Human gut Bacteroides capture vitamin B12 via cell surface-exposed lipoproteins
Figures
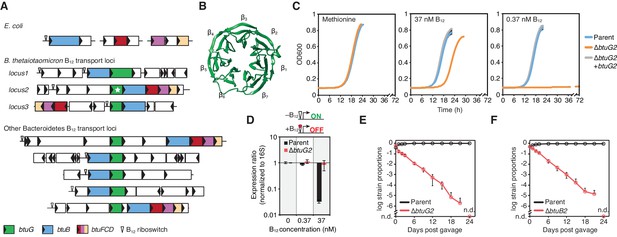
BtuG homologs are exclusively found among the Bacteroidetes, facilitate the acquisition of cyanocobalamin in vitro and confer a fitness advantage in gnotobiotic mice.
(A) Genetic loci encoding corrinoid transport components in E. coli, B. thetaiotaomicron and other Bacteroidetes. (B) BtuG2 (PDB 3DSM) adopts a seven-bladed β-propeller fold. (C) Growth curves for the B. thetaiotaomicron parent strain, btuG2 deletion strain or complemented strain grown in minimal media supplemented with methionine or vitamin B12. Data are representative of three independent trials; error bars indicate ±SD from three technical replicates. (D) Gene expression ratios for B. thetaiotaomicron strains grown in minimal media with methionine and indicated concentrations of vitamin B12. Expression of locus2 (BT1956) was normalized first to 16S rRNA and then to each strain’s expression in 0 nM vitamin B12. Data are representative of two independent trials; error bars indicate ± SD from three biological replicates. (E, F) B. thetaiotaomicron strain ratios determined from gDNA extracted from fecal samples collected over time from gnotobiotic mice. (n.d., not detected; n = 4 mice/group; error bars indicate ± SD).

A BtuG homolog is required for B. thetaiotaomicron fitness in gnotobiotic mice.
B. thetaiotaomicron parent, ∆btuG2 and complemented (∆btuG2 + btuG2) strain ratios determined from gDNA extracted from fecal samples of gnotobiotic mice collected over time. (n = 5 mice; error bars indicate ± SD). The fitness defect of a B. thetaiotaomicron strain lacking btuB2 (Figure 1F) is also complemented by expression of btuB2 in single copy from a heterologous chromosomal locus (Degnan et al., 2014a).
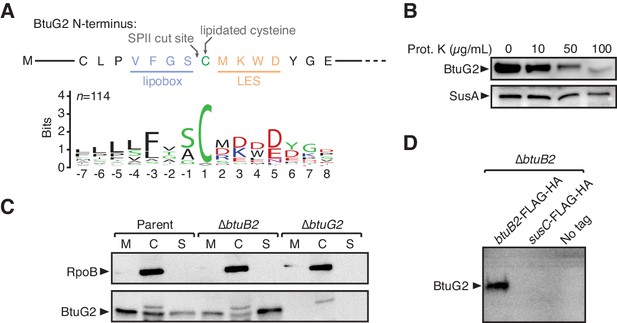
BtuG2 is a cell surface-exposed lipoprotein that interacts with the outer membrane transporter BtuB2.
(A) N-terminus of BtuG2 and sequence logo of 114 BtuG homologs reveal sequence signatures indicative of a surface-exposed lipoprotein, including a lipobox, adjacent cysteine residue and lipoprotein export signal (LES). (B) Protease degradation of BtuG2 on whole B. thetaiotaomicron cells suggests BtuG2 is surface-exposed. SusA is a periplasmic control. Data are representative of three independent trials. (C) B. thetaiotaomicron cells separated into membrane (M), cytoplasm/periplasm (C), and supernatant (S) fractions reveal that BtuG2 is predominantly associated with the membrane in parent cells, but predominantly associated with the supernatant in ∆btuB2 cells. Data are representative of four independent trials. (D) In vivo pull-down of BtuG2 by TAP-tagged BtuB2 suggests an interaction with BtuB2, but not with an unrelated outer membrane β-barrel protein SusC. Data are representative of two independent trials.
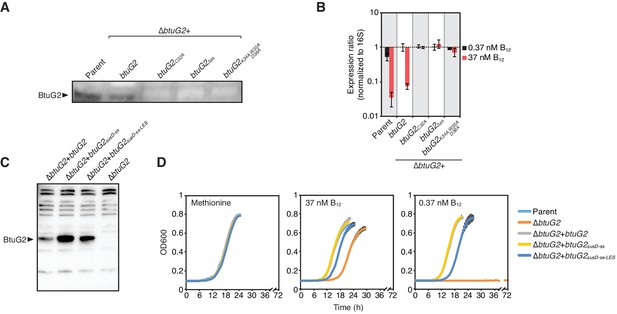
BtuG2 lipoprotein sequence signatures are required for protein production and can be functionally replaced with N-terminal sequences from the unrelated cell surface lipoprotein SusD.
(A) Western blot of B. thetaiotaomicron whole cell lysates of parent strain and a panel of btuG2 complemented strains probed with anti-BtuG2. Data are representative of three independent trials. (B) qRT-PCR of B. thetaiotaomicron strains grown in minimal media supplemented with methionine and either 37 nM or 0.37 nM vitamin B12. Data are normalized first to 16S rRNA and then to expression of ∆btuG2 + btuG2 in 0.37 nM vitamin B12. Data are representative of two independent trials; error bars indicate ± SD from three biological replicates. (C) Western blot of B. thetaiotaomicron whole cell lysates of a panel of btuG2 complemented strains probed with anti-BtuG2. Data are representative of two independent trials. (D) Growth curves of B. thetaiotaomicron strains grown in minimal media supplemented with methionine or indicated concentrations of vitamin B12. Data are representative of two independent trials; error bars indicate ± SD from three technical replicates; ss, signal sequence; LES, lipoprotein export signal.
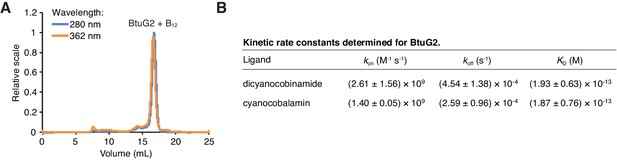
BtuG2 binds cyanocobalamin and its corrinoid precursor with femtomolar affinity.
(A) SEC-MALS traces for recombinant BtuG2 incubated with vitamin B12. BtuG2 (protein) absorbance is measured at 280 nm; vitamin B12 absorbance is measured at 362 nm. Data are representative of three independent trials. (B) Kinetic rate constants and equilibrium dissociation constant for BtuG2 binding to dicyanocobinamide and cyanocobalamin determined by SPR. Data are representative of three independent trials; error represents ± SD from rate constants measured across three Biacore chip cells.
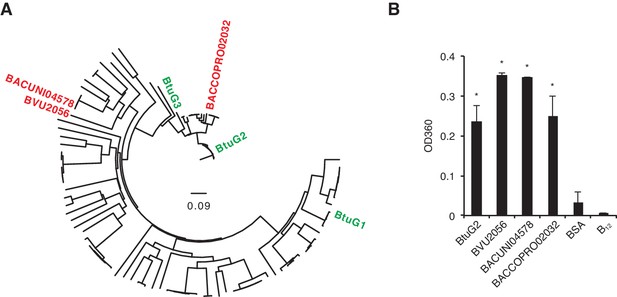
Cyanocobalamin binding by diverse BtuG homologs.
(A) A gene tree assembled from 114 BtuG homologs (Supplementary file 1) indicates the relative homology of three BtuG homologs from B. vulgatus, B. uniformis and B. coprophilus (BVU2056, BACUNI04578 and BACCOPRO02032, respectively; shown in red). (B) Purified proteins (and a no-protein control) were incubated with B12, filtered to remove free vitamin, and quantified for absorbance at 360 nm (corresponding to vitamin B12 absorbance). BSA, bovine serum albumin; B12, no-protein control after B12 incubation and filtering. *p<0.05 for samples compared against no-protein control (B12); error bars indicate ± SD from two technical replicates.
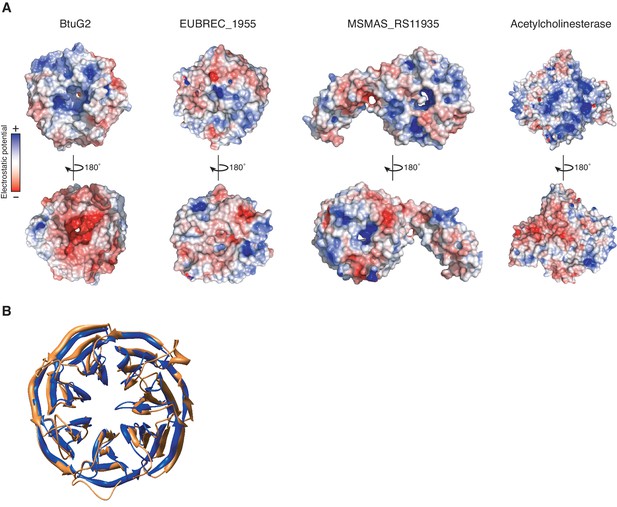
Surface electrostatic profiles of the seven-bladed β-propeller proteins BtuG2, EUBREC_1955 and MSMAS_RS11935, and the globular enzyme acetylcholinesterase.
(A) Model of surface electrostatic profile based on the crystal structure of BtuG2 (PDB 3DSM; left) reveals a largely positive electrostatic potential (blue) on the face of BtuG2 harboring the C-terminal 6xHis tag, and a largely negative electrostatic potential (red) on the opposing face, unlike the unrelated seven-bladed β-propellers EUBREC_1955 from the human gut anaerobe Eubacterium rectale (PDB 3S25; center left) and MSMAS_RS11935 from the methanogenic archeon Methanosarcina mazei (PDB 1L0Q; center right). Opposing electrostatic charge surfaces on enzymes like acetylcholinesterase (PDB 1GQR; right) are thought to aid in ligand orientation and binding during catalysis (Tan et al., 1993; Getzoff et al., 1983; Ripoll et al., 1993). (B) Structural similarity between BtuG2 (PDB 3DSM; gold) and the β-propeller domain of MSMAS_RS11935 (PDB 1L0Q; blue) (Jing et al., 2002). These domains are superimposed with a root mean square deviation of 1.75 Å.
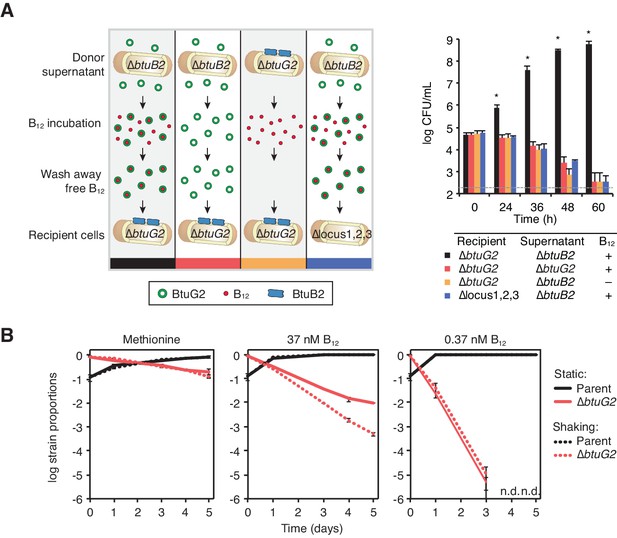
BtuG2 can function extracellularly as a corrinoid delivery protein and confers a fitness advantage to BtuG2 producer cells.
(A) Schematic (left) and measured CFUs (right) from an experiment in which B. thetaiotaomicron recipient cultures (∆btuG2 or ∆locus1,2,3) received donor supernatant from B. thetaiotaomicron donor strains (∆btuB2 or ∆btuG2), with or without vitamin B12. Recipient cultures were plated for CFUs over time. Data are representative of four independent trials; *p<0.05 for black bars compared against red, yellow or blue bars; error bars indicate ± SD from two biological replicates. (B) B. thetaiotaomicron parent and ∆btuG2 strains were co-cultured in minimal media supplemented with methionine or the indicated concentrations of vitamin B12 and incubated at 37 ˚C anaerobically either statically (solid lines) or shaking (dotted lines). Cells were passaged into fresh media daily and strain abundances were determined by qRT-PCR using barcode-specific primers. n.d., not detected; data are representative of two independent trials; error bars indicate ± SD from three technical replicates.
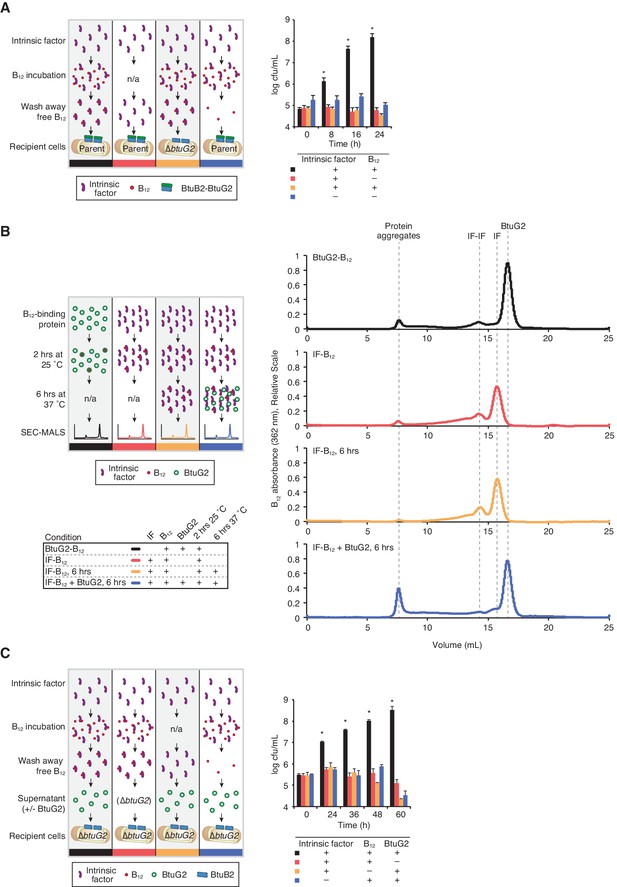
BtuG2 mediates vitamin B12 piracy from host intrinsic factor.
(A) Schematic (left) and measured CFUs (right) from an experiment in which B. thetaiotaomicron recipient cultures (parent or ∆btuG2) received recombinant human IF with or without vitamin B12, or the filtrate from the last IF–B12 wash. Recipient cultures were plated for CFUs over time. Data are representative of two independent trials; *p<0.05 for black bars compared against red, yellow or blue bars; error bars indicate ± SD from two biological replicates. (B) Schematic (left) and SEC-MALS traces at 362 nm absorbance (right) of recombinant human IF and/or recombinant BtuG2 incubated with vitamin B12. 362 nm absorbance measures B12-associated proteins and each trace represents one of the four conditions illustrated in the schematic. Data are representative of two independent trials. (C) Schematic (left) and CFUs (right) from an experiment in which B. thetaiotaomicron recipient cultures (∆btuG2) received recombinant human IF with or without vitamin B12 or the filtrate from the last IF–B12 wash, and donor supernatant from B. thetaiotaomicron strains (∆btuB2 or ∆btuG2). Recipient cultures were plated for CFUs over time. Data are representative of two independent trials; *p<0.05 for black bars compared against red, yellow or blue bars; error bars indicate ± SD from two biological replicates.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (E. coli) | S17-1 lambda pir | PMID_6340113 | thi pro hdsR hdsM + recA, chromosomal insertion of RP4-2(Tc::Mu Km::Tn7), AmpS | |
| Strain, strain background (E. coli) | BL21 Rosetta (DE3) | Novagen | F- ompT hsdSB(rB- mB-) gal dcm (DE3) pRARE (CamR) | |
| Strain, strain background (Bacteroides thetaiotaomicron) | VPI-5482 ∆tdk | PMID_18611383 | ||
| Strain, strain background (B. thetaiotaomicron) | VPI-5482 ∆tdk ∆locus1 ∆locus3 | PMID_24439897 | ||
| Strain, strain background (B. thetaiotaomicron) | VPI-5482 ∆tdk ∆locus1 ∆btuG2 ∆locus3 | This paper | ||
| Strain, strain background (B. thetaiotaomicron) | VPI-5482 ∆tdk ∆locus1 ∆btuB2 ∆locus3 | PMID_24439897 | ||
| Strain, strain background (B. thetaiotaomicron) | VPI-5482 ∆tdk ∆locus1 ∆locus2 ∆locus3 | PMID_24439897 | ||
| Strain, strain background (B. thetaiotaomicron) | VPI-5482 ∆tdk ∆locus1 ∆locus3 att::pNBU2_tet_BC01 | This paper | ||
| Strain, strain background (B. thetaiotaomicron) | VPI-5482 ∆tdk ∆locus1 ∆btuG2 ∆locus3 att::pNBU2_tet_BC14 | This paper | ||
| Strain, strain background (B. thetaiotaomicron) | VPI-5482 ∆tdk ∆locus1 ∆btuB2 ∆locus3 att::pNBU2_tet_BC14 | This paper | ||
| Strain, strain background (B. thetaiotaomicron) | VPI-5482 ∆tdk ∆locus1 ∆btuG2 ∆locus3 att::pNBU2_tet_BC16_us1957_btuG2 | This paper | ||
| Strain, strain background (E. coli) | BL21 Rosetta (DE3) pET21_NESG_btuG2 | This paper | ||
| Strain, strain background (E. coli) | BL21 Rosetta (DE3) pET21_NESG_btuG2_10xHis | This paper | ||
| Strain, strain background (E. coli) | BL21 Rosetta (DE3) pET21_NESG_BVU2056 | This paper | ||
| Strain, strain background (E. coli) | BL21 Rosetta (DE3) pET21_NESG_BACUNI04578 | This paper | ||
| Strain, strain background (E. coli) | BL21 Rosetta (DE3) pET21_NESG_BACCOPRO02032 | This paper | ||
| Recombinant DNA reagent | pExchange-tdk | PMID: 18611383 | plasmid | |
| Recombinant DNA reagent | pExchange_tdk_∆btuG2 | plasmid | ||
| Recombinant DNA reagent | pNBU2_ermG | PMID: 18611383 | plasmid | |
| Recombinant DNA reagent | pNBU2_ermG_us1957 | This paper | plasmid | |
| Recombinant DNA reagent | pNBU2_tet_BC01 | PMID: 18996345 | plasmid | |
| Recombinant DNA reagent | pNBU2_tet_BC14 | PMID: 18996345 | plasmid | |
| Recombinant DNA reagent | pNBU2_tet_BC16 | PMID: 24439897 | plasmid | |
| Recombinant DNA reagent | pNBU2_erm_us1957_btuG2_C32A | This paper | plasmid | |
| Recombinant DNA reagent | pNBU2_erm_us1957_btuG2_∆ss | This paper | plasmid | |
| Recombinant DNA reagent | pNBU2_erm_us1957 _btuG2_K34A W35A D36A | This paper | plasmid | |
| Recombinant DNA reagent | pNBU2_erm_us1957_btuG2_susD-ss | This paper | plasmid | |
| Recombinant DNA reagent | pNBU2_erm_us1957_btuG2_susD-ss-LES | This paper | plasmid | |
| Recombinant DNA reagent | pNBU2_erm_us1957_btuB2_FLAG_HA | This paper | plasmid | |
| Recombinant DNA reagent | pNBU2_erm_us1957 _BT1763_FLAG_HA | This paper | plasmid | |
| Recombinant DNA reagent | pNBU2_erm_us1957 _BT3704_HA | This paper | plasmid | |
| Recombinant DNA reagent | pET21_NESG_btuG2 | Northeast Structural Genomics Consortium; PDB 3DSM | plasmid | |
| Recombinant DNA reagent | pET21_NESG_btuG2_10xHis | This paper | plasmid | |
| Recombinant DNA reagent | pET21_NESG_BVU2056 | This paper | plasmid | |
| Recombinant DNA reagent | pET21_NESG_ BACUNI04578 | This paper | plasmid | |
| Recombinant DNA reagent | pET21_NESG_ BACCOPRO02032 | This paper | plasmid |
Additional files
-
Supplementary file 1
(Table S1) Identified BtuB-BtuG pairs from BlastP and Phyre2 searches of BT1490, BT1954 and BT2095.
(Table S2) Amino acid sequences of 114 BtuG homologs used to create the sequence logo in Figure 2A. (Table S3) Bacterial strains, plasmids and oligonucleotide primers used in this study.
- https://doi.org/10.7554/eLife.37138.012
-
Transparent reporting form
- https://doi.org/10.7554/eLife.37138.013





