Induction of human somatostatin and parvalbumin neurons by expressing a single transcription factor LIM homeobox 6
Figures
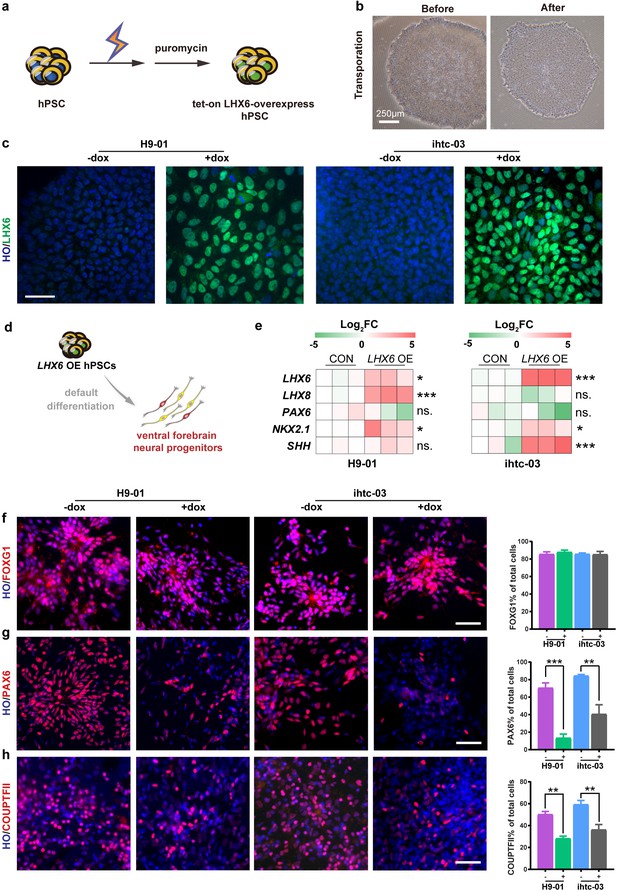
Construction of inducible LHX6 OE hPSCs.
(a) Schematic representation of electroporation to establish inducible LHX6 overexpressing (OE) hPSCs. (b) Bright-field images of hPSC colonies before and after electroporation. (c) After doxycycline induction, two inducible LHX6 OE hPSC cell lines expressed LHX6. Scale bar, 50 μm. (d) Schematic showing the differentiation of transgenic hPSC lines into dorsal neurons without adding morphogens. CON: default control group (−dox), LHX6 OE: LHX6 OE group (+dox). (e) mRNA expression levels for two transgenic hPSC-derived neurospheres and each control at day 17; n ≥ 3 for each cell line. (f–h) Representative images and quantification of transcription factors FOXG1 (f), PAX6 (g) and COUPTFII (h) expressed in CON and LHX6 OE neural precursors from two cell lines.
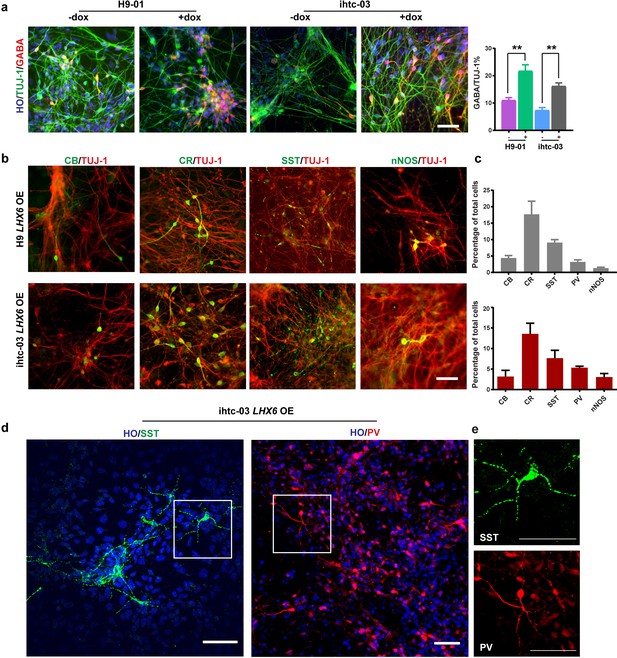
LHX6 is sufficient to convert hPSC-derived dorsal neurons to GIN subtypes.
(a) At day 35, immunostaining of TUJ-1 showed a similar efficiency of neuronal differentiation in all LHX6 OE groups and controls and a higher percentage of GABA+ cells in LHX6 OE cells. Scale bar, 50 μm. (b) The GIN subtypes calbindin (CB, day 35), calretinin (CR, day 45), somatostatin (SST, day 50), nNOS (day 80) were presented in the LHX6OE cells from two cell lines. Scale bar, 50 μm. (c) Quantification of CB+, CR+, SST+, and PV+ cells among the TUJ-1+ cells. Upper, H9-01 LHX6 OE; below, ihtc-03 LHX6 OE group. (d) SST and PV expression in the ihtc-03 LHX6 OE group. (e) Neurons expressing SST and PV interneurons showed a characteristic morphological structure with more than five secondary branches.
Generation of SST and PV subtypes by overexpressing LHX6 and ventral patterning.
(a) Schematic showing the differentiation of ventral cells from transgenic hPSC lines after treatment with SAG. V-CON, ventral control group (−dox); V-LHX6 OE, ventral LHX6 OE group (+dox). (b) The mRNA expression levels of ventral transcriptional markers in three transgenic hPSC lines and in each control at 17 days post-differentiation; n ≥ 3 biological replicates. (c) The proportions of LHX6+ cells in the control and LHX6 OE groups. The NKX2.1, GABA, TUJ-1 groups all displayed high expression of LHX6 in both the H9-01 and ihtc-03 cell lines with or without dox. Scale bar, 50 μm. (d–e) Quantification of NKX2.1+ cells (d) and GINs (e). (f) Percentage of GINs in the ventral ihtc-03 LHX6 OE group. At least 1500 cells were counted from random selected fields in each cell line, n ≥ 3 for each cell line. (g) Immunostaining of GIN subtypes in the ventral ihtc-03 control (V-CON) and LHX6 OE groups. (h) Representative tracing images of three different types PV+ neurons (type I, 130/369; type II, 137/369; type III multipolar cells, 102/369).
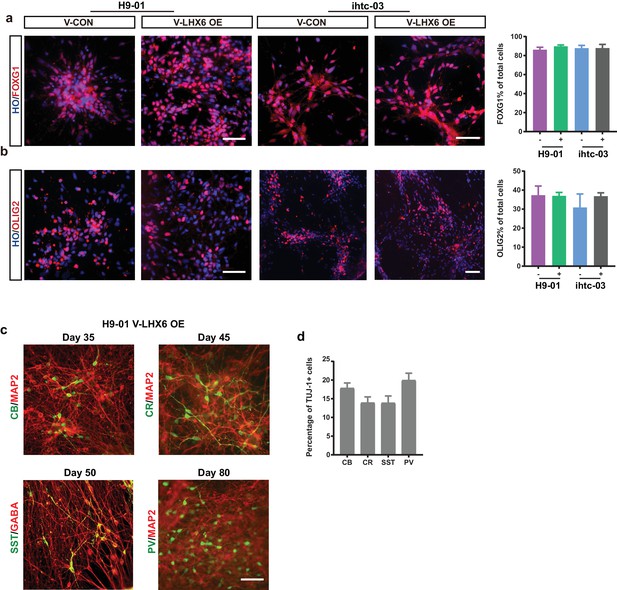
GIN progenitors identity and subtype distribution.
(a–b) The expression and quantification of FOXG1 (a) and OLIG2 (b) in the control group and the LHX6 OE group. Scale bar, 50 μm. (c) Representative images of GABA interneuron subtype proteins expressed in the H9-01 cell line. Scale bar, 50 μm. (d) Quantification of GABA interneuron subtypes in TUJ1+ cells in the H9-01 cell line.
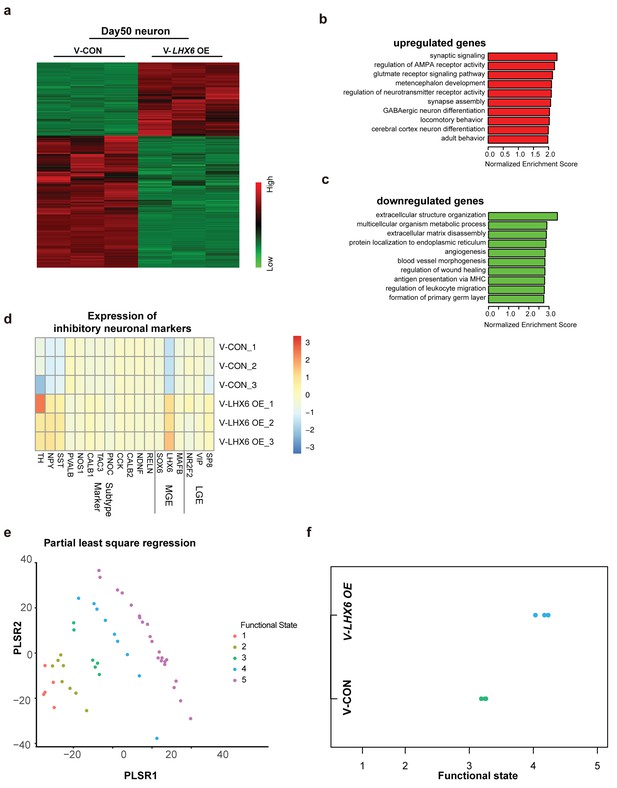
Integrative transcriptomic analyses of hPSC-derived GINs at day 50.
(a) Heatmap showing relative expression of 3467 differentially expressed genes in V-LHX6 OE cells compared to controls. (b) Bar plot presenting the top 10 non-overlapping enriched gene ontology (GO) terms in upregulated genes. (c) Bar plot presenting the top 10 non-overlapping enriched GO terms in downregulated genes. (d) Heapmap of genes associated with lateral ganglionic eminence (LGE), MGE or subtype interneuron genes in our cells (Lake et al., 2016). (e) Functional order (1–3, immature; 4, transitional and 5, highly functional neurons) displayed by Patch-Seq single cell population samples when projected on the first and second partial least-square (PLS) regression components of single cell RNA-seq profiles. (f) Functional state of our samples predicted by the PLSC1 and 2 derived linear model.
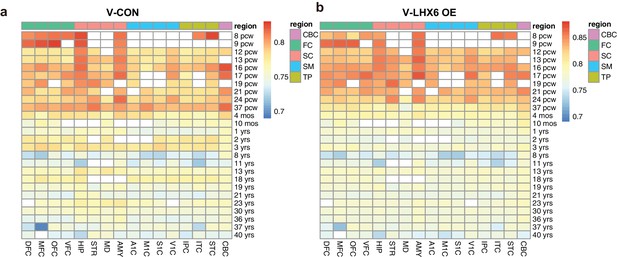
Comparison of hPSC-derived 50-day-old neurons with human fetal brain.
(a–b) Gene expression of V-CON (a) and V-LHX6 OE (b) human-induced pluripotent stem cell (hiPSC) 50-day-old neurons, which most closely resemble human first trimester brain cortex. The heatmaps display the Spearman correlation of our samples with data from regions of the human brain taken from the Brainspan Atlas of the Developing Human Brain (http://www.brainspan.org). White-filled cells indicate missing data.
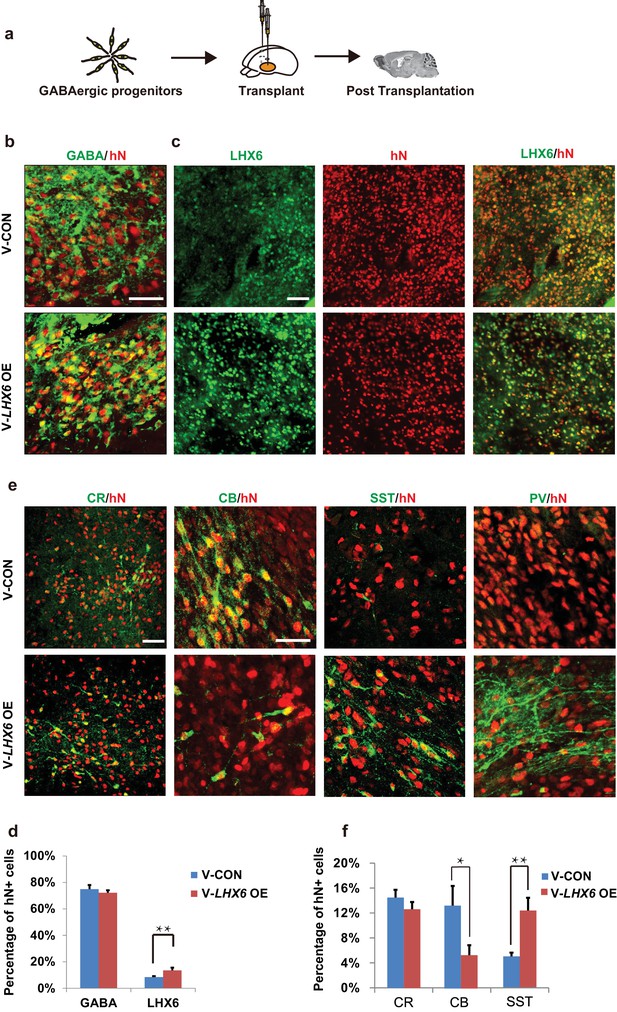
Neonatal transplantation showed that overexpression of LHX6 increased the differentiation of SST and PV neurons.
(a) Schematic showing the transplantation of hPSC-derived GABAergic progenitors into the basal forebrain of neonatal mice. (b–c) Immunostaining of GABA (b) and LHX6 (c) co-labeled with hN in grafted cells at 3 months after transplantation. Scale bar, 50 μm. (d) Quantification of GABA+ and LHX6+ cells among hN+ cells. Over 6000 hN+ cells were counted; n = 4 for V-CON and n = 5 for V-LHX6 OE. (e) At 3 months after transplantation, all four GIN subtypes (CR, CB, SST, and PV) could be detected. Scale bar, 50 μm. (f) Quantification of GIN subtypes at 3 months after transplantation. Over 3000 hN+ cells were counted for each subtype; n = 4 for V-CON and n = 5 for V-LHX6 OE.

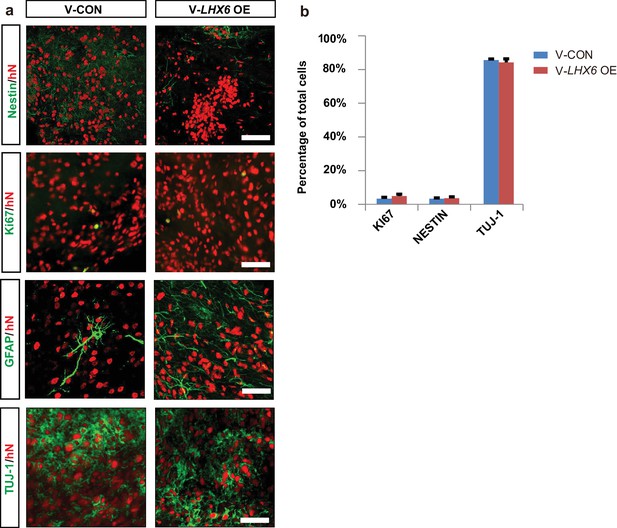
Electrophysiological characteristic of grafted human neurons.
(a) Immunostaining of brain slices for Nestin, Ki67, GFAP, and TUJ-1 that co-labeled with hN in the control and LHX6 OE groups. Scale bar, 50 μm. (b) Percentage of KI67, NESTIN and TUJ-1 in hN+ cells. Over 3000 hN+ cells were counted for each bar; n = 4 for V-CON and n = 5 for V-LHX6 OE.
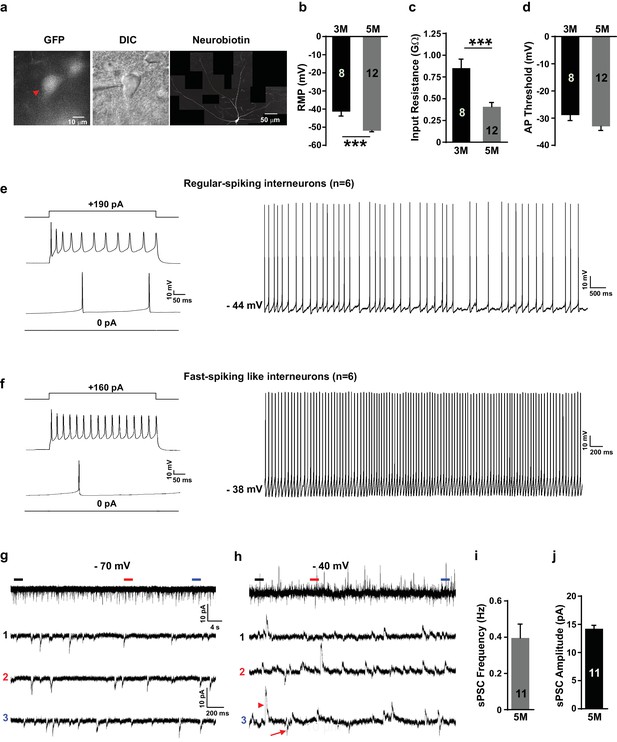
Grafted neurons show functional maturation and fast-spiking-like interneuron high-frequency firing in vivo.
(a) Representative image of an LHX6-EGFP OE cell at 5 months after in vivo transplantation. The cell was recorded with whole-cell configuration and then filled with neurobiotin. The recorded cell was visualized with EGFP (left, red arrow) and differential interference contrast microscopy (DIC) (middle) and was reconstructed via neurobiotin staining (right). (b–d) Summary of resting membrane potential (RMP) (b), input resistance (c), and action potential (AP) threshold (d) from LHX6-EGFP OE cells at 3 months and 5 months after transplantation. ***, p<0.001. (e) Sample traces of voltage changes in an LHX6-EGFP OE cell. Changes of membrane potential were evoked by current injection in 0 pA and 190 pA, respectively (left). Spontaneous firings of an LHX6-EGFP OE cell at a subthreshold holding of −44 mV. (f) Sample traces of voltage changes in a LHX6-EGFP OE cell. Changes of membrane potential were evoked by current injection in 0 pA and 160 pA (left). The recorded cell could fire action potentials at a maximus rate of 40 Hz. Spontaneous firings of an LHX6-EGFP OE cell at a subthreshold holding of −38 mV (right). (g–h) Sample traces of spontaneous postsynaptic currents (sPSCs) at a holding potential of −70 mV (g) and −40 mV (h) from an LHX6-EGFP OE cell at 5 months after transplantation. The bottom three traces are enlarged from the top traces. (i–j) Summary of frequency (i) and amplitude (j) of sPSCs recorded at a holding potential of –70 mV from LHX6-EGFP OE cells at 5 months after transplantation.
Immunostaining of garfted cells.
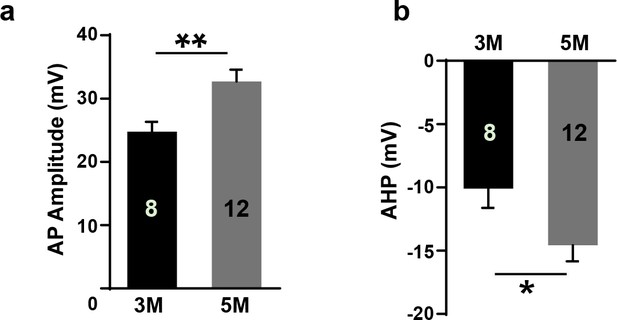
(a–b) Summary of AP amplitude (a) and AHP (b) of GIN from control and LHX6 OE groups at 3 months and 5 months post-transplantation.
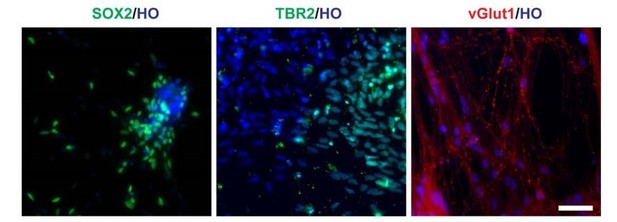
Cells in LHX6 OE group expressing SOX2, TBR2 and vGlut1 (scale bar, 100μm) (related to Figure 2 in manuscript).
https://doi.org/10.7554/eLife.37382.015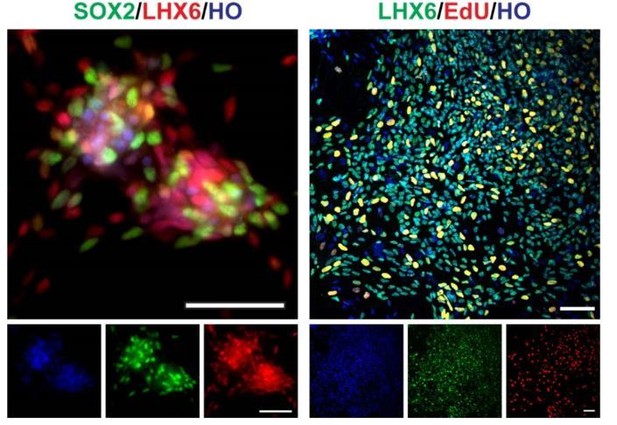
SOX2, LHX6, EdU staining of day35 V-LHX6 OE cells (scale bar, 100μm).
https://doi.org/10.7554/eLife.37382.016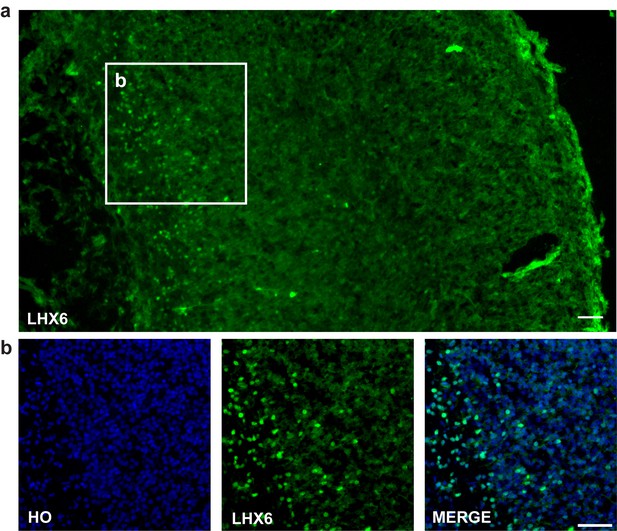
LHX6 staining.
(a–b) LHX6 staining of E18.5 mouse forebrain cortex (scale bar, 100μm).
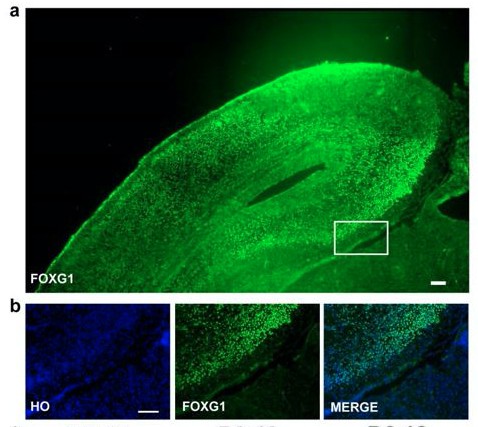
FOXG1 staining.
(a–b) FOXG1 staining of E18.5 mouse forebrain (scale bar,100μm).
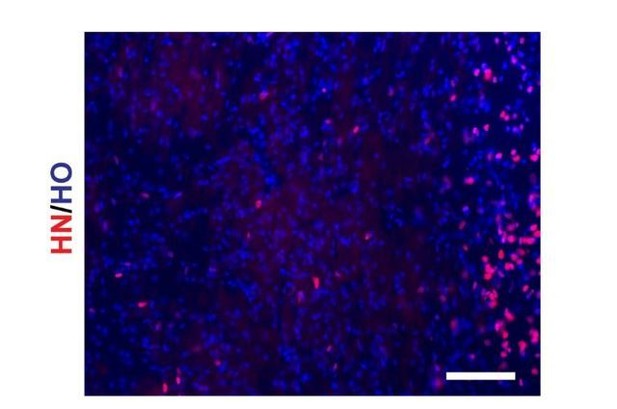
hN staining of transplanted mouse brain slice (scale bar, 100μm).
https://doi.org/10.7554/eLife.37382.019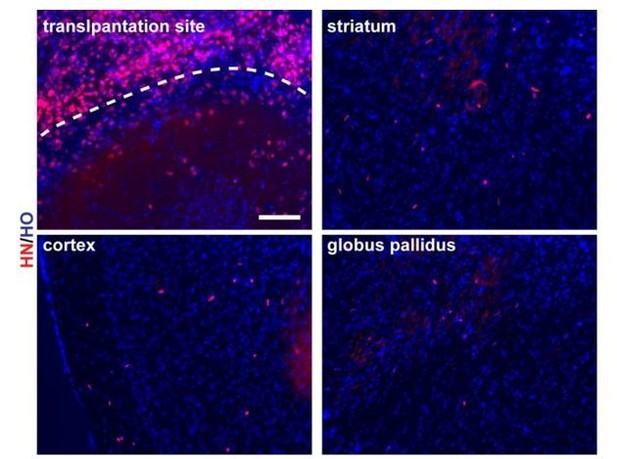
3 months mouse brain slices (scale bar, 100μm).
https://doi.org/10.7554/eLife.37382.020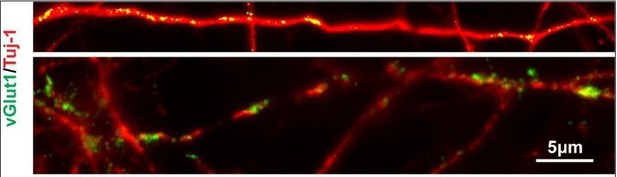
V-LHX6 OE neurons also received a small amount of excitatory inputs by vGlut1.
https://doi.org/10.7554/eLife.37382.021Additional files
-
Supplementary file 1
Supplementary Table 1: Summary of PV and SST neurons deriverd from hPSCs.
Supplementary Table 2: Antibodies used in this study.
Supplementary Table 3: Primers used in this study.
Supplementary Table 4: Mycoplasma contamination testing of cell lines.
- https://doi.org/10.7554/eLife.37382.012
-
Transparent reporting form
- https://doi.org/10.7554/eLife.37382.013





