | rs138420351 | 208566 | 2.02E-02 | 4.30E-02 | 1 |
| T | UGT2A1 | ENSG00000173610.7 | 19 | rs1587766 | 112959 | 2.06E-02 | 4.31E-02 | 1 |
| T | RP4- 607I7.1 | ENSG00000255521.1 | 34 | rs2553783 | −82085 | 2.66E-02 | 4.41E-02 | 1 |
| N | SENP3 | ENSG00000161956.8 | 5 | rs78378222 | 106559 | 2.19E-02 | 4.45E-02 | 1 |
| N | KDM6B | ENSG00000132510.6 | 5 | rs143094271 | −280121 | 2.16E-02 | 4.54E-02 | 1 |
| T | SKIL | ENSG00000136603.9 | 82 | rs34194057 | −525916 | 1.13E-02 | 4.63E-02 | 1 |
| T | EPHB2 | ENSG00000133216.12 | 52 | . | −680104 | 6.04E-03 | 4.97E-02 | 1 |
cis-meQTLs in candidate genes where FDR < 0.1.
The statistics were calculated with an additive linear model. Here B is used as the effect allele. Full Table of cis-meQTLs with nominal significance (p-value<0.05) is given in Supplementary file 5.
| Tissue | SNP | CpG* | Beta | t-stat | p-value | FDR | Gene | CpG island | Promoter | AA | AB | BB |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 13_ 41179798 | 13_ 41189143 | −0.413 | −19.837 | 6.57E-20 | 4.88E-15 | FOXO1 | no | no | 15 | 16 | 4 |
| T | 13_ 41179798 | 13_ 41189143 | −0.333 | −8.296 | 3.29E-11 | 7.74E-07 | FOXO1 | no | no | 28 | 23 | 5 |
| T | 5_1283755 | 5_ 1285974 | −0.427 | −7.167 | 2.21E-09 | 3.46E-05 | TERT | no | no | 36 | 17 | 3 |
| N | 5_ 1283755 | 5_ 1285974 | −0.461 | −7.46 | 1.42E-08 | 1.11E-04 | TERT | no | no | 18 | 14 | 3 |
| T | 5_ 1286516 | 5_ 1277576 | −0.289 | −6.358 | 4.53E-08 | 3.99E-04 | TERT | yes | no | 7 | 29 | 20 |
| T | 1_ 22450487 | 1_ 22456326 | −0.123 | −5.507 | 1.04E-06 | 6.36E-03 | WNT4 | no | no | 40 | 15 | 1 |
| N | 5_ 1286516 | 5_ 1285974 | 0.426 | 5.477 | 4.51E-06 | 1.76E-02 | TERT | no | no | 6 | 22 | 7 |
| T | 5_ 1286516 | 5_ 1285974 | 0.299 | 4.739 | 1.61E-05 | 4.82E-02 | TERT | no | no | 7 | 29 | 20 |
| N | 13_ 41179798 | 13_ 41223110 | −0.174 | −4.957 | 2.10E-05 | 5.36E-02 | FOXO1 | no | no | 15 | 16 | 4 |
| N | 5_ 1283755 | 5_ 1282413 | −0.17 | −4.816 | 3.16E-05 | 7.12E-02 | TERT | no | no | 18 | 14 | 3 |
| T | 11_ 225196 | 11_ 195431 | 0.222 | 4.452 | 4.31E-05 | 9.21E-02 | BET1L | no | ODF3 | 48 | 8 | 0 |
-
* Minimum CpG coverage of ≥2 required in ≥90% of samples. CpG in 1 Mb flank from the SNP.
Replication of previously suggested UL predisposition loci.
For study details, see the literature references in the main text. The population, locus, gene and risk allele (RA) information were collected from the original studies. The associations, P (UKBB) column, were taken from the UKBB summary statistics for 15,453 UL cases. The bolded values pass FWER (Bonferroni for seven independent loci; p<0.05/7).
| Study | Population | Locus | Suggested gene | SNP | RA | Method | P (UKBB) |
|---|---|---|---|---|---|---|---|
| Cha et al. | Japanese | 22q13.1 | TNRC6B | rs12484776 | G | GWAS | 1.0E-10 |
| Edwards et al. | European Americans | 22q13.1 | TNRC6B | rs12484776 | G | GWAS | 1.0E-10 |
| Cha et al. | Japanese | 11p15.5 | BET1L | rs2280543 | G | GWAS | 2.2E-08 |
| Edwards et al. | European Americans | 11p15.5 | BET1L | rs2280543 | G | GWAS | 2.2E-08 |
| Zhang et al. | African Americans | 1q42.2 | PCNXL2 | rs7546784 | - | Admixture | 3.6E-02 |
| Zhang et al. | African Americans | 2q32.2 | PMS1 | rs256552 | - | Admixture | 1.8E-01 |
| Cha et al. | Japanese | 10q24.33 | SLK | rs7913069 | A | GWAS | 3.4E-01 |
| Hellwege et al. | African American | 22q13.1 | CYTH4 | rs5995416 | C | GWAS | 5.3E-01 |
| Hellwege et al. | African American | 22q13.1 | CYTH4 | rs739187 | C | GWAS | 6.2E-01 |
| Hellwege et al. | African American | 22q13.1 | CYTH4 | rs713939 | C | GWAS | 8.0E-01 |
| Hellwege et al. | African American | 22q13.1 | CYTH4 | rs4821628 | G | GWAS | 8.1E-01 |
| Eggert et al. | Multiple | 17q25.3 | FASN | rs4247357 | A | Linkage and GWAS | 8.1E-01 |
GWAS catalog and references to earlier literature on GRS SNPs.
The GRS SNPs rs10936600, rs11674184, rs2235529, rs2736100, rs2853676, rs72709458 and rs78378222 were found in the GWAS catalog (version 1.0.1; www.ebi.ac.uk/gwas). The numbers for allele frequency (AF), odds-ratio (OR) and association (P) follow those reported in the GWAS catalog.
| Pubmed | Date | Journal | Gene symbol | Risk allele | AF | OR | P | |
|---|---|---|---|---|---|---|---|---|
| 18835860 | 2008-10-01 | J Med Genet | Idiopathic pulmonary fibrosis | TERT | rs2736100-A | 0.41 | 2.11 | 3.00E-08 |
| 19578367 | 2009-07-05 | Nat Genet | Glioma | TERT | rs2736100-G | 0.49 | 1.27 | 2.00E-17 |
| 19578367 | 2009-07-05 | Nat Genet | Glioma | TERT | rs2853676-A | 0.73 | 1.26 | 4.00E-14 |
| 19836008 | 2009-10-15 | Am J Hum Genet | Lung adenocarcinoma | TERT | rs2736100-G | 0.5 | 1.12 | 2.00E-10 |
| 20139978 | 2010-02-07 | Nat Genet | Red blood cell count | TERT | rs2736100-G | 0.4 | 0.07 | 3.00E-08 |
| 20543847 | 2010-06-13 | Nat Genet | Testicular germ cell cancer | TERT | rs2736100-T | 0.49 | 1.33 | 8.00E-15 |
| 20700438 | 2010-08-05 | PLoS Genet | Lung adenocarcinoma | TERT | rs2736100-G | 0.39 | 1.46 | 2.00E-22 |
| 20871597 | 2010-09-26 | Nat Genet | Lung adenocarcinoma | TERT | rs2736100-C | 0.39 | 1.27 | 3.00E-11 |
| 21531791 | 2011-04-29 | Hum Mol Genet | Glioma | TERT | rs2736100-? | - | 1.25 | 1.00E-14 |
| 21725308 | 2011-07-03 | Nat Genet | Lung cancer | TERT | rs2736100-C | 0.41 | 1.27 | 1.00E-27 |
| 21827660 | 2011-08-09 | BMC Med Genomics | Glioma | TERT | rs2736100-? | - | - | 7.00E-09 |
| 21946351 | 2011-09-25 | Nat Genet | Basal cell carcinoma | TP53 | rs78378222-C | - | 2.16 | 2.00E-20 |
| 22886559 | 2012-08-11 | Hum Genet | Glioma | TERT | rs2736100-G | 0.494 | 1.30 | 4.00E-09 |
| 23143601 | 2012-11-11 | Nat Genet | Lung cancer | TERT | rs2736100-G | 0.4 | 1.38 | 4.00E-27 |
| 23472165 | 2013-03-05 | PLoS One | Endometriosis | WNT4 | rs2235529-T | 0.152 | 1.28 | 7.00E-09 |
| 23472165 | 2013-03-05 | PLoS One | Endometriosis | WNT4 | rs2235529-C | 0.709 | 1.19 | 6.00E-06 |
| 23472165 | 2013-03-05 | PLoS One | Endometriosis | WNT4 | rs2235529-T | 0.134 | 1.25 | 8.00E-07 |
| 23472165 | 2013-03-05 | PLoS One | Endometriosis | WNT4 | rs2235529-A | 0.153 | 1.30 | 3.00E-09 |
| 23583980 | 2013-04-14 | Nat Genet | Interstitial lung disease | TERT | rs2736100-A | 0.49 | 1.37 | 2.00E-19 |
| 24403052 | 2014-01-08 | Hum Mol Genet | Basal cell carcinoma | TP53 | rs78378222-G | - | 2.24 | 4.00E-22 |
| 24465473 | 2014-01-21 | PLoS One | Telomere length | TERT | rs2736100-C | 0.08 | 4.00E-06 | |
| 24908248 | 2014-06-08 | Nat Genet | Glioma (high-grade) | TERT | rs2736100-C | 0.51 | 1.39 | 1.00E-15 |
| 24945404 | 2014-06-19 | PLoS Genet | Bone mineral density (paediatric, upper limb) | WNT4 | rs2235529-C | 0.85 | 0.12 | 1.00E-08 |
| 24945404 | 2014-06-19 | PLoS Genet | Bone mineral density (paediatric, upper limb) | WNT4 | rs2235529-C | - | 0.12 | 3.00E-07 |
| 25855136 | 2015-04-09 | Nat Commun | Basal cell carcinoma | TP53 | rs78378222-G | 0.018 | 2.07 | 1.00E-20 |
| 26424050 | 2015-10-01 | Nat Commun | Glioblastoma | - | rs72709458-T | - | 1.68 | 6.00E-24 |
| 27363682 | 2016-07-01 | Nat Commun | Multiple myeloma | - | rs10936600-A | - | 1.20 | 6.00E-15 |
| 27501781 | 2016-08-09 | Nat Commun | EGFR mutation-positive lung adenocarcinoma | TERT | rs2736100-G | 0.387 | 1.42 | 2.00E-31 |
| 27539887 | 2016-08-19 | Nat Commun | Basal cell carcinoma | TP53 | rs78378222-G | 0.01 | 1.41 | 2.00E-10 |
| 27863252 | 2016-11-17 | Cell | Mean corpuscular hemoglobin | TP53 | rs78378222-G | 0.0121 | 0.10 | 6.00E-09 |
| 27863252 | 2016-11-17 | Cell | Mean corpuscular hemoglobin | TERT | rs2736100-A | 0.4982 | 0.04 | 5.00E-34 |
| 27863252 | 2016-11-17 | Cell | Platelet count | TERT | rs2736100-A | 0.4984 | 0.03 | 3.00E-20 |
| 28017375 | 2016-12-22 | Am J Hum Genet | Mean corpuscular volume | TERT | rs2736100-A | - | 0.00 | 3.00E-06 |
| 28017375 | 2016-12-22 | Am J Hum Genet | Mean corpuscular volume | TERT | rs2736100-? | - | - | 2.00E-11 |
| 28017375 | 2016-12-22 | Am J Hum Genet | Mean corpuscular hemoglobin | TERT | rs2736100-? | - | - | 1.00E-08 |
| 28135244 | 2017-01-30 | Nat Genet | Pulse pressure | TP53 | rs78378222-T | 0.99 | 0.90 | 2.00E-10 |
| 28346443 | 2017-03-27 | Nat Genet | Glioma | TP53 | rs78378222-G | 0.013 | 2.53 | 9.00E-38 |
| 28346443 | 2017-03-27 | Nat Genet | Glioblastoma | TP53 | rs78378222-G | 0.013 | 2.63 | 5.00E-29 |
| 28346443 | 2017-03-27 | Nat Genet | Non-glioblastoma glioma | TP53 | rs78378222-G | 0.013 | 2.73 | 5.00E-27 |
| 28537267 | 2017-05-24 | Nat Commun | Endometriosis | GREB1 | rs11674184-T | 0.61 | 1.13 | 3.00E-17 |
| 28537267 | 2017-05-24 | Nat Commun | Endometriosis | GREB1 | rs11674184-T | 0.61 | 1.12 | 3.00E-14 |
| 28604728 | 2017-06-12 | Nat Genet | Testicular germ cell tumor | TERT | rs2736100-A | 0.51 | 1.28 | 9.00E-25 |
| 28604732 | 2017-06-12 | Nat Genet | Testicular germ cell tumor | TERT | rs2736100-A | 0.5 | 1.29 | 8.00E-20 |
Additional files
-
Supplementary file 1
Summary statistics for the UKBB and Helsinki cohorts.
For each of the 57 GRS SNPs, we report the allele frequency, association effect size and P-value, together with the heterogeneity estimates Cochrane's Q-value and I2 index.
- https://doi.org/10.7554/eLife.37110.009
-
Supplementary file 2
Summary statistics for UKBB, Helsinki and NFBC.
For each of the 57 GRS SNPs, we report the allele frequency, association effect size and P-value, together with the heterogeneity estimates Cochrane's Q-value and I2 index.
- https://doi.org/10.7554/eLife.37110.010
-
Supplementary file 3
Summary statistics for the five UKBB follow-up cohorts.
For each of the 57 GRS SNPs, we report the allele frequency, association effect size and P-value, together with the heterogeneity estimates Cochrane's Q-value and I2 index.
- https://doi.org/10.7554/eLife.37110.011
-
Supplementary file 4
All the cis-eQTL summary statistics.
Tumors (T) and myometrium normal tissues (N) were analyzed separately. For each SNP, we report the local permutation test results from FastQTL (details in the Methods section).
- https://doi.org/10.7554/eLife.37110.012
-
Supplementary file 5
All the cis-meQTL summary statistics and annotation for their genomic context.
Tumors and myometrium normal tissues were analyzed separately. The association statistics are based on MatrixEQTL (details in the Methods section).
- https://doi.org/10.7554/eLife.37110.013
-
Supplementary file 6
Pathway-based analysis of the genome-wide significant SNPs.
Includes target gene prioritization, pathway enrichment and tissue-specific expression profiling results from the DEPICT framework (details in the Methods section).
- https://doi.org/10.7554/eLife.37110.014
-
Transparent reporting form
- https://doi.org/10.7554/eLife.37110.015





