KCNE1 tunes the sensitivity of KV7.1 to polyunsaturated fatty acids by moving turret residues close to the binding site
Figures
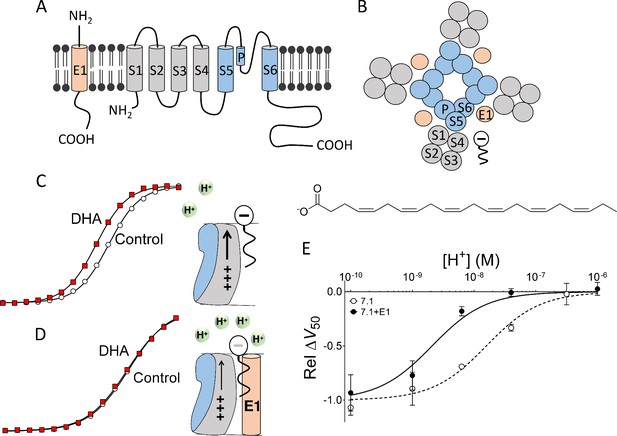
Concept of DHA-induced shift in KV7.1 channel voltage dependence.
(A) Schematic side view of one subunit of KCNE1 and KV7.1. KCNE1 is in light orange. KV7.1 is in grey (transmembrane helices S1-S4 forming the voltage-sensing domain) and blue (transmembrane helices S5 and S6 forming the pore domain). P denotes pore helix. (B) Schematic top-down view of the KV7.1+E1 channel complex. Same coloring as in A. The putative localization of a polyunsaturated fatty acid between neighboring voltage-sensing domains is included. (C) Cartoon and representative example of previously published key data showing that at pH 7.4 70 µM DHA facilitates KV7.1 channel opening by electrostatically facilitating outward S4 movement, seen as a shift of G(V) curve towards more negative voltages (modified data from Liin et al. [2015b]). (D) Cartoon and representative example of previously published key data showing that at pH 7.4, co-expression with KCNE1 decreases the local pH at the PUFA-binding site, which renders DHA uncharged and ineffective. As a consequence, 70 µM DHA fails to facilitate KV7.1+E1 channel opening at pH 7.4, seen as a lack of shift of G(V) curve toward more negative voltages (modified data from Liin et al. [2015b]). (E) KCNE1 changes the pH dependence of the DHA-induced shift in the voltage dependence of channel opening, ΔV50. [DHA]=70 µM. The ΔV50 values were normalized to the fitted maximum ΔV50 (using Equation 2) at very basic pH for each channel type to better visualize the different pKa values for the different channel constructs. Relative ΔV50 are expressed as mean ± SEM (modified and complemented data from Liin et al. [2015b]). Best fit of Equation (2): pKa = pH 8.7 for KV7.1+E1 and 7.8 for KV7.1 alone. n = 3–6 per data point.
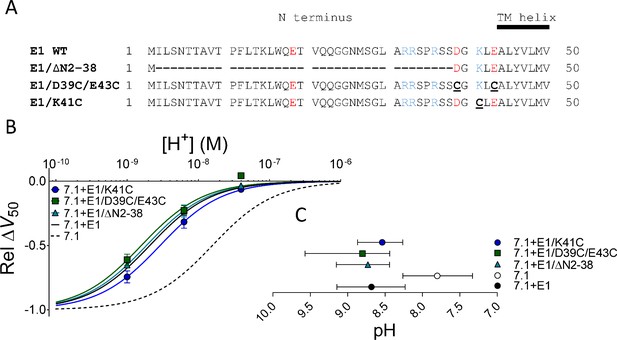
Removal of KCNE1 extracellular residues did not restore KV7.1-like DHA effect.
(A) Sequences of the N-terminal segments of WT human KCNE1 and three different KCNE1 mutants. Acidic residues colored red and basic residues colored blue. (B) pH dependence of the DHA effect (70 µM) on the relative ΔV50 for each KCNE1 mutant co-expressed with WT KV7.1. Data shown as mean ± SEM. n = 3–6 per data point. Fits for KV7.1 alone (black dashed line) and KV7.1+E1 (solid black line) from data in Figure 1E are shown for comparison. (C) Apparent pKa for the DHA effect on indicated mutants. Data shown as mean ± asymmetrical 95% confidence interval determined using Equation 2 on the data in B (best fit with Equation 2: pKa = pH 8.7 for KV7.1+E1/∆N2-38, 8.8 for KV7.1+E1/D39C/E43C, and 8.5 for KV7.1+E1/K41C).
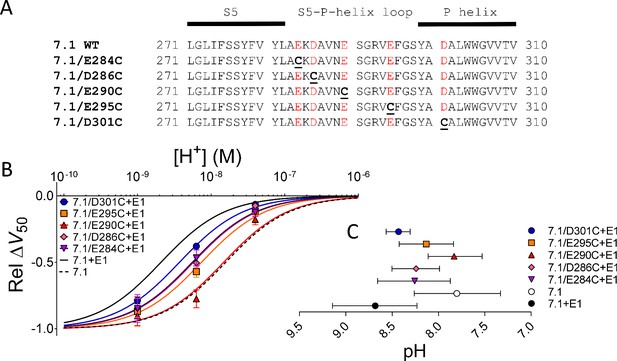
Removal of negative charges in the S5-P-helix loop partially or completely restored KV7.1-like DHA effect.
(A) Sequences of the S5-P-helix loop of WT human KV7.1 and five KV7.1 mutants. Acidic residues colored red. (B) pH dependence of the DHA effect (70 µM) on the relative ΔV50 for each KV7.1 mutants co-expressed with WT KCNE1. Data shown as mean ± SEM. n = 3–7 for each data point. Fits for KV7.1 alone (black dashed line) and KV7.1+E1 (solid black line) from data in Figure 1E are shown for comparison. (C) Apparent pKa for the DHA effect on indicated mutants. Data shown as mean ± asymmetrical 95% confidence interval determined using Equation 2 (best fit with Equation 2: pKa = pH 8.3 for KV7.1/E284C + E1, 8.2 for KV7.1/D286C + E1, 7.8 for KV7.1/E290C + E1, 8.1 for KV7.1/E295C + E1, and 8.4 for KV7.1/D301C + E1). Figure 3—figure supplement 1 is associated with Figure 3.
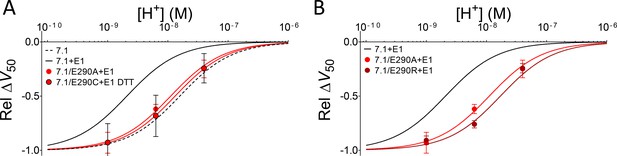
Alternative substitutions at position 290 support the hypothesis that the negative charge at position 290 is important for DHA protonation in the presence of KCNE1.
(A) pH dependence of the DHA effect (70 µM) on the relative ΔV50 for KV7.1/E290A + E1 and KV7.1/E290C + E1 in the presence of DTT (0.5 mM DTT was added to the incubation solution and control solution to prevent disulphide bond formation during oocyte incubation and experiment). Data shown as mean ± SEM. n = 4–7 for each data point. Fit for KV7.1+E1 (solid black line) and KV7.1 alone (black dashed line) are shown for comparison. Best fit with Equation 2: pKa = pH 8.0 for KV7.1/E290A + E1, and 7.9 for KV7.1/E290C + E1 in DTT. (B) pH dependence of the DHA effect (70 µM) on the relative ΔV50 for KV7.1/E290A + E1 and KV7.1/E290R + E1. Data shown as mean ± SEM. n = 3–7 for each data point. Fit for KV7.1+E1 (solid black line) is shown for comparison. Best fit with Equation 2: pKa = pH 8.0 for KV7.1/E290A + E1, and 7.7 for KV7.1/E290R + E1.
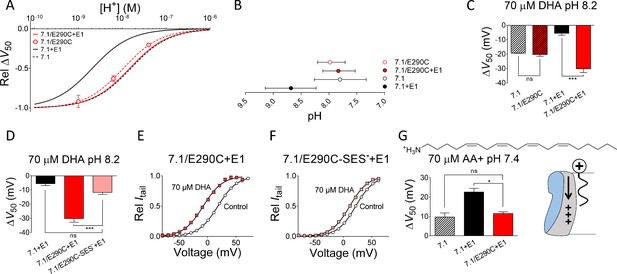
The charge at E290 is important for the PUFA effect.
(A) pH dependence of the DHA effect (70 µM) on the relative ΔV50 for KV7.1/E290C expressed without KCNE1. Data shown as mean ± SEM. n = 4–14 for each data point. Fits for KV7.1 alone (black dashed line), KV7.1+E1 (solid black line), and KV7.1/E290C co-expressed with KCNE1 (red solid line) are shown for comparison. (B) Apparent pKa for the DHA effect on indicated mutants. Data shown as mean ± asymmetrical 95% confidence interval determined using Equation 2 (best fit with Equation 2: pKa = pH 8.0 for KV7.1/E290C). (C) Summary of ∆V50 induced by 70 µM DHA at pH 8.2 for indicated mutants. The E290C mutation only increases the DHA effect when KCNE1 is co-expressed. Data shown as mean ± SEM. n = 3–5. One-way ANOVA with Tukey’s multiple comparison test. *** denotes p<0.001 and ns denotes p>0.05. (D) Summary of ∆V50 induced by 70 µM DHA at pH 8.2 for indicated mutants. MTSES—modification of E290C is denoted by -SES—. MTSES—modification of E290C restores WT KV7.1+E1 like response to DHA. Data shown as mean ± SEM. n = 3–7. One-way ANOVA with Dunnett’s multiple comparison test and KV7.1/E290C-SES—+E1 set as control. *** denotes p<0.001 and ns denotes p>0.05. (E–F) Representative effect of DHA (70 µM) on the G(V) curve at pH 8.2 for (E) KV7.1/E290C + E1 and (F) KV7.1/E290C-SES—+E1. Control data in black and DHA data in red. (G) Summary of effect of arachidonoyl amine (AA+, 70 µM, structure on top) on V50 of indicated mutants at pH 7.4. The E290C mutation decreases the AA+ effect on KV7.1+E1. Data shown as mean ± SEM (modified WT data from Liin et al. [2015b]). n = 3–10. One-way ANOVA with Dunnett’s multiple comparison test and KV7.1/E290C + E1 set as control. * denotes p<0.05 and ns denotes p>0.05. Schematic illustration describes electrostatic AA+-induced prevention of outward S4 movement (right). Figure 4—figure supplement 1 is associated with Figure 4.
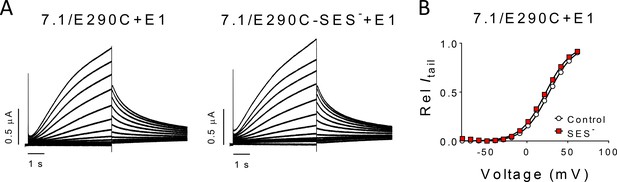
Lack of effect of MTSES— modification on intrinsic properties of KV7.1/E290C + E1.
(A) Representative current family for KV7.1/E290C + E1 before (left) and after (right) MTSES— (10 mM) modification at pH 7.4. Activating voltage steps from −80 mV to + 60 mV from a holding voltage of −80 mV. Tail voltage to −20 mV. (B) Representative normalized G(V) curve for KV7.1/E290C + E1 before (open symbols) and after (red symbols) MTSES— (10 mM) modification. The average ∆V50 induced by MTSES— modification was −4.0 ± 2.5 mV (n = 3).
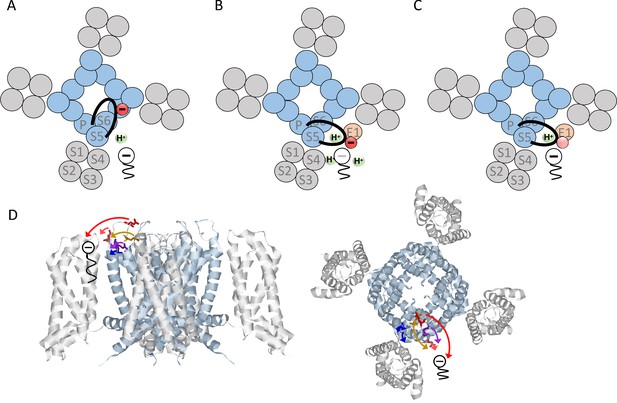
Proposed model of how KCNE1 induces protonation of PUFAs.
(A) PUFA (black) binds to KV7.1 in the lipid-filled space between two voltage-sensing domains (grey). The negatively charged PUFA head attracts the positively charges in S4 to facilitate channel activation. In KV7.1 alone, the negatively charged S5-P-helix loop (black with red negative charge) belonging to the pore region (blue) is not close to the PUFA-binding site and will therefore not induce PUFA protonation. (B) KCNE1 (light orange) co-expression with KV7.1 induces a conformational change that moves the S5-P-helix loop closer to the PUFA. The negative charges in the S5-P-helix loop, especially E290, attract protons to the PUFA binding site, which decreases the local pH. Decreased local pH causes PUFA protonation, which renders the PUFA uncharged and ineffective. (C) When negative charges in the S5-P-helix loop are neutralized, the ability of the S5-P-helix loop to attract protons is reduced. This tends to preserve the negative charge and the activating effect of the PUFA even in the presence of KCNE1. (D) Localization of indicated residues in the S5-P-helix loop of human KV7.1, side view (left) and top-down view (right) (homology model based on the Cryo EM structure of Xenopus KV7.1 [Sun and MacKinnon, 2017] and KV1.2/2.1 [Long et al., 2007]). Arrows indicate the suggested translocation of the turret region towards the PUFA binding site. Same color coding as in Figure 3 panel B and C (E284 in purple, D286 in pink, E290 in red, E295 in orange, and D301 in blue).
Tables
| Resource | Designation | Source | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (H. sapiens) | KCNQ1 | NA | GenBank Acc.No. NM_000218 | |
| Gene (H. sapiens) | KCNE1 | NA | GenBank Acc.No. NM_000219 | |
| Chemical compound, drug | Docosahexaenoic acid (DHA) | Sigma | Cat#: D2534 | |
| Chemical compound, drug | Sodium [2-sulfonatoethyl] methanethiosulfonate (MTSES-) | Toronto Research Chemicals | Cat#: S672000 |
Additional files
-
Supplementary file 1
- https://doi.org/10.7554/eLife.37257.010
-
Transparent reporting form
- https://doi.org/10.7554/eLife.37257.011





