Peptidoglycan precursor synthesis along the sidewall of pole-growing mycobacteria
Figures
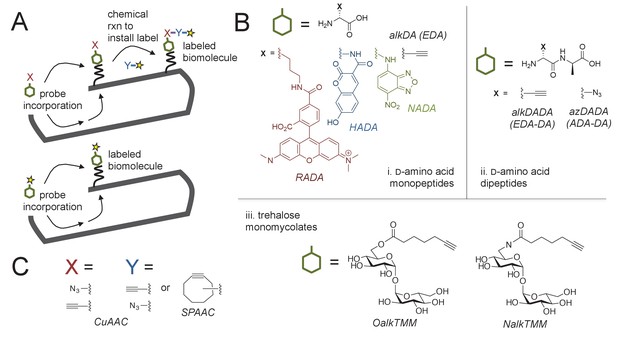
Cell envelope metabolic labeling in mycobacteria.
(A) Schematic of one- and two-step metabolic labeling. Top, a cell envelope precursor or ‘probe’ bearing a reactive group is incorporated into the envelope by the endogenous enzymatic machinery of the cell. The presence of the probe is then revealed by a chemical reaction with a label that bears a complementary reactive group. Bottom, in some cases the probe can be pre-labeled, bypassing the chemical ligation step and embedding the detection moiety directly into the macromolecule. Yellow star, fluorophore. See (Siegrist et al., 2015) for more details. (B) Probes used in this work to mark the mycobacterial envelope. See text for details. Colored and black chemical structures denote probes used in one- and two-step labeling, respectively. C, X and Y reactive partners used in this work for two-step labeling as shown in A. CuAAC, copper-catalyzed azide-alkyne cycloaddition; SPAAC, strain-promoted azide-alkyne cycloaddition.
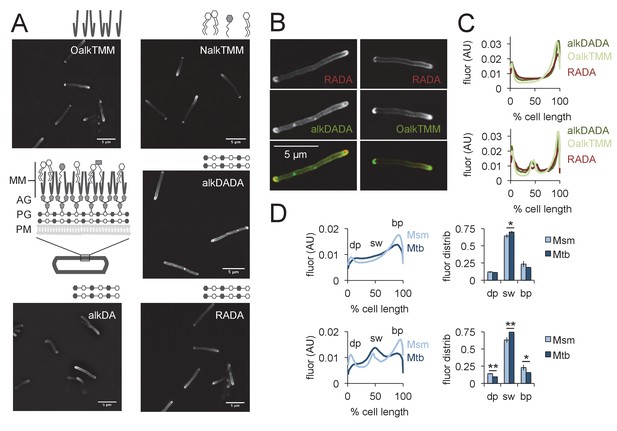
Asymmetric polar gradients of cell envelope metabolic labeling in mycobacteria.
(A) M. smegmatis was incubated for 15 min (~10% generation) in the indicated probe, then washed and fixed. Alkynyl probes were detected by CuAAC with azido-CR110 and cells were imaged by structured illumination microscopy. MM, mycomembrane; AG, arabinogalactan; PG, peptidoglycan; PM, plasma membrane. (B) M. smegmatis dual labeled with RADA and alkDADA, left, or RADA and OalkTMM, right, and imaged by conventional fluorescence microscopy. (C) M. smegmatis was labeled as in B and cellular fluorescence was quantitated for cells without (top; 77 < n < 85) or with (bottom; 9 < n < 51) visible septa for RADA, OalkTMM and alkDADA. Signal was normalized to cell length and to total fluorescence intensity. Cells were oriented such that the brighter pole is on the right hand side of the graph. (D) M. smegmatis (Msm; light blue) or M. tuberculosis (Mtb; dark blue) was labeled with HADA for 15 min or 2 hr (~10% generation), respectively, then washed and fixed. Fluorescence was quantitated as in C for cells without (top; 118 < n < 332) and with (bottom; 55 < n < 85) visible septa. We defined the dim pole (dp) as the sum of the fluorescence intensity over the first 15% of the cell; the sidewall (sw) as the sum of the middle 70%; and the bright pole (bp) as the sum over the final 15% of the cell. Fluor distrib, average fluorescence distribution. AU, arbitrary units. Error bars, ±standard deviation. Statistical significance between M. smegmatis (five biological replicates) and M. tuberculosis (three biological replicates) was assessed for the dim pole, sidewall and bright pole by two-tailed Student’s t test. *p<0.05; **p<0.005.
-
Figure 2—source data 1
Conventional fluorescence and structured illumination microscopy images of mycobacteria labeled with peptidoglycan and trehalose monomyocolate probes.
- https://doi.org/10.7554/eLife.37243.008
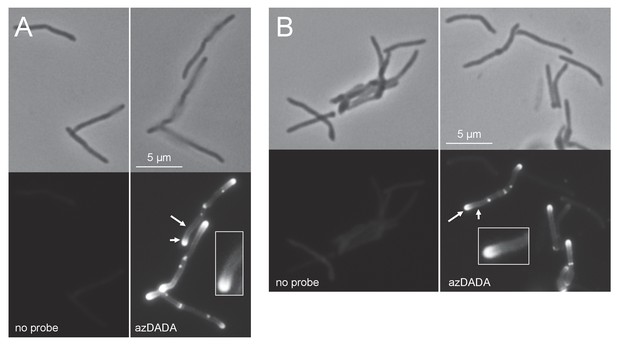
Metabolic labeling by azDADA comprises polar gradients in live M. smegmatis.
Bacteria were labeled with azDADA for 15 min and subjected to either A, low copper CuAAC (with BTTP ligand), or B, SPAAC. Longer arrow, polar labeling. Short arrow, sidewall labeling.
-
Figure 2—figure supplement 1—source data 1
Conventional fluorescence microscopy images of AzDADA-labeled M. smegmatis revealed by SPAAC or CuAAC.
- https://doi.org/10.7554/eLife.37243.005
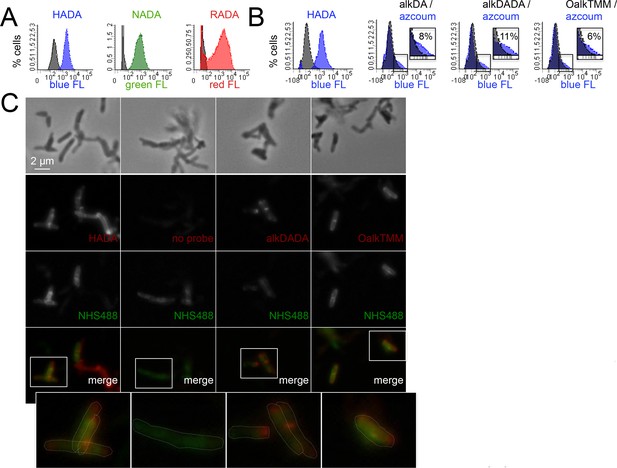
Heterogeneous envelope probe labeling in M. tuberculosis.
(A) M. tuberculosis was incubated or not for 20 hr in HADA (blue), NADA (green) or RADA (red), washed and fixed. Fluorescence was assessed by flow cytometry. Grey, unlabeled control. (B) M. tuberculosis was incubated or not for 2 hr with HADA, alkDA, alkDADA or OalkTMM (all blue). After washing and fixing as in A, cells labeled with alkynyl probes were subjected to CuAAC with azido-coumarin. Fluorescence was assessed by flow cytometry. Grey, no probe control. Non-overlapping regions of the histograms that represent bacteria with probe-derived fluorescence are magnified and quantitated as % of total bacteria counted. (C) Fluorescence microscopy of M. tuberculosis briefly treated with a fluorescent amine-reactive dye (NHS488; green) then incubated or not for 2 hr with HADA, alkDADA or OalkTMM (red). Alkynyl probes detected as in B. Cell envelope labeling is apparent both along the sidewall, where it colocalizes with NHS488, as well as at the cell poles, where it does not. Dotted white lines highlight cell contours.
-
Figure 2—figure supplement 2—source data 1
Flow cytometry and conventional fluorescence microscopy images of M. tuberculosis labeled with different probes.
- https://doi.org/10.7554/eLife.37243.007
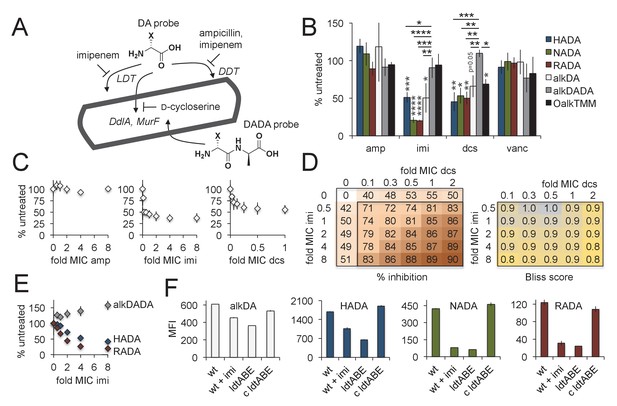
Multiple pathways of d-amino acid probe incorporation in M. smegmatis.
(A) Schematic of the theoretical routes of d-amino acid (DA) and d-alanine d-alanine (DADA) probe incorporation. LDT, l,d-transeptidase, DDT, d,d-transpeptidase (DDTs). For more details see Figure 3—figure supplement 1. (B) Sensitivity of HADA (blue), NADA (green), RADA (red), alkDA (light grey), alkDADA (dark grey) and OalkTMM (black) to antibiotics. Imi, imipenem + clavulanate; amp, ampicillin + clavulanate; dcs, d-cycloserine; vanc, vancomycin. M. smegmatis was pretreated or not with the indicated antibiotics at 2X MIC for 30 min then incubated an additional 15 min in the presence of probe. The bacteria were then washed and fixed. The alkyne-bearing probes were detected by CuAAC with azido-CR110 and quantitated by flow cytometry. Experiment was performed three to four times in triplicate. For each biological replicate, the averaged median fluorescence intensities (MFI) of the drug-treated samples were divided by the MFI of untreated bacteria. Data are expressed as the average percentage of untreated labeling across the biological replicates. Error bars, ±standard deviation. Statistical significance compared to alkDADA was assessed by two-tailed Student’s t test of percentages for biological replicates. Horizontal black stars: *p<0.05; **p<0.005; ***p<0.0005; ****p<0.00005. Statistical significance compared to probe-matched controls that were not treated with antibiotic was assessed by two-tailed Student’s t test of log10 MFI data for biological replicates. Vertical dark grey stars: *p<0.05; **p<0.005; ***p<0.0005; ****p<0.00005. (C) Effect of antibiotic dose on alkDA-derived fluorescence. M. smegmatis was pretreated or not with drugs at the fold-MIC indicated and labeled as in B. Experiment was performed three times in triplicate. For each biological replicate, the averaged MFI of the control (no drug, no alkDA but subjected to CuAAC) was subtracted from the averaged MFI of the drug-treated sample. This was then divided by the averaged MFI of untreated control (no drug but incubated in alkDA and subjected to CuAAC) from which the control MFI had also been subtracted. Data are expressed as the average percentage of untreated labeling across the biological replicates. Error bars, ±standard deviation. (D) Left, combined effects of imipenem and d-cycloserine on alkDA-derived fluorescence. M. smegmatis was pretreated or not with the drugs at the fold-MIC indicated and labeled as in B. Experiment was performed twice in triplicate with similar results. One data set is shown. The percent of untreated labeling was calculated as in C and subtracted from 100 to obtain the percent inhibition. Right, Bliss interaction scores for each pair of doses in left-hand graph were calculated as (EI + ED EIED)/EI,D where EI is the effect of imipenem at dose i, ED is the effect of d-cycloserine at dose d and EI,D is the observed effect of the drugs at dose i and dose d. Combinations that produce Bliss scores greater than, equal to, or less than one are, respectively, interpreted as antagonistic, additive, or synergistic interactions. (E) Dose-dependent effect of imipenem on alkDADA (grey), HADA (blue) and RADA (red) labeling. M. smegmatis was pretreated or not with imipenem at the fold-MIC indicated and labeled as in B. Experiment was performed three times in triplicate and one representative data set is shown. For each technical replicate, the averaged median fluorescence intensities (MFI) of the drug-treated samples were divided by the averaged MFI of untreated bacteria. Data are expressed as the average percentage of untreated labeling across the technical replicates. Error bars, ±standard deviation. (F) Wildtype M. smegmatis pre-treated or not with 2X MIC imipenem and untreated ΔldtABE and complement (c ldtABE) were labeled with alkDA (white), HADA (blue), NADA (green) or RADA (red) and processed as in B. Experiment was performed two to ten times in triplicate. Representative data from one of the biological replicates is shown here. Error bars, ±standard deviation.
-
Figure 3—source data 1
Flow cytometry data of mycobacteria incorporating peptidoglycan probes in different conditions.
- https://doi.org/10.7554/eLife.37243.013
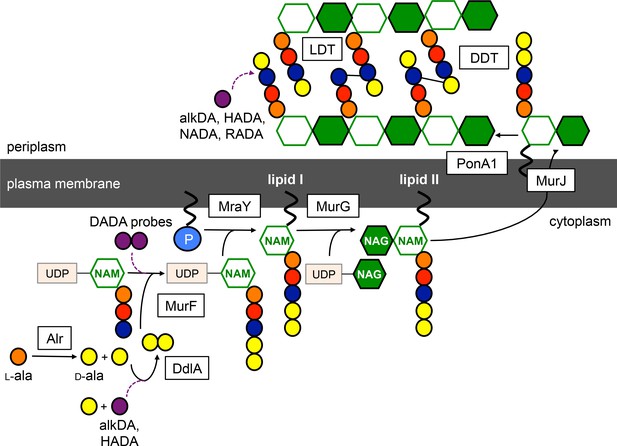
Schematic of peptidoglycan synthesis in mycobacteria.
Purple dashed lines, routes of probe incorporation proposed in this work. d,d-transpeptidases (DDT) and l,d-transpeptidases (LDT) respectively catalyze 4–3 and 3–3 crosslinks and can perform exchange reactions with free d-amino acids. Our data support l,d-transpeptidase-mediated exchange as the primary extracellular route of monopeptide probe incorporation into mycobacterial peptidoglycan.
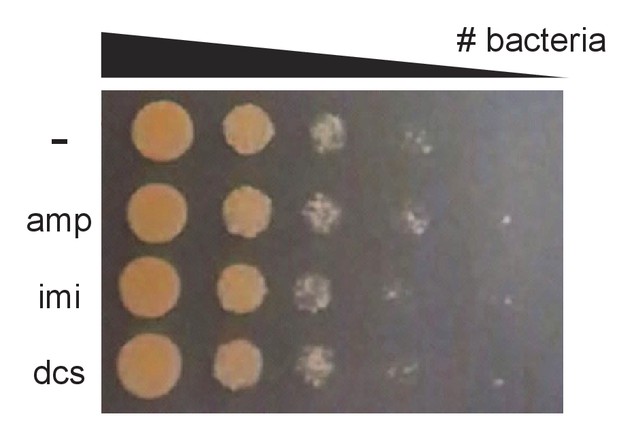
Antibiotics do not cause obvious cell death in 45 min.
M. smegmatis cultures were treated with 2X MIC of indicated antibiotics for 45 min, washed, and 10-fold serial dilutions were spotted onto LB agar.
-
Figure 3—figure supplement 2—source data 1
Growth of M. smegmatis in presence of antibiotics.
- https://doi.org/10.7554/eLife.37243.011
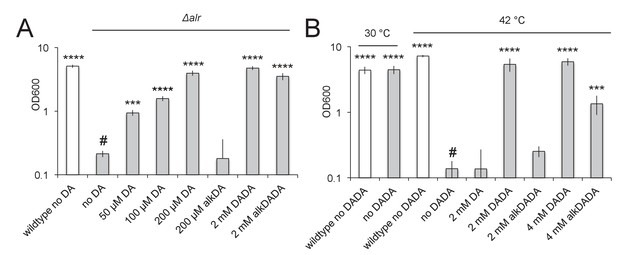
alkDADA rescues the growth of d-alanine racemase (Alr) and ligase (DdlA) mutants.
alkDADA probe supports the growth of Δalr, A, or temperature-sensitive ddlAts, B. Wildtype (white bars) and Δalr (grey bars), B, or ddlAts (grey bars), were grown in the presence or absence of exogenous d-alanine, d-alanine- d-alanine or alkynyl derivatives thereof. Error bars, ±standard deviation. Significant differences compared to Δalr no d-alanine (#, second bar from left), A, or ddlAts at 42˚C no d-alanine-d-alanine (#, fourth bar from left), B, One-way ANOVA with Dunnett’s test, are shown for 3–4 biological replicates. ***p<0.005, ****p<0.0005.
-
Figure 4—source data 1
OD600 measurements of growth in different mycobacterial strains and conditions.
- https://doi.org/10.7554/eLife.37243.020
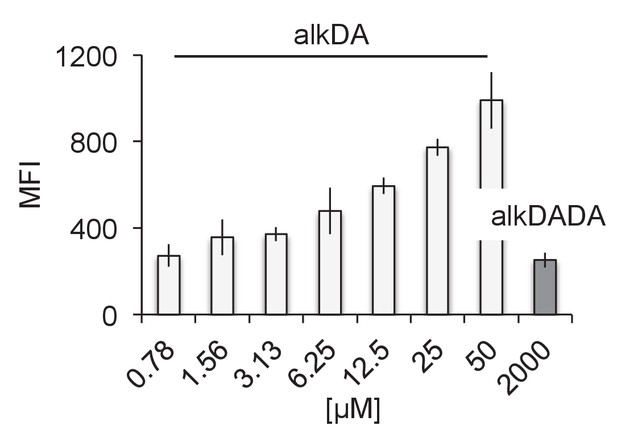
alkDA labels at much lower concentrations than alkDADA.
M. smegmatis were labeled with 2 mM of alkDADA (dark grey) or different concentrations of alkDA (light grey) for 15 min then fixed and subjected to CuAAC. MFI, median fluorescence intensity from which the control (no probe but subjected to CuAAC) was subtracted. Error bars, ± standard deviation.
-
Figure 4—figure supplement 1—source data 1
Flow cytometry data of M. smegmatis incorporating AlkDa and AlkDADA.
- https://doi.org/10.7554/eLife.37243.016
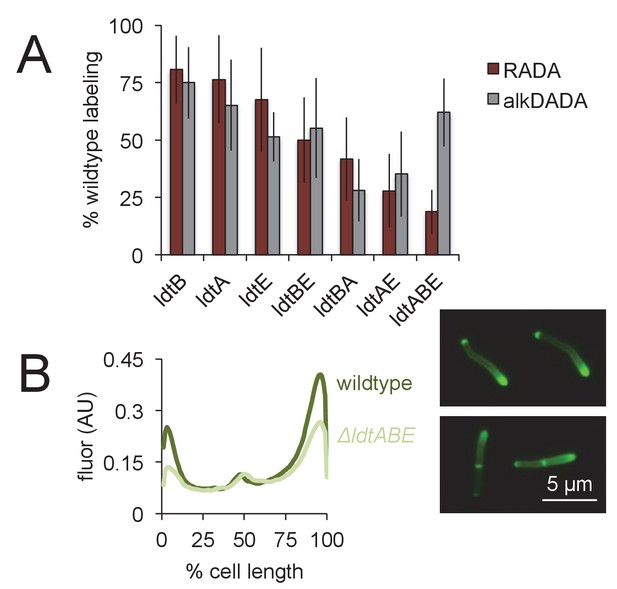
Loss of LdtA, LdtB and/or LdtE decreases both RADA and alkDADA labeling.
(A) Wildtype and ldt strains were labeled with RADA (red) or alkDADA (grey) for 30 min, then washed and fixed. alkDADA detected by CuAAC with azido-CR 110. Fluorescence was quantitated by flow cytometry. Data are expressed as percentage of untreated wildtype labeling as in Figure 3B. Experiment was performed 4–11 times in triplicate. Error bars, ± standard deviation. Differences within RADA- and alkDADA-labeled strains are significant at p<0.005, one-way ANOVA comparison of log10 data for biological replicates. (B) Wildtype (dark green) and ΔldtABE (light green) M. smegmatis were labeled with alkDADA for 15 min, then washed, fixed and subjected to CuAAC with azido-CR110 prior to imaging. Fluorescence was quantitated for 40 < n < 70. Signal was normalized by cell length and brighter poles oriented to the right-hand side of the graph. AU, arbitrary units. Experiment repeated twice with similar results. Representative images at right.
-
Figure 4—figure supplement 2—source data 1
Flow cytometry and conventional fluorescence images of Ldt mutants labeled with peptidoglycan probes.
- https://doi.org/10.7554/eLife.37243.018
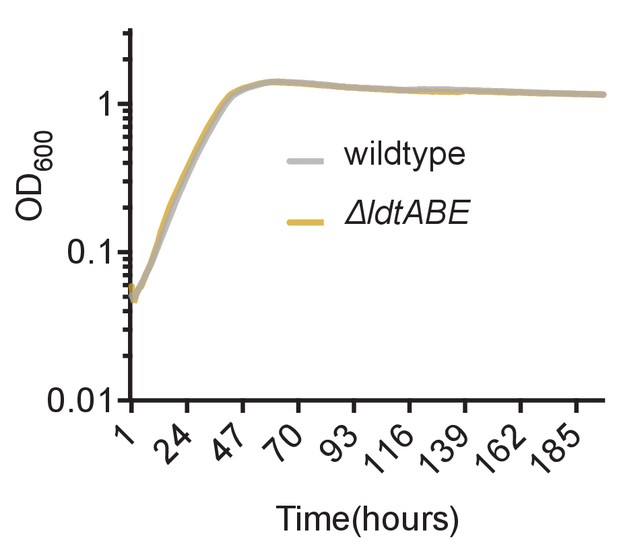
Loss of LdtA, LdtB and LdtE do not cause a growth defect in M. smegmatis.
Culture turbidity increases at the same rate for wildtype and ΔldtABE growing in 7H9 medium.
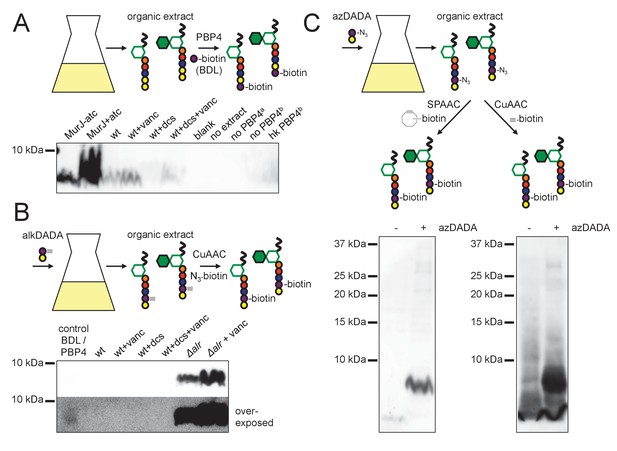
alkDADA and azDADA incorporate into lipid-linked peptidoglycan precursors.
(A) Detection of lipid-linked peptidoglycan precursors from organic extracts of M. smegmatis. Endogenous d-alanines (yellow) were exchanged for biotin-d-lysine (BDL; purple) via purified S. aureus PBP4. Biotinylated species detected by blotting with streptavidin-HRP. MurJ (MviN) depletion strain was incubated in anhydrotetracycline (atc) to induce protein degradation. Other strains were treated with vancomycin (vanc), d-cycloserine (dcs) or a combination prior to harvesting. Wt, wildtype; blank, no sample run; no extract, BDL and PBP4 alone a, organic extract from MurJ - atc; b, organic extract from MurJ + atc; hk, heat-killed. (B) Detection of lipid-linked peptidoglycan precursors labeled by alkDADA in vivo. Wildtype and Δalr strains were incubated in alkDADA (purple and yellow) and treated or not with the indicated antibiotics prior to harvest. Alkyne-tagged species from organic extracts were ligated to picolyl azide biotin via CuAAC then detected as in A. BDL/PBP4, endogenous precursors from MurJ + atc were subjected to in vitro exchange reaction as in A. (C) Detection of lipid-linked peptidoglycan precursors labeled by azDADA in vivo. Δalr was incubated in azDADA (purple and yellow). Azide-tagged species from organic extracts were ligated to DIFO-biotin via SPAAC, left, or to alkyne biotin via CuAAC, right, then detected as in A.
-
Figure 5—source data 1
Blots showing dipeptide probes in endogenous mycobacterial peptidoglycan precursors.
- https://doi.org/10.7554/eLife.37243.030
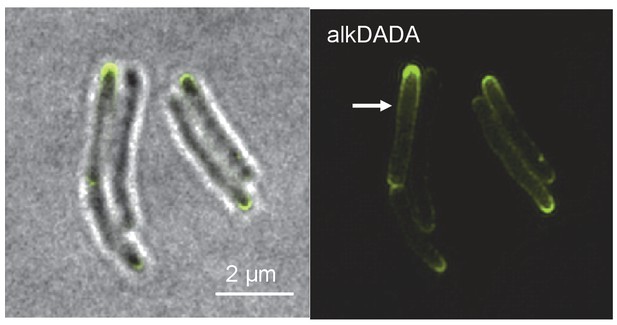
Short incubation in alkDADA results in polar and sidewall labeling.
Structured illumination microscopy of M. smegmatis incubated in alkDADA for 2 min then washed, fixed and subjected to CuAAC with azido-CR110. Arrows highlight sidewall signal.
-
Figure 5—figure supplement 1—source data 1
Structured illumination microscopy images of M. smegmatis labeled with AlkDADA.
- https://doi.org/10.7554/eLife.37243.023
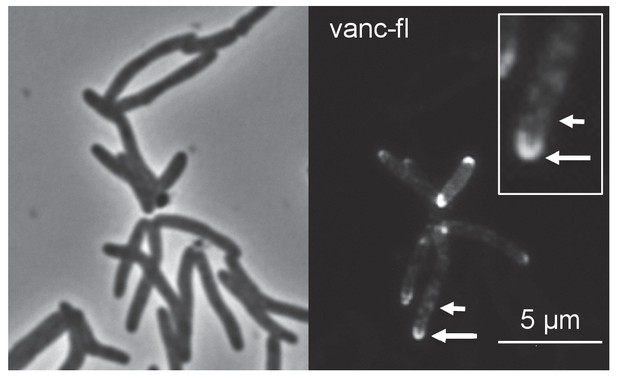
Fluorescent vancomycin (vanc-fl) labeling at poles and sidewall.
M. smegmatis were incubated in the fluorescent antibiotic for 3 hr or ~one doubling. Longer arrow, polar labeling. Short arrow, sidewall labeling.
-
Figure 5—figure supplement 2—source data 1
Conventional fluorescence microscopy images of M. smegmatis labeled with fluorescent vancomycin.
- https://doi.org/10.7554/eLife.37243.024
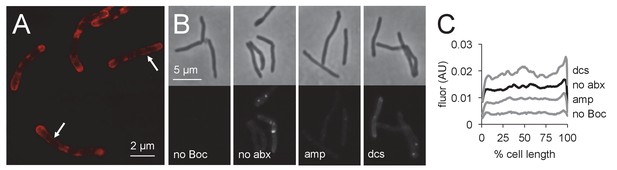
Penicillin-binding proteins are present along mycobacterial cell periphery.
(A) M. smegmatis merodiploid expressing PonA1-mRFP imaged by structured illumination microscopy. Arrows highlight sidewall signal. (B) Bocillin labeling of live M. smegmatis pre-treated or not with ampicillin or d-cycloserine. No Boc, autofluorescence; no abx, no antibiotic. (C) Quantitation of cellular fluorescence for B. 60 < n < 180. Signal was normalized by cell length and brighter poles oriented to the right-hand side of the graph. AU, arbitrary units.
-
Figure 5—figure supplement 3—source data 1
Structured illumination microscopy images of PonA1-mRFP and conventional fluorescence microscopy images of M. smegmatis labeled with Bocillin-Fl.
- https://doi.org/10.7554/eLife.37243.025
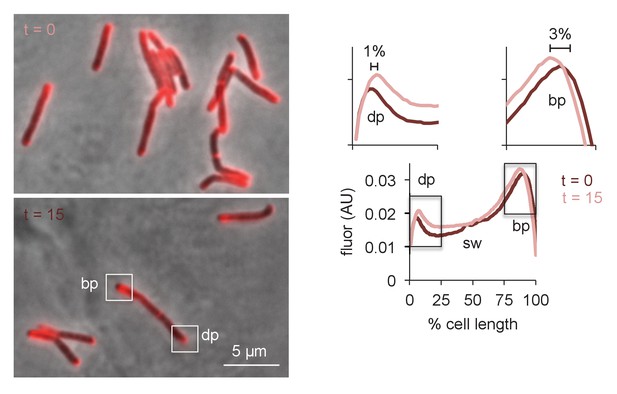
Physical expansion of the mycobacterial cell is confined to the poles and occurs more rapidly at the RADA-bright tip.
M. smegmatis were incubated in RADA for 15 min, washed and grown for 0 or 15 min in the absence of probe. Images obtained by conventional fluorescence microscopy, left, were quantified for 24 < n < 41 cells, right, as in Figure 5—figure supplement 3C. Magnification of the dim and bright pole quantitation are shown above the main graph. Distances between local maxima are expressed as percentage of total, normalized cell length. Experiment was performed twice with similar results. dp, dim pole; sw, sidewall; bp, bright pole. AU, arbitrary units.
-
Figure 5—figure supplement 4—source data 1
Pulse-chase of M. smegmatis labeled with RADA.
- https://doi.org/10.7554/eLife.37243.026
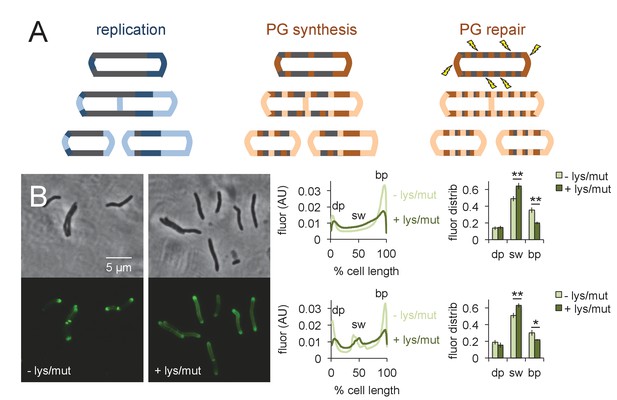
Peptidoglycan synthesis is redistributed to the sidewall upon cell wall damage.
(A) Model for the spatial organization of peptidoglycan (PG) synthesis and repair with respect to mycobacterial growth and division. Left, regions of cell surface that expand are highlighted in blue. Middle and right, areas of peptidoglycan precursor synthesis are highlighted in orange. (B) M. smegmatis was pretreated (dark green) or not (light green) with lysozyme (lys) and mutanolysin (mut) for 30 min then incubated an additional 15 min in the presence of alkDADA. The bacteria were then washed and fixed and subjected to CuAAC with azido-CR110. Fluorescence was quantitated as in Figure 2 for cells without (top; 232 < n < 236) and with (bottom; 29 < n < 55) visible septa. Dim pole (dp), sidewall (sw) and bright pole (bp) defined as in Figure 2. Fluor distrib, average fluorescence distribution. AU, arbitrary units. Error bars, ±standard deviation. Statistical significance between untreated (four biological replicates) and lysozyme/mutanolysin-treated (three biological replicates) was assessed for the dim pole, sidewall and bright pole by two-tailed Student’s t test. *p<0.05; **p<0.005.
-
Figure 6—source data 1
Conventional fluorescence microscopy images of M. smegmatis challenged with peptidoglycan-digesting enzymes.
- https://doi.org/10.7554/eLife.37243.032
Tables
| Reagent type (species) or resource | Desig-nation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain (M. smegmatis mc2155) | M. smegmatis | NC_008596 in GenBank | Wildtype M. smegmatis | |
| Genetic reagent (M. smegmatis) | ∆alr | This paper | The mutant was generated by recombineering protocols described in DOI: 10.1038/nmeth996 and DOI: 10.1007/978-1-4939-2450-9_10; see the methods section for further detail. | |
| Genetic reagent (M. smegmatis) | ΔldtA | doi:10.1101/291823 | Obtained from Dr. Eric Rubin (Harvard SPH) and Dr. Hesper Rego (Yale Med) | |
| Genetic reagent (M. smegmatis) | ΔldtB | doi:10.1101/291823 | Obtained from Dr. Eric Rubin (Harvard SPH) and Dr. Hesper Rego (Yale Med) | |
| Genetic reagent (M. smegmatis) | ΔldtE | doi:10.1101/291823 | Obtained from Dr. Eric Rubin (Harvard SPH) and Dr. Hesper Rego (Yale Med) | |
| Genetic reagent (M. smegmatis) | ΔldtAE | doi:10.1101/291823 | Obtained from Dr. Eric Rubin (Harvard SPH) and Dr. Hesper Rego (Yale Med) | |
| Genetic reagent (M. smegmatis) | ΔldtBE | doi:10.1101/291823 | Obtained from Dr. Eric Rubin (Harvard SPH) and Dr. Hesper Rego (Yale Med) | |
| Genetic reagent (M. smegmatis) | ΔldtBA | doi:10.1101/291823 | Obtained from Dr. Eric Rubin (Harvard SPH) and Dr. Hesper Rego (Yale Med) | |
| Genetic reagent (M. smegmatis) | ΔldtABE | doi:10.1101/291823 | Obtained from Dr. Eric Rubin (Harvard SPH) and Dr. Hesper Rego (Yale Med) | |
| Genetic reagent (M. smegmatis) | MurJ (MviN) | doi: 10.1126/scisignal.2002525. | Obtained from Dr. Chris Sassetti (U Mass Med) | |
| Genetic reagent (M. smegmatis) | ddlAts | doi: 10.1128/JB.182.23. 6854–6856.2000 | Obtained from Dr. Graham Hatfull (U Pitt) | |
| Genetic reagent (M. smegmatis) | pTetOldtA | doi: 10.1101/291823 | Obtained from Dr. Eric Rubin (Harvard SPH) and Dr. Hesper Rego (Yale Med) | |
| Genetic reagent (M. tuberculosis) | ΔRD1 ΔpanCD | doi: 10.1016/j.vaccine.2006.05.097 | Obtained from Dr. Bill Jacobs (Einstein Med) | |
| Other | RADA | doi: 10.1002/anie.201206749; doi: 10.1038/nprot.2014.197 | Synthesized by Tocris Bioscience (Bristol, United Kingdom) following referenced protocols | |
| Other | NADA | doi: 10.1002/anie.201206749; doi: 10.1038/nprot.2014.197 | Synthesized by Tocris Bioscience (Bristol, United Kingdom) following referenced protocols | |
| Other | HADA | doi: 10.1002/anie.201206749; doi: 10.1038/nprot.2014.197 | Synthesized by Tocris Bioscience (Bristol, United Kingdom) following referenced protocols | |
| Other | alkyne-d-alanine (alkDA;EDA) | Thermo Fisher, Waltham, MA | Cat # AC441225000 | |
| Other | alkyne-d-alanine-d-alanine (alkDADA; EDADA) | doi: 10.1038/nature12892 | Synthesized by the Chemical Synthesis Core Facility at Albert Einstein College of Medicine (NY, USA) following the referenced protocols | |
| Other | azido-d-alanine-d-alanine (azDADA; ADADA) | doi: 10.1038/nature12892 | Synthesized by the Chemical Synthesis Core Facility at Albert Einstein College of Medicine (NY, USA) following the referenced protocols | |
| Other | O-alkyne-trehalose monomycolate (OalkTMM) | doi: 10.1002/anie.201509216 | ||
| Other | N-alkyne-trehalose monomycolate (NalkTMM) | doi: 10.1002/anie.201509216 | ||
| Software, algorithm | MATLAB codes | This paper | Scripts designed for MATLAB to analyze the fluorescence profiles along a cell body from data collected in Oufti (doi: 10.1111/mmi.13264). | |
| Chemical compound, drug | Fmoc-D-Lys(biotinyl)- OH (BDL precursor) | Chem-Impex International | Cat # 16192 | Deprotected as described in doi: 10.1021/ja508147s to yield BDL |
| DNA reagent | PBP4 plasmid | doi: 10.1021/ja508147s | Obtained from Dr. Suzanne Walker (Harvard Med) |
Additional files
-
Source code 1
MatLab code to plot the fluorescence distribution of mycobacterial cells.
- https://doi.org/10.7554/eLife.37243.033
-
Transparent reporting form
- https://doi.org/10.7554/eLife.37243.034





