Pharmacologic ATF6 activating compounds are metabolically activated to selectively modify endoplasmic reticulum proteins
Figures
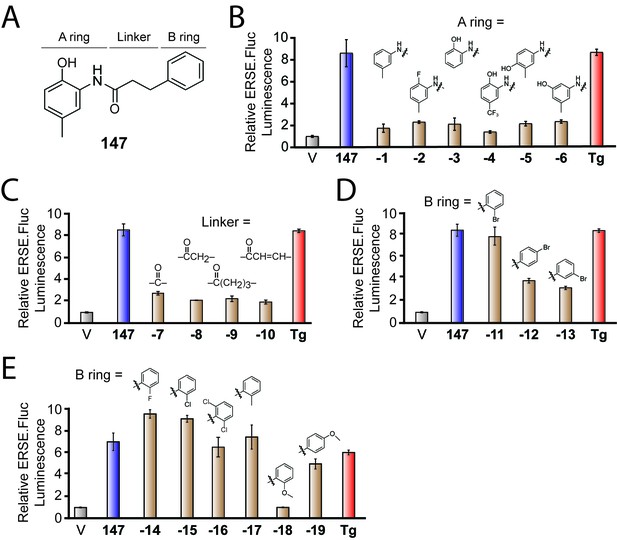
Activities of 147 analogs toward ATF6 activation.
(A) Structure of 147 with its three components – the A-ring, B-ring, and linker – indicated. (B) Bar graph showing the relative activation of the ERSE.FLuc ATF6 transcriptional reporter in HEK293T cells treated with 147 (10 µM; 18 hr), the ER stressor thapsigargin (Tg; 0.5 µM; 18 hr), or the indicated analogs of 147 where the A-ring was varied (10 µM; 18 hr). Error bars show SEM for three technical replicates. (C) Bar graph as in (B) where HEK293T cells were treated with the indicated 147 analogs where the linker was varied (10 µM; 18 hr). Error bars show SEM for three technical replicates. (D) Bar graph as in (B) where HEK293T cells were treated with the indicated 147 analogs in which a bromine atom was placed in the ortho, para, and meta positions of the B-ring (10 µM; 18 hr). Error bars show SEM for three technical replicates. (E) Bar graph as in (B) where HEK293T cells were treated with the indicated 147 analogs with a variety of substituents incorporated onto the B-ring. Error bars show SEM for three technical replicates.
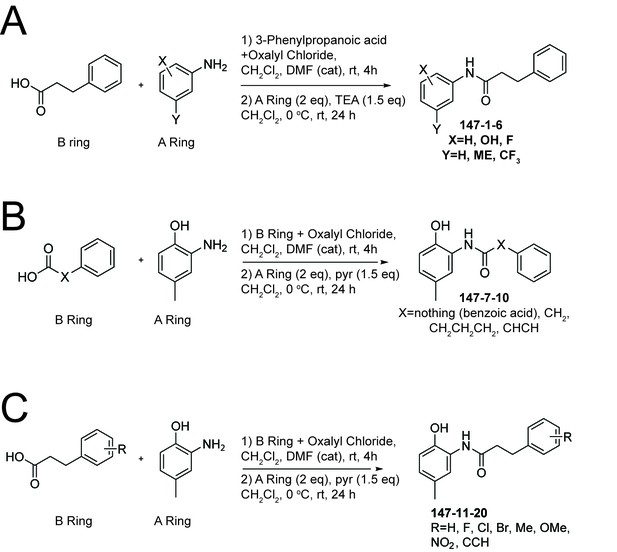
Synthetic schemes used to prepare analogs of 147.
(A) Synthetic scheme for analogs with A-Ring modifications. (B) Synthetic scheme for analogs with chain linker modifications. (C) Synthetic scheme for analogs with B-Ring modifications.
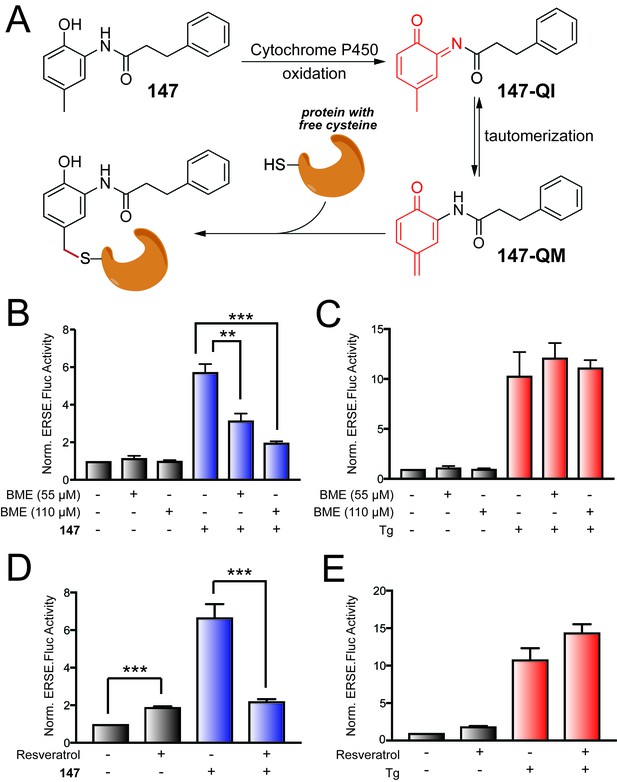
147-dependent ATF6 activation is sensitive to increases in cellular free thiols and resveratrol.
(A) Illustration showing a mechanism whereby 147 is converted to an o-quinone imine (147-QI) followed by tautomerization to a quinone methide (147-QM), which can then react with cellular proteins through nucleophiles such as free Cys side chains. (B) Bar graph showing the activation of the ERSE.FLuc ATF6 reporter in HEK293T cells treated with 147 (10 µM) and/or β-mercaptoethanol (BME; 55 μM or 110 μM) for 18 hr. Error bars show SEM for 3 independent experiments. *p<0.05. (C) Bar graph showing the activation of the ERSE.FLuc ATF6 reporter in HEK293T cells treated with thapsigargin (Tg; 0.5 µM) and/or β-mercaptoethanol (BME; 55 μM or 110 μM) for 18 hr. Error bars show SEM for 3 independent experiments. (D) Bar graph showing the activation of the ERSE.FLuc ATF6 reporter in HEK293T cells treated with 147 (10 µM) and/or resveratrol (10 µM) for 18 hr. Error bars show SEM for 4 independent experiments. ***p<0.005. (E) Bar graph showing the activation of the ERSE.FLuc ATF6 reporter in HEK293T cells treated with Tg (500 nM) and/or resveratrol (10 µM) for 18 hr. Error bars show SEM for 4 independent experiments.
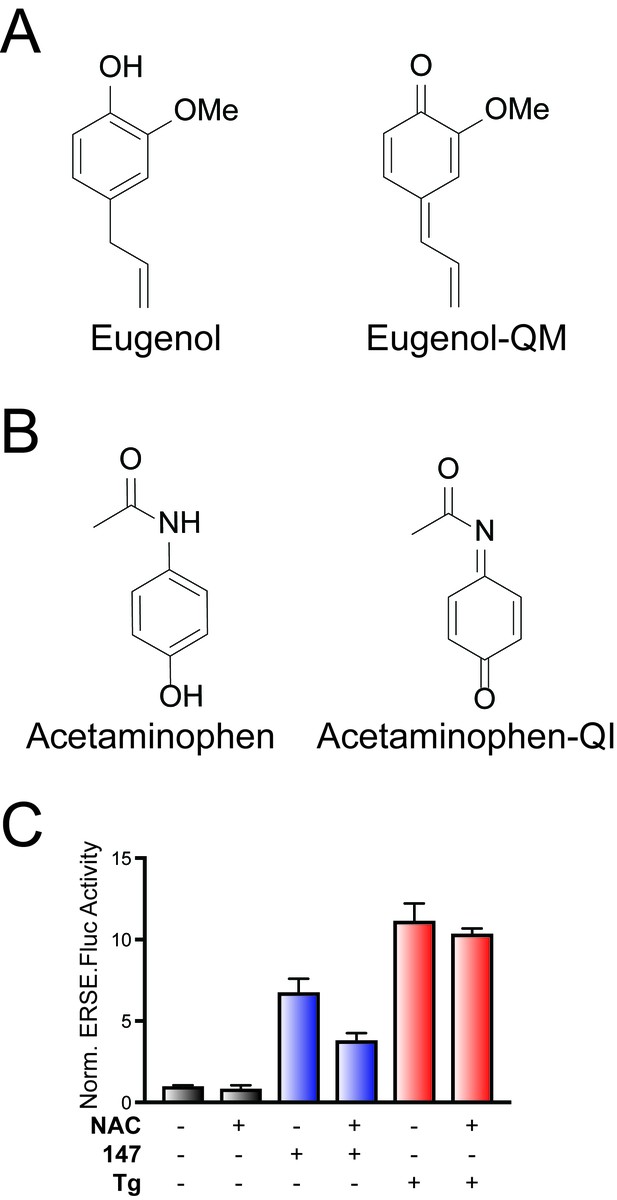
147-dependent ATF6 activation is sensitive to increases in cellular free thiols and resveratrol.
(A) Structure of Eugenol, which can be metabolically oxidized to create a p-quinone methide similar to that predicted to be formed in the mechanism of 147-dependent ATF6 activation (see Figure 2A). (B) Structure of acetaminophen, which can be metabolically activated to an N-acetyl-p-benzoquinone imine, similar to that which may form in the oxidation of 147 (see Figure 2A). (C) Bar graph showing the activation of the ERSE.FLuc ATF6 reporter in HEK293T cells treated with 147 (10 µM), thapsigargin (Tg; 0.5 µM) and/or N-acetyl cysteine (NAC; 5 mM), added 4 hr prior to addition of 147 or Tg, for 18 hr. Error bars show SEM for three technical replicates.
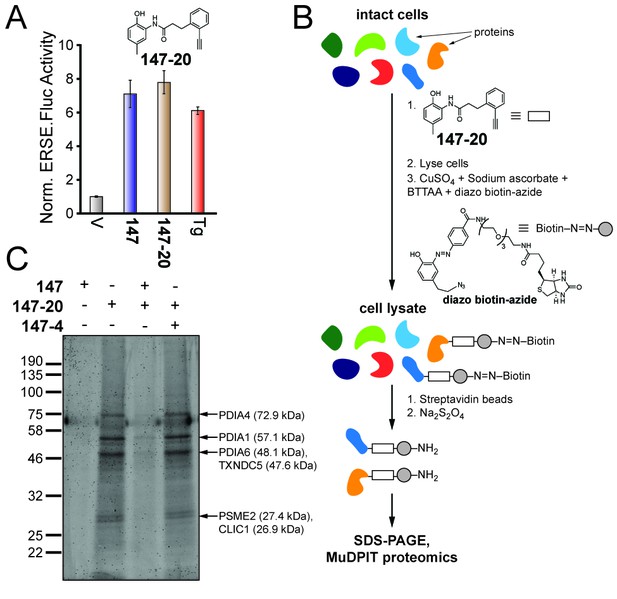
Compound 147 covalently modifies cellular proteins.
(A) Bar graph showing the relative activation of the ERSE.FLuc ATF6 reporter in HEK293T cells treated with 147 (10 µM), 147–20 (10 µM), or thapsigargin (Tg; 0.5 µM) for 18 hr. Error bars show SEM for three technical replicates. (B) Schematic showing the protocol for affinity purification of proteins covalently modified by 147–20. (C) Coomassie-stained SDS-PAGE of affinity purified proteins from ALMC-2 cells treated with 147 (10 µM), or 147–20 (10 µM), or the combination of the 147–20 (10 µM) and 147 (50 µM), or 147–20 (10 µM) and 147–4 (50 µM) in combination for 18 hr. Specific proteins identified by mass spectrometry of excised bands are indicated.
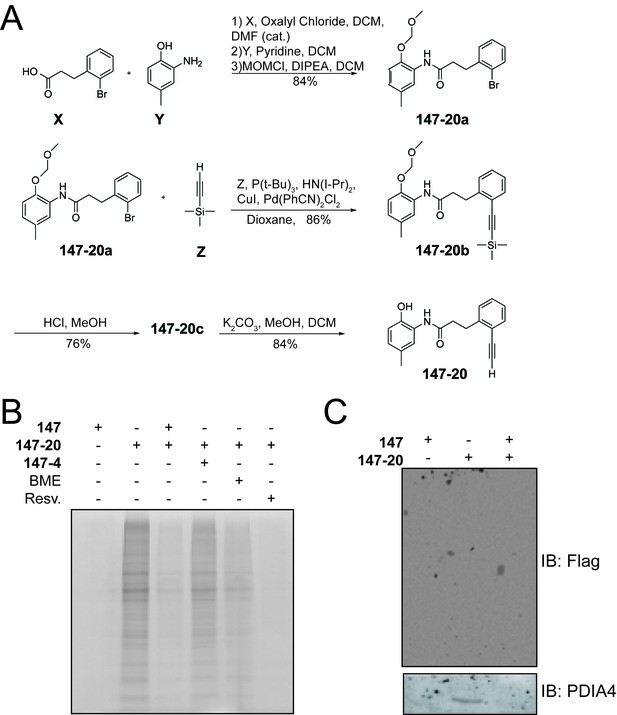
Compound 147 covalently modifies cellular proteins.
(A) Synthetic scheme for the preparation of 147–20. (B) Coomassie-stained SDS-PAGE of affinity purified proteins from ALMC-2 cells treated with 147 (10 µM), or 147–20 (10 µM), or the combination of 147–20 (10 µM) and 147 (50 µM), or the combination of 147–20 (10 µM) and 147–4 (50 µM),or the combination of 147 (10 µM) and β-mercaptoethanol (BME; 110 μM), or the combination of 147 (10 µM) and resveratrol (10 µM) for 18 hr. (C) Immunoblot of affinity purified FLAG-tagged ATF6 from HEK293T cells stably expressing FLAG-tagged ATF6 treated with 147 (10 µM), 147–20 (10 µM), or 147–20 (10 µM) and 147 (50 µM) in combination for 18 hr.
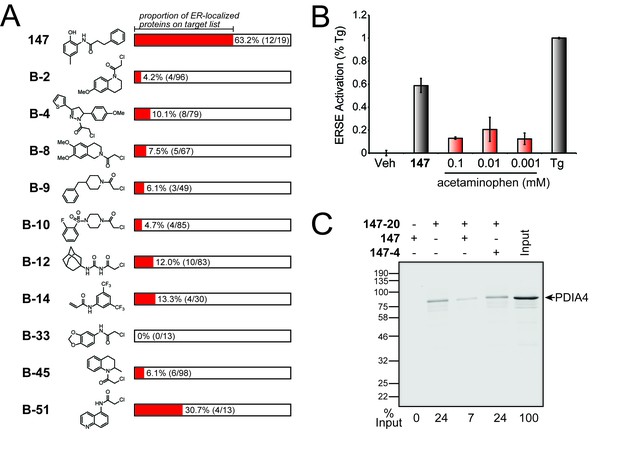
Comparison of S-reactive electrophiles demonstrating that compound 147 selectively modifies ER-localized proteins.
(A) Proportion of ER-localized proteins covalently labeled by 147 or fragment electrophiles reported in (Backus et al., 2016). The ER-localized proportion for each electrophile indicated in red. The numbers in parentheses indicate the number of ER-localized proteins in a given electrophile’s target list on the left and the total number of proteins in the target list on the right. The electrophiles from (Backus et al., 2016) are denoted with a B- followed by the compound numbers used in that work. (B) Bar graph showing activation of the ERSE.FLuc reporter in HEK293T cells treated with 147 (10 µM), thapsigargin (Tg; 0.5 µM) or the indicated dose of acetaminophen for 18 hr. Error bars show SEM for 3 independent experiments. (C) Immunoblot of PDI4 in 147–20 affinity purified proteins from HEK293T cells treated with 147 (10 µM), or 147–20 (10 µM), or the combination of 147–20 (10 µM) and 147 (50 µM), or the combination of 147–20 (10 µM) and 147–4 (50 µM) for 18 hr. The relative recovery of PDIA4 under these different conditions is indicated below the immunoblot.
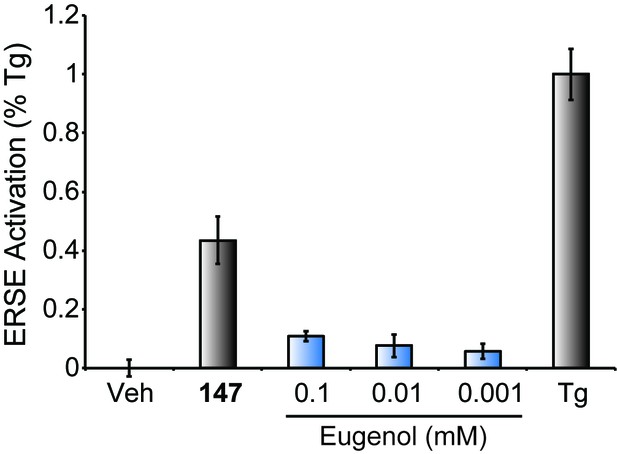
Compound 147 selectively modifies ER-localized proteins.
Graph showing activation of the ERSE.FLuc reporter in HEK293T cells treated with 147 (10 µM), thapsigargin (Tg; 0.5 µM) or the indicated dose of eugenol for 18 hr. Error bars show SEM for 3 independent experiments.
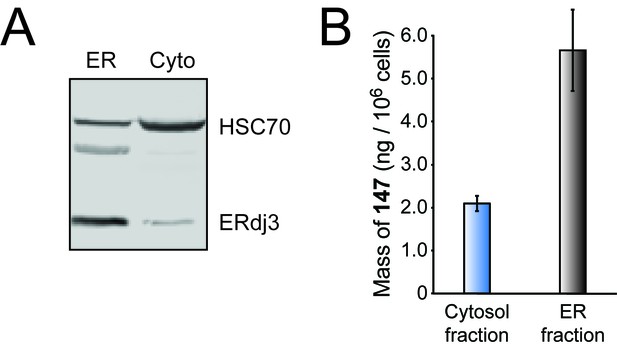
Compound 147 preferentially partitions to the ER.
(A) Western blot for ERdj3 (an ER-resident protein) and HSC70 (a cytosolic protein) in the ER and cytosolic fractions from a digitonin extraction of HEK293T cells after 15 min of treatment with 147 (10 μM). (B) Enrichment of 147 as determined by mass spectrometry in the cytosolic fraction versus the ER fraction of HEK293T cells after 15 min of treatment with 147 (10 µM). Error bars show SEM for 3 independent experiments.
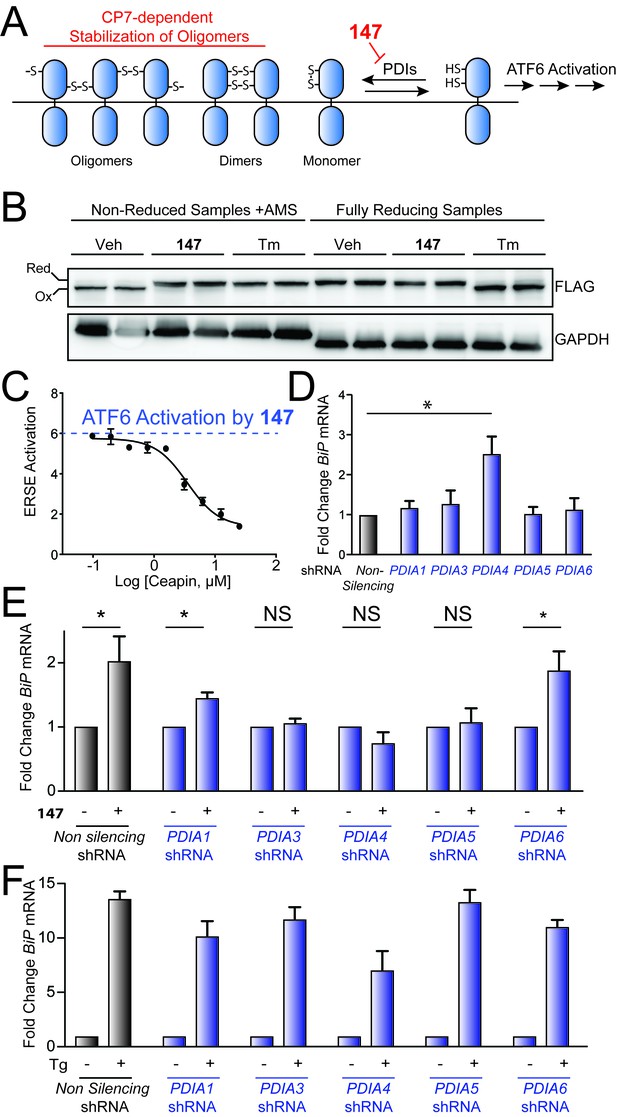
Genetic depletion of select PDIs alters 147-dependent ATF6 activation.
(A) Mechanistic model of 147-dependent ATF6 activation, as described in the main text. The red component of the figure shows how pharmacologic modulation of ATF6 by 147 or CP7 influences ATF6 activity. (B) Immunoblot showing lysates prepared from HeLa cells stably expressing FLAG-tagged ATF6 treated for 6 hr with 147 (10 µM) or tunicamycin (Tm; 1 mg/mL). Lysates were separated by non-reducing or reducing SDS-PAGE prior to immunoblotting. The bands representing oxidized and reduced ATF6 are indicated. Note the increased ATF6 migration observed in reducing gels for Tm-treated cells, reflecting the inhibited N-linked glycosylation of ATF6 in these samples. (C) Graph showing relative activation of the ERSE.Fluc ATF6 reporter in HEK293T cells co-treated for 18 hr with 147 (10 µM) and increasing concentrations of CP7, as indicated. Error bars show SEM for three technical replicates. (D) Bar graph showing BiP expression in HEK293T cells stably expressing non-silencing, PDIA1, PDIA3, PDIA4, PDIA5, or PDIA6 shRNA, as indicated. BiP expression in the PDI-depleted cells is shown relative to cells expressing non-silencing shRNA. Error bars show SEM for 3 independent experiments. *indicates p<0.05. (E) Bar graph showing BiP expression in HEK293T cells expressing non-silencing, PDIA1, PDIA3, PDIA4, PDIA5, or PDIA6 shRNA treated for 6 hr with or without 147 (10 µM). BiP expression levels for samples treated with 147 are normalized to the corresponding vehicle-treated controls. Un-normalized data are shown in Figure 5—figure supplement 1B. Error bars show SEM for 3 independent experiments. *indicates p<0.05. (F) Bar graph showing BiP expression in HEK293T cells expressing non-silencing, PDIA1, PDIA3, PDIA4, PDIA5, or PDIA6 shRNA treated for 6 hr with or without thapsigargin (Tg; 0.5 µM). BiP expression levels for samples treated with Tg are normalized to the corresponding vehicle-treated controls. Un-normalized data are shown in Figure 5—figure supplement 1C Error bars show SEM for 3 independent experiments.
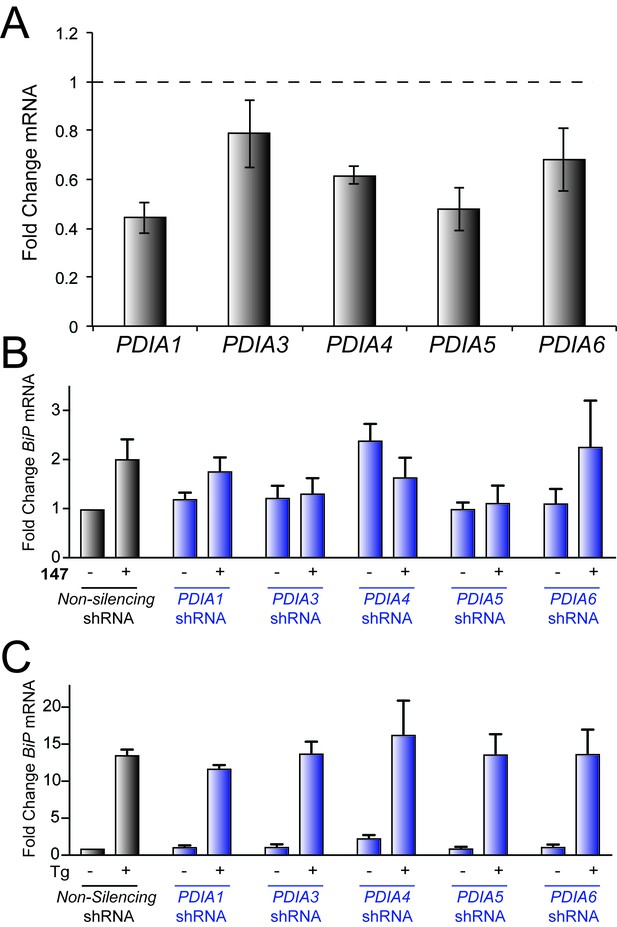
Genetic depletion of select PDIs disrupts 147-dependent ATF6 activation.
(A) Bar graph showing expression of PDIA1, PDIA3, PDIA4, PDIA5, or PDIA6 in HEK293T cells depleted for the same PDI. The dashed line reflects expression of each PDI in control cell lines expressing non-silencing shRNA. Error bars show ± 95% confidence interval. (B), (C) Un-normalized data for Figure 5E and Figure 5F, respectively. Error bars show SEM for 3 independent experiments.
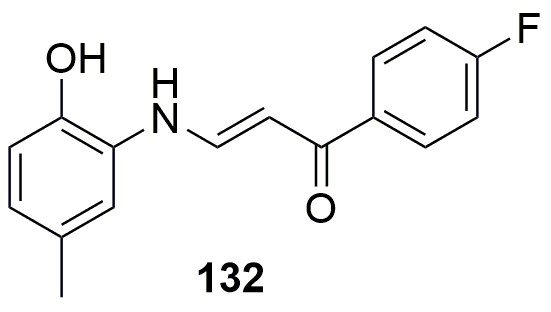
Structure of 132.
https://doi.org/10.7554/eLife.37168.016Tables
Protein targets that are covalently modified by 147.
https://doi.org/10.7554/eLife.37168.008| Protein name | Symbol | Uniprot accession | Competition Ratio* | Redox?† | ER-localization?‡ |
|---|---|---|---|---|---|
| Cytoskeleton-associated protein 4 | CKAP4 | Q07065 | 2.8 (1.7, 3.5) | N | Y |
| Protein disulfide isomerase A4 | PDIA4 | P13667 | 2.4 (1.8, 3.3) | Y | Y |
| Protein disulfide isomerase A6 | PDIA6 | Q15084 | 2.4 (2.0, 3.0) | Y | Y |
| Protein disulfide isomerase A1 (a.k.a. Prolyl 4-hydroxylase subunit beta) | PDIA1 (a.k.a. P4HB) | P07237 | 2.3 (2.1, 3.1) | Y | Y |
| Protein disulfide isomerase A3 | PDIA3 | P30101 | 2.2 (1.8, 3.1) | Y | Y |
| Thioredoxin | TXN | P10599 | 2.2 (1.2, 3.5) | Y | N |
| Thioredoxin domain-containing protein 5 | TXNDC5 | Q8NBS9 | 2.2 (2.0, 2.8) | Y | Y |
| Heme oxygenase 2 | HMOX2 | P30519 | 2.2 (1.4, 4.3) | N | Y |
| Chloride intracellular channel 1 | CLIC1 | O00299 | 1.9 (1.5, 2.3) | N | N |
| Thioredoxin-related transmembrane protein 1 | TMX1 | Q9H3N1 | 1.9 (1.5, 4.3) | Y | Y |
| Voltage dependent anion channel 2 | VDAC2 | P45880 | 1.6 (1.2, 1.9) | N | N§ |
| Poly(rC)-binding protein 1 | PCBP1 | Q15365 | 1.6 (1.4, 1.8) | N | N |
| Ribosome-binding protein 1 | RRBP1 | Q9P2E9 | 1.5 (1.3, 1.8) | N | Y |
| Proteasome subunit alpha type 1 | PSMA1 | P25786 | 1.5 (1.1, 1.7) | N | N |
| d-3-phosphoglycerate kinase | PHGDH | O43175 | 1.5 (1.4, 1.9) | N | N |
| Histocompatibility minor 13 | HM13 | Q8TCT9 | 1.5 (1.2, 2.0) | N | Y |
| Polyadenylate-binding protein 4 | PABPC4 | Q13310 | 1.5 (1.1, 1.7) | N | N |
| Ribophorin 1 | RPN1 | P04843 | 1.4 (1.3, 1.8) | N | Y |
| ER resident protein 29 | ERP29 | P30040 | 1.4 (1.0, 1.8) | N | Y |
-
*Competition ratio = Ratio of TMT reporter ions for the protein in question between samples treated with 147–20 and those co-treated with 147–20 and 147. The median value across all cell lines and replicates are shown, with the values at the 1st and 3rd quartiles shown in parenthesis.
†Indicates whether the protein in question has the ‘cell redox homeostasis’ GO annotation (GO:0045454).
-
‡Indicates whether the protein in question has the ‘endoplasmic reticulum part’ GO annotation (GO:0044432).
§VDAC2 is a mitochondrial membrane protein but has been shown to localize to sites of mitochondria that are associated with the ER.
-
Table 1—source data 1
Excel spreadsheet showing the competition ratio data for all 90 proteins that were identified as targets of 147 in every replicate and every cell type.
- https://doi.org/10.7554/eLife.37168.009
-
Table 1—source data 2
Excel spreadsheet showing all the proteins identified among the affinity-purified targets of 147.
- https://doi.org/10.7554/eLife.37168.010
Additional files
-
Supplementary file 1
Supplementary experimental methods, including lentivirus production, qPCR primers and synthetic methods and characterization data for small molecules.
- https://doi.org/10.7554/eLife.37168.017
-
Transparent reporting form
- https://doi.org/10.7554/eLife.37168.018





