Species and cell-type properties of classically defined human and rodent neurons and glia
Figures
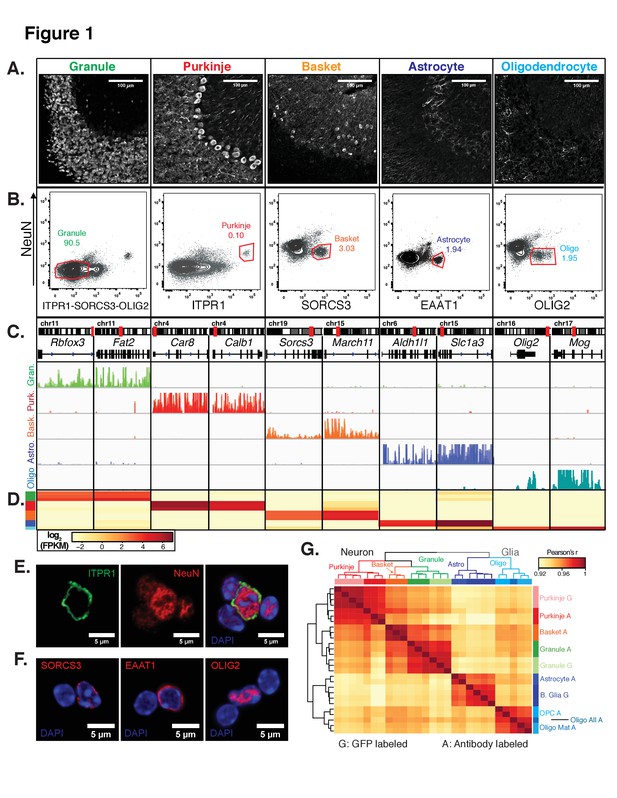
Generation of gene expression profiles for distinct cell types from the cerebella of wild-type mice.
(A) Immunofluorescence staining of five distinct cell types in the cerebellum. Antibodies used to label each cell type: NeuN for granule cells, ITPR1 for Purkinje cells, SORCS3 for basket cells, GFAP labels the cell bodies and process of astrocytes, MOG labels the cell bodies and process of oligodendrocyte. (B) Fluorescence activated sorting of stained nuclei from five cell types. Antibodies used for staining are indicated on the x- and y-axes. Percentage of each cell type based on the positive population is indicated. (C) Browser view showing examples of gene that are specifically expressed in each of the five cell types. (D) Heatmap of FPKM levels for example genes in each of the five cell types (granule – green, Purkinje – red, basket – orange, astrocyte – blue, oligodendrocytes - cyan). (E-F) Examples of labeled cerebellar nuclei using cell-type specific antibodies. Nuclei are counterstained with DAPI, a marker for heterochromatin. (E) Nuclei labeled with antibodies against ITPR1 and NeuN. Itpr1, an endoplasmic reticulum membrane protein, is localized at the nuclear membrane, while NeuN, a splicing factor, is localized at euchromatin inside the nucleus. (F) Antibodies against the basket cell marker SORCS3 and astrocyte marker EAAT1, two cellular membrane proteins, show labeling of the nuclear membrane. Antibodies against the oligodendrocyte marker and transcription factor OLIG2 show labeling in euchromatin. (G) Heatmap showing the pairwise Pearson’s correlation coefficient of GFP- (G) and antibody- (A) sorted nuclei. Hierarchical clustering is performed on the 250 most variable genes across all conditions. See also Figure 1—figure supplement 1, Figure 1—figure supplement 2.
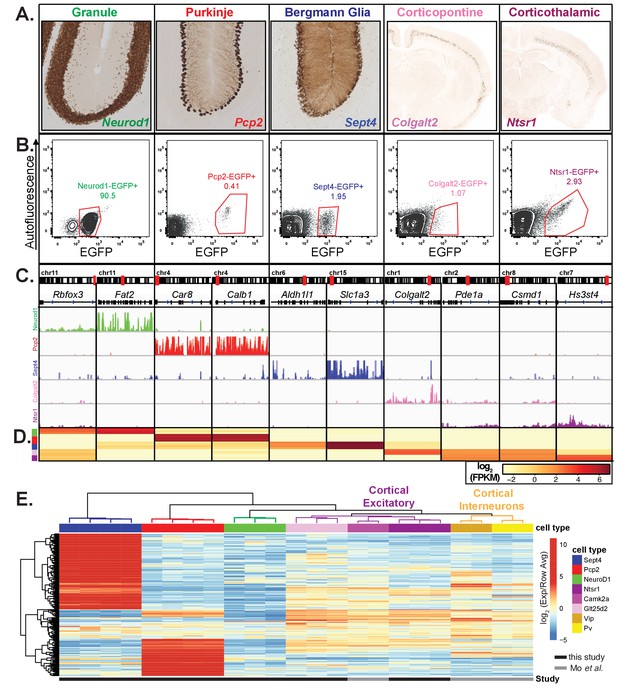
Nuclear RNA profiles can specify cell-type identity, related to Figure 1.
(A) EGFP-L10a expression from five bacTRAP animals. Neurod1, Pcp2, and Sept4 drive expression in granule cells, Purkinje cells, and Bergmann glia of the cerebellum. Colgalt2 and Ntsr1 drive expression in corticopontine and corticothalamic pyramidal cells of the cortex. All images are from the GENSAT Project. (B) Fluorescence activated sorting for EGFP+ nuclei from each of the five bacTRAP lines. The percentage of GFP+ nuclei is indicated. (C) Browser view showing nuclear expression of genes that are specific to each of the five cell types, including genes that are shared between two cell types (Pde1a and Csmd1 are expressed in both cortical pyramidal cell types). (D) Heatmap of FPKM levels for example genes in each of the five cell types (granule – green, Purkinje – red, Bergmann glia – blue, corticopontine – plum, corticothalamic - purple). (E) Heatmap showing expression of the 250 most variable genes across eight cell types. In addition to the five cell types described in (A–C), nuclear expression from three cortical cell types – excitatory neurons, PV interneurons, and VIP interneurons – from (Mo et al., 2015) are shown. Expression is normalized to the average expression across all samples for each gene. Hierarchical clustering is performed on both samples and genes.
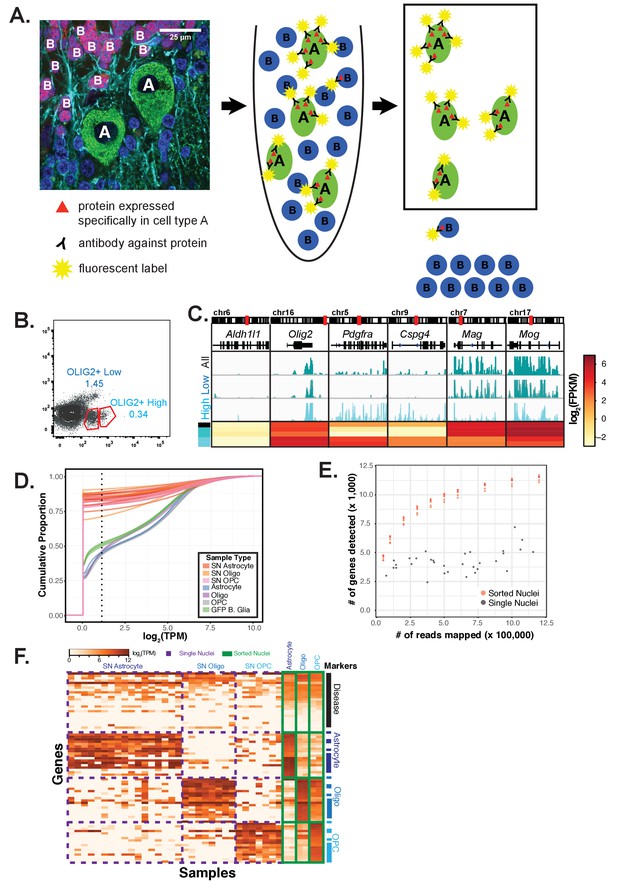
Overview of nuclei labeling and sorting strategy and comparison to single nuclei sequencing, related to Figure 1.
(A) Schematic showing the strategy for cell-type specific nuclei purification and gene expression profiling. Starting from whole tissue containing heterogeneous cell populations intermingled together, nuclei are isolated. Nuclei from a cell type of interest are fluorescently labeled using antibodies against proteins found specifically in that cell type, and then separated using fluorescent activated nuclear sorting. RNA isolated from these nuclei is used for genome-wide expression profiling by RNA-seq. (B) The OLIG2+ population can be separated into two populations. (C) Expression profiling of the two populations by RNA-seq reveals that OLIG2+ low nuclei come from mature oligodendrocytes and OLIG2+ high nuclei come from both oligodendrocyte precursor cells (OPCs) and mature oligodendrocytes. Browser view and heatmap shows gene expression from OLIG2+ low (Mature) and OLIG2+ high (OPC+ Mature) populations. Markers: Aldh1l1 (astrocytes), Olig2 (all oligodendrocytes), Pdgra and Cspg4 (OPCs), Mag and Mog (mature oligodendrocytes). (D–E) Comparative analysis of mouse cerebellar glial nuclear RNA-seq data from this study compared to mouse hippocampal single nuclei sequencing (sNuc-Seq) data from (Habib et al., 2016). (D) Cumulative distribution function plot of normalized read counts per gene in log transcripts per million (log2(TPM)) for sNuc-Seq (red/orange/pink) and antibody sorted nuclear RNA-seq datasets (blue/purple/grey). Also shown are 4 replicates of Bergmann glia nuclear RNA-seq datasets obtained from GFP+ sorted nuclei from Sept4-EGFP-L10a BacTRAP mice (green). The dotted line at 1.1 log2(TPM) is the threshold used by Habib et al. for defining expressed genes. (E) Scatterplot showing number of reads sequenced versus number of genes detected for sNuc-Seq (grey) and down-sampled nuclear RNA-seq datasets (orange). For each sNuc-Seq dataset, the number of aligned reads were extracted using samtools. For each nuclear RNA-seq dataset, samtools was used to randomly sample 50, 100, 200, 300, 400, 500, 600, 800, 1,000, and 1200 thousand reads. The number of genes detected was defined as the number of genes with a log2(TPM) of greater than 1.1. (F) Heatmap showing normalized gene expression in sNuc-Seq (purple dashed boxes) and nuclear RNA-seq datasets (green boxes) for neurological disease, astrocyte, oligodendrocyte, and OPC marker genes. Disease genes were taken from a review of Alzheimer’s disease genes (Karch and Goate, 2015). Astrocyte, oligodendrocyte, and OPC marker genes were derived from Supplementary file 1 from (Habib et al., 2016) or from Supplementary file 3 from this paper. The top ten genes from each group were used: highest by TPM value for the Habib et al. genes or lowest by Specificity Index for nuclear RNA-seq data.
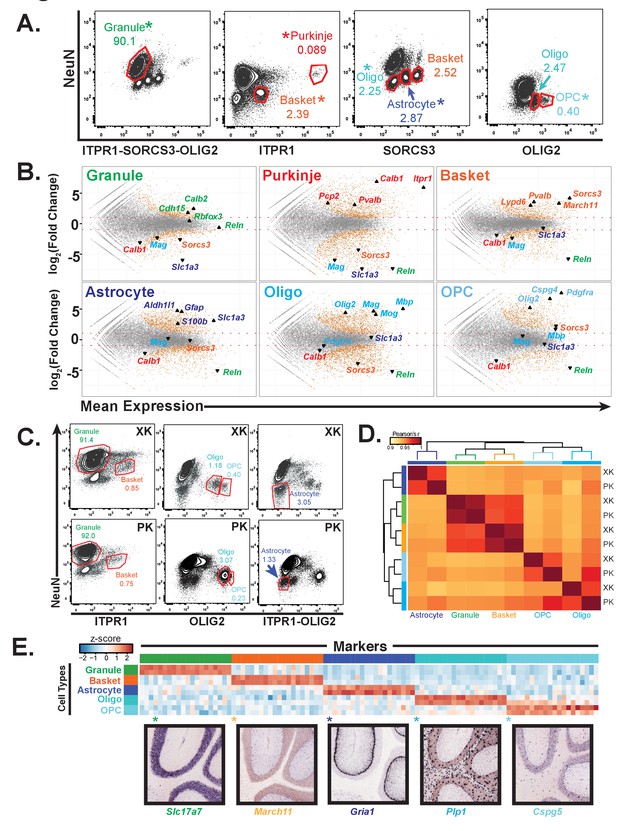
Generation of gene expression profiles for distinct cell types from rat and human cerebella.
(A) Fluorescence activated sorting of stained nuclei from six cell types in the rat cerebellum. Antibodies are indicated on the x and y axis. When a cell type can be isolated from more than one staining scheme, the population used for downstream analysis is indicated with (*). Percentage of population in each gate is indicated. (B) Differential expression analysis of antibody sorted nuclei for six cell types compared to unsorted nuclei from the rat cerebellum. Known markers for each cell type are highlighted: granule (Cdh15, Calb2, Rbfox3, Reln), Purkinje (Pcp2, Pvalb, Cabl1, Itpr1), Basket (Lypd6, Pvalb, Kit, Sorcs3), Astrocyte (Aldh1l1, Gfap, S110b, Slc1a3), all oligodendrocytes (Olig2), mature oligodendrocyte, (Labeled Oligo: Mag, Mog, Mbp), oligodendrocyte precursor cells, (Labeled OPC: Cspg4, Pdgfra). (C) Fluorescence activated sorting of granule, basket, astrocyte, mature oligodendrocyte, and OPC nuclei from the cerebellum of two human samples (XK and PK). Percentage of population in each gate is indicated. (D) Heatmap showing Pearson’s correlation coefficient between human samples. Hierarchical clustering is performed using the 250 most variable genes across samples. (E) Heatmap showing the 20 most specific genes for each human cell type as identified by the Specificity Index algorithm. Rows: sorted nuclei from XK, PK samples; columns: genes enriched in each cell type. Color represents the z-score of gene expression compared to all samples. Lower panels show examples of gene expression from the Allen Mouse Brain Atlas for genes identified as highly specific based on the Specificity Index analysis of human cell types. Allen Brain Atlas example gene is with (*) on heatmap. See also Figure 2—figure supplement 1.
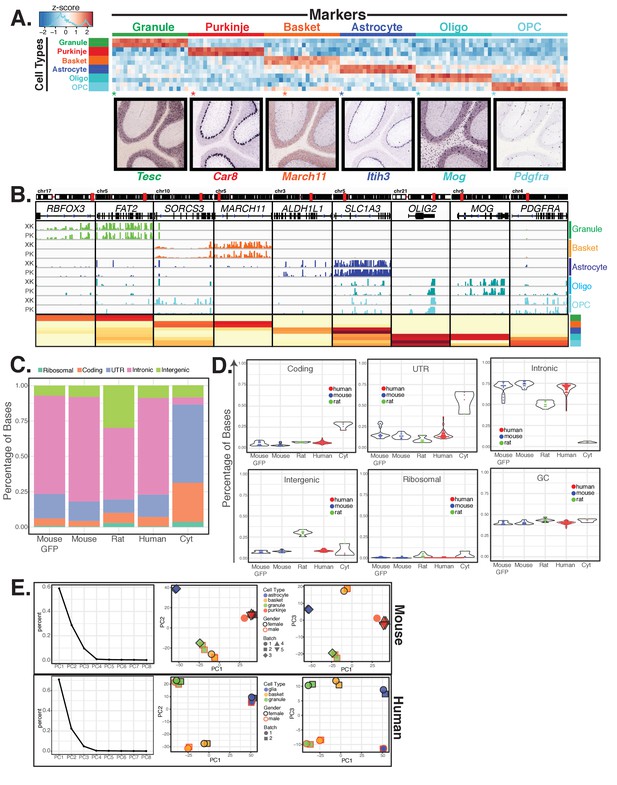
Analysis of cell type specific gene expression profiles generated from rat and human cerebella identifies known mouse marker genes for each cell type, related to Figure 2.
(A) Heatmap showing the 20 most specific genes for each rat cell type as identified by the Specificity Index algorithm. Rows: cell-type specific samples; columns: genes. Color represents the z-score of gene expression compared to all samples. Lower panels show examples of gene expression from the Allen Mouse Brain Atlas for highly specific genes identified by analysis of the rat data. Allen Brain Atlas example gene is with (*) on heatmap. Distribution of astrocyte-specific gene Itih3 is characteristic of both Bergmann glia, a type of astrocyte, and non-Bergmann glia astrocytes of the cerebellum. The percentage of top 20 rat SI genes evident in Allen Mouse Brain Atlas mouse ISH database that show expected distribution for each cell type: granule 93%, Purkinje 100%, basket 100%, astrocyte 100%, oligodendrocyte 85%, OPC 82%. (B) Browser view and heatmap showing gene expression of cell-type specific markers across human samples from five cell types. (C–D) Metrics of RNA-seq reads from mouse GFP-sorted nuclei, antibody sorted nuclei from mouse, rat, or human, and cytoplasmic RNA from mouse, rat, and human. (C) Stacked bar chart showing the mean fraction of bases that align to ribosomal, coding, UTR, intronic, or intergenic features from all experiments. (D) Violin plots showing the distribution across all experiments of reads that map to ribosomal, coding, UTR, intronic, and interenic features. Also shown is the distribution of GC content of reads for each experiment. Values for each individual sample are overlaid. (E) Analysis of batch effects from mouse and human experiments using principal components analysis. Left panel: contribution to variance for the first eight principal components (PC). Middle panel: plot of the first against the second principal components. Right panel: plot of the first against the third principal components. Each sample is color filled according to cell type. Border color indicates gender: black for females, red for males. Shape indicates batch.
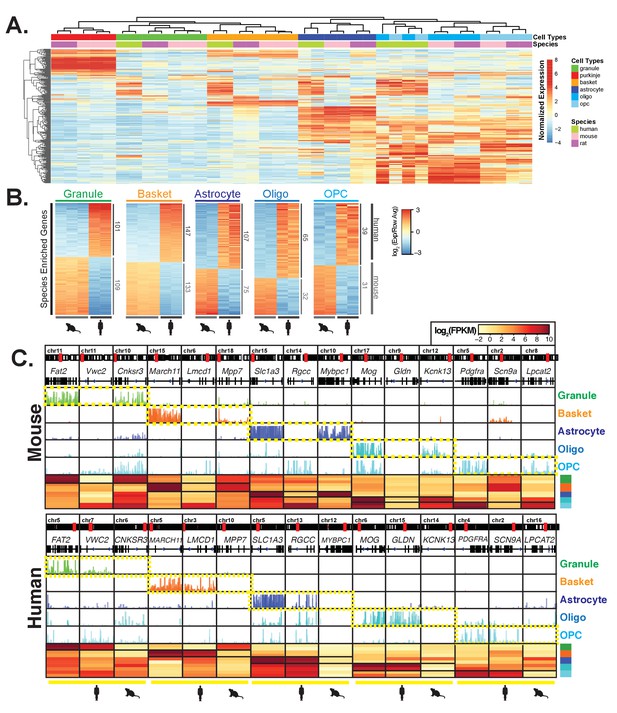
Comparative analysis of gene expression across species reveals cell-type and species specific differences.
(A) Heatmap showing normalized expression of the 250 most variable genes across all samples. Hierarchical clustering is performed using these genes and reveals that samples cluster primarily by cell type and secondarily by species. (B) Heatmap showing mouse- and human-enriched genes for each cell type, excluding any genes that are mouse- or human-enriched across all cell types. The number of genes that are significantly human or mouse enriched is indicated. (C) Browser view and heatmaps showing for each cell type, an example of a shared marker gene, a human-enriched gene, and a mouse-enriched gene. See also Figure 3—figure supplement 1, Figure 3—figure supplement 2, Figure 3—figure supplement 3.
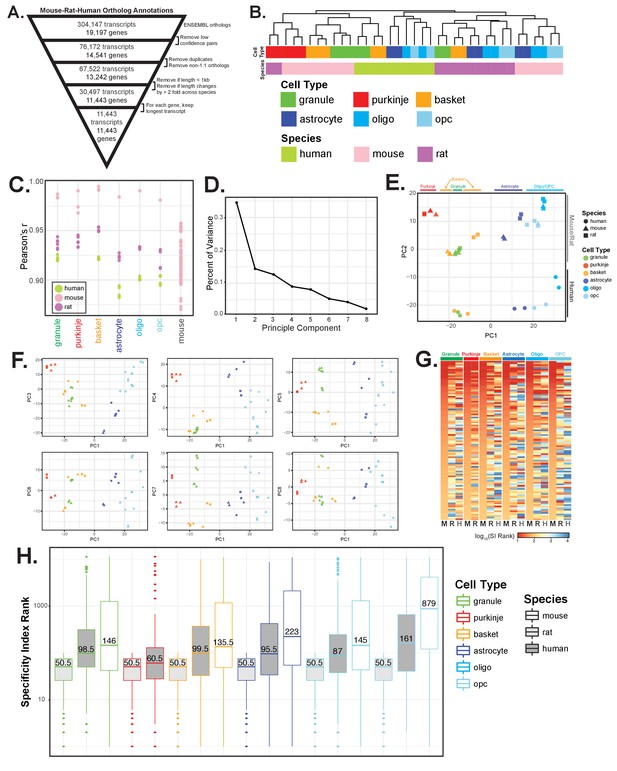
Comparative analysis of gene expression in specific cell types of mouse, rat, and human, related to Figure 3.
(A) Schematic for defining ortholog annotation for mouse, rat, and human genes. Ortholog annotation across species were downloaded from ENSEMBL, filtered to include only high confidence pairs, 1:1 orthologs, genes greater than 1 kb in total length, and genes that change by less than 2-fold in length across species. For each gene, the longest orthologous transcript was used for annotation. (B) Hierarchical clustering of mouse, rat, and human cell-type specific datasets based on expression of all genes instead of the top 250 most variable genes reveals clustering primarily by species. (C) Dot plot showing pairwise Pearson’s correlation coefficient for each sample, broken down by cell type. Last column shows the pairwise Pearson’s correlation coefficient between all cell types in mouse. (D–F) Principal components analysis of all datasets. (D) Plot showing the percent of variance explained by each of the first eight principal components. (E) Scatterplot showing values of the first two principal components (PC1, PC2). PC1 separates samples based on cell type while PC2 separates mouse and rat from human samples. (F) Scatterplots showing the first principal component versus components three through eight. (G-H) Specificity index (SI) analysis of cell-type specific genes in mouse, rat, and human. SIs for each species were computed separately using the set of genes with high confidence 1:1 orthologs across all three species as defined in (A). For Purkinje neurons, SIs were computed in mouse and rat using all 6 cell types. For the other five cell types, SIs were computed in mouse, rat, and human using only these 5 cell types. (G) Heatmap showing SI calculated ranks. Shown are the top 100 mouse SI genes for each cell type. Colors indicate log10 rank position of these 100 genes in mouse, rat, and human. (H) Boxplot representation of SI ranks from (G).
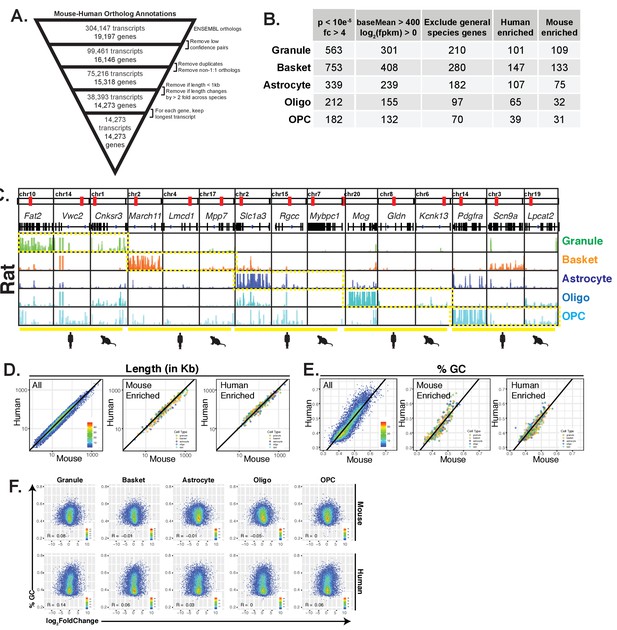
Detailed comparative analysis of gene expression in five cerebellar cell types in mouse and human, related to Figure 3.
(A) Schematic for defining ortholog annotations for mouse and human genes. Ortholog annotation across species were downloaded from ENSEMBL, filtered to include only high confidence pairs, 1:1 orthologs, genes greater than 1 kb in total length, and genes that change by less than 2-fold in length across species. For each gene, the longest orthologous transcript was used for annotation. (B) Table showing for each cell type, the number of genes that are differentially expressed (adjusted p<10e-5, fold change >4) between mouse and human, the number that are left after filtering for expression levels (baseMean >400, log2(FPKM) >4), and the number that are left after filtering for genes that are differentially expressed across all cell types between mouse and human. This number is then broken down into human and mouse enriched genes. (C) Browser views showing expression in rat for marker, human-enriched, and mouse-enriched genes. (D) Scatterplots of mouse versus human gene lengths (in Kb, log10 scale) for all genes (left), mouse-enriched genes (middle), or human-enriched genes (right). Mouse- and human-enriched genes are color coded by cell type. Line in all panels is the least squares regression with equation: log10(gene length human)=1.015 * log10(gene length mouse) – 0.0154. (E) Scatterplot of mouse versus human GC content for all genes (left), mouse-enriched genes (middle), or human-enriched genes (right). Mouse- and human-enriched genes are color coded by cell type. Line in all panels is the least squares regression with equation: GChuman = 1.299 * GCmouse – 0.138. (F) Scatterplot of fold change (between mouse and human) versus GC content for each gene. GC content is derived from mouse genes (top row) or human genes (bottom row). The Pearson correlation coefficient (R) is shown for each graph.
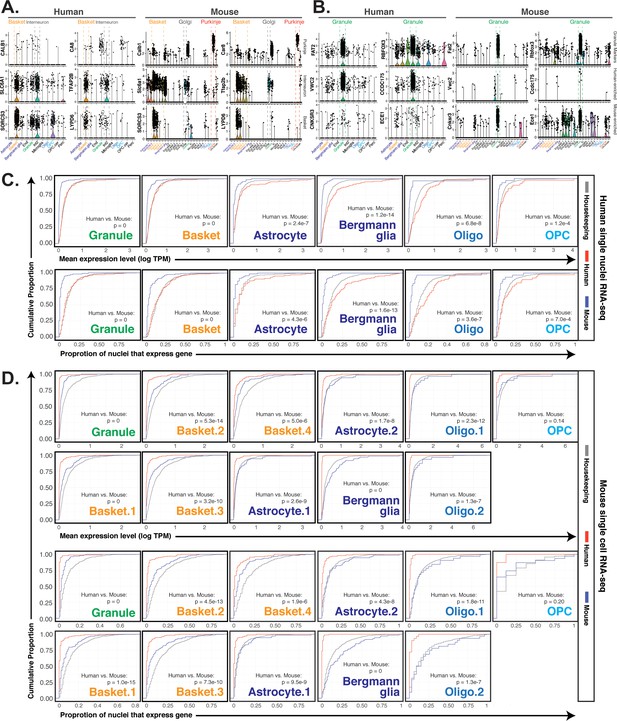
Expression of species-enriched genes in published cerebellar single nuclei/cell RNA-seq data, related to Figure 3.
For all figures, human single nuclei RNA-seq data is from (Lake et al., 2018) and mouse single cell RNA-seq data is from (Saunders et al., 2018). Author designations for cell types are used except that Purk1 and Purk2 clusters from (Lake et al., 2018) have been renamed Basket and Interneuron2 (Int2). (A) Violin plots showing expression of Purkinje neuron marker genes CALB1/Calb1 and CA8/Car8 (top), cerebellar interneuron marker genes SLC6A1/Slc6a1 and TFAP2B/Tfap2b (middle), and basket neuron marker genes SORCS3/Sorcs3 and LYPD6/Lypd6 (bottom) in human and mouse cerebellar cell types. (B–D) Expression of mouse and human-enriched genes identified in this study in cell nuclei/cell data. (B) Violin plots showing expression values for the granule marker genes FAT2/Fat2 and RBFOX3/Rbfox3, human-enriched granule cell genes VWC2/Vwc2 and CCDC175/Ccdc175, and mouse-enriched granule cell genes CNKSR3/Cnksr3 and ECE1/Ece1. (C) For each cell type in human cerebellar single nuclei RNA-seq data, empirical cumulative distribution function (ECDF) plots show expression of housekeeping, human-enriched, or mouse-enriched genes in single nuclei data. Top row: distributions of mean expression levels; bottom row: proportion of nuclei with expression greater than one log2(TPM). Kolmogorov Smirnov test p-values comparing the distribution of human-enriched versus mouse-enriched genes are shown. Distribution of human- and mouse-enriched astrocyte genes are shown in both astrocytes and Bergmann glia. (D) For each cell type in mouse cerebellar single nuclei RNA-seq data, ECDF plots show expression of housekeeping, human-enriched, or mouse-enriched genes in single nuclei data. Top two rows: distributions of mean expression levels; bottom two rows: proportion of nuclei with expression greater than one log2(TPM). Kolmogorov Smirnov test p-values comparing the distribution of human-enriched versus mouse-enriched genes are shown. Distribution of human- and mouse-enriched astrocyte genes are shown in astrocytes and Bergman glia, basket cell genes are shown in basket cell subtypes 1 through 4, and oligodendrocyte genes are shown in oligodendrocyte subtypes 1 and 2.
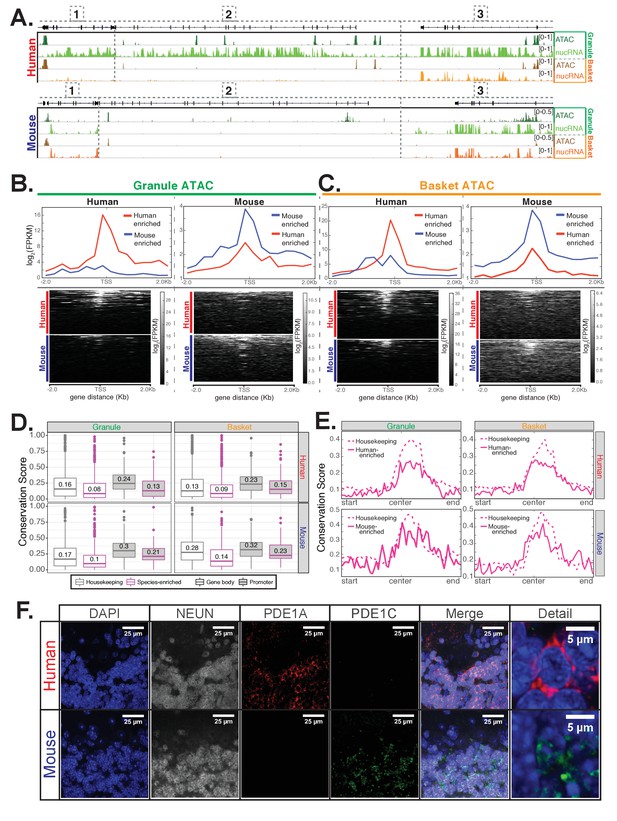
Epigenetic and immunofluorescence validation of gene expression differences between mouse and human cerebellar granule cells.
(A) Browser views showing a homologous region of approximately 150 kb from chr14 in human and chr12 in mouse. Minimum and maximum data range values are indicated for each track. Genes located in this region are: [1] JKAMP (human)/Jkamp (mouse) [2] CCDC175/Ccdc175 [3] RTN1/Rtn1. For each species, four tracks are shown: ATAC-seq DNA accessibility from granule nuclei (dark green), nuclear RNA levels from granule nuclei (green), ATAC-seq from basket nuclei (dark orange), nuclear RNA levels from basket nuclei (orange). The merged profile of two biological replicates is shown for all tracks. All three genes in the locus are strongly expressed in human granule cells and are associated with the presence of ATAC DNA accessibility sites. JKAMP/Jkamp and RTN1/Rtn1 are also expressed in human basket cells and mouse granule and basket cells and are also associated with DNA accessibility peaks. CCDC175/Ccdc175 is not expressed in human basket cells or mouse granule or basket cells; correspondingly, the promoter and gene body of this gene are depleted for ATAC DNA accessibility sites in these three cell types compared to human granule cells. (B-C) Analysis of ATAC-seq DNA accessibility assay from human (left) or mouse (right) from sorted cerebellar granule (B) or basket (C) cell nuclei. (B) Top: metagene analysis showing the median log2(FPKM) of reads from the promoter regions of 101 human-enriched or 109 mouse-enriched granule cell species specific genes. Bottom: read density of ATAC-seq reads over the promoter of each gene individually. (C) Top: metagene analysis showing the median log2(FPKM) of ATAC-seq reads from the promoter regions of 147 human-enriched for 133 mouse-enriched basket cell species specific genes. Bottom: read density of ATAC peaks over the promoters of each gene individually. (D-E) Analysis of DNA sequence conservation for accessible chromatin regions as defined by ATAC-seq peaks. (D) Analysis of DNA sequence conservation in promoter (magenta) or gene body (grey) ATAC peaks. Top: boxplots of 100-way human PhastCons scores for human cerebellar granule (left) or basket (right) cell ATAC peaks that are associated with 101 human-enriched granule cell genes (magenta outline, left), 147 human-enriched basket cell genes (magenta outline, right), or 472 genes that are not significantly differentially expressed between mouse and human (housekeeping, grey outline, left and right). Bottom: boxplots of 60-way mouse PhastCons scores for mouse cerebellar granule (left) or basket (right) cell ATAC-seq defined peaks that are associated with 109 mouse-enriched granule cell genes (magenta outline, left), 133 mouse-enriched basket cell genes (magenta outline, right), or 472 genes that are not significantly differentially expressed between mouse and human (housekeeping, grey outline, left and right). Promoter peaks are filled in grey; gene body peaks are unfilled. Median values for each group are indicated. T-test (one-sided, unequal variance) p-values comparing conservation scores for species-specific versus housekeeping genes: human granule promoter (0.0047), human granule gene body (1.0e-6), human basket promoter (0.017), human basket gene body (1.1e-14), mouse granule promoter (0.13), mouse granule gene body (2.8e-8), mouse basket promoter (0.25), mouse basket gene body(1.4e-8). (E) Metagene analysis showing mean 100-way human PhastCons scores (top) or 60-way mouse PhastCons scores (bottom) across the length of ATAC-seq peaks located in the promoters of species-enriched genes (solid magenta lines) or housekeeping genes (dashed magenta lines). (F) Immunofluoresence confirming the expression of PDE1A and PDE1C in mouse and human cerebellar slices. In mouse, PDE1C (green) is present specifically in granule cells. PDE1A (red) is not expressed above the background of the assay. In human cerebellum, PDE1A (red) is evident specifically in granule cells, and background labeling is observed for PDE1C (green). NeuN (grey) is a marker for granule cells. Staining was performed at least two times each using sections from two separate mice and two human donors. Images shown are representative of all data collected. See also Figure 4—figure supplements 1–2.
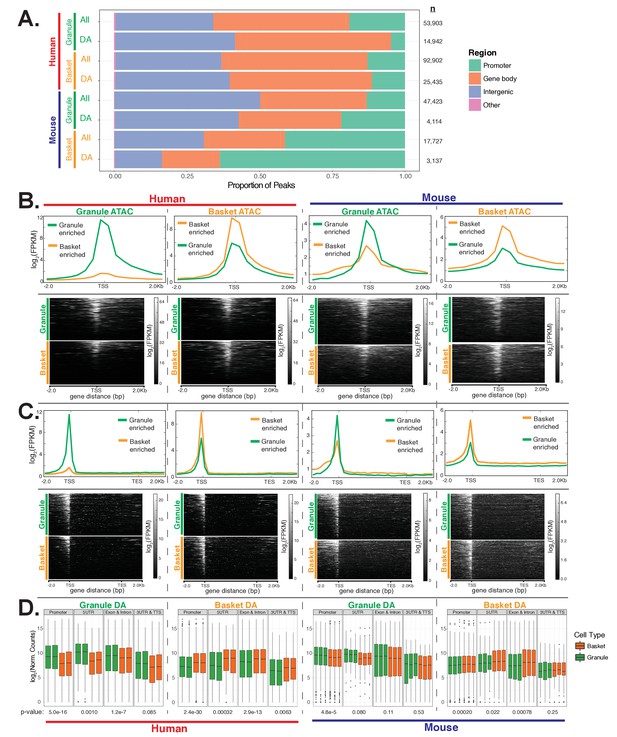
Analysis of cell-type specific ATAC peaks and examples of mouse- and human-enriched genes in granule cells, related to Figure 4.
(A-B) Browser view and heatmap showing chromatin accessibility by ATAC-seq (dark green) and nuclear gene expression levels by RNA-seq (green) from granule cells from human (red) and mouse (blue). (A) Examples of housekeeping, human-enriched, and mouse-enriched genes. (B) Example of gene usage switch between species. Expression of two Pde1 family members – PDE1A/Pde1 a and PDE1C/Pde1 c – in granule cells from mouse and human. Mouse granule cells express Pde1c but do not express Pde1a, as evidenced by the presence of ATAC DNA accessible sites and granule cell nuclear RNA for Pde1c but not Pde1a. In contrast, granule cells from human cerebellum express high levels of PDE1A, as demonstrated by the presence of nuclear transcripts and ATAC peaks indicating DNA accessibility in the promoter and gene body. PDE1C is expressed in human granule cells, but at much lower levels, as indicated by the presence of DNA accessible sites but lower levels of nuclear transcripts. Note: the annotated gene for Pde1c in mouse is around 300 kb while the annotated gene for PDE1C in human is around 600 kb. However, the longest human orthologous transcript to mouse Pde1c is around 300 kb. Shown in this figure is the promoter and 5’ end of this longest orthologous transcript. (C) All images are from the Allen Mouse Brain Atlas, unless noted. Because of the density of granule cells in the cerebellum, some light staining is usually observed in the granule cell layer in these images even when there is no expression. Top: expression of the granule-specific marker Etv1 and mouse-enriched genes Cnksr3, Ece1, and Pde1c in the granule layer of the cerebellum (indicated by arrow). Ece1 expression is also observed in the Purkinje layer of the cerebellum. Pde1c expression image is from the GENSAT Project. Bottom: expression of the human enriched genes Ccdc175, Clvs2, Vwc2, and Pde1a in the mouse brain. No staining was detected in any region for Ccdc175 and Vwc2. Arrows point to strong staining of Clvs2 in the dentate gyrus and of Pde1a in layers 5 and 6 of the cortex and CA1-3 of the hippocampus. (D) Brower view and heatmap showing the expression of the seven GATA transcription factors (TRPS1 and GATA1 – 6) across all cerebellar cell types in human (top) and mouse (bottom). Arrows indicate expression of TRPS1 in granule cells. (E) Browser view showing chromatin accessibility and nuclear gene expression in human cerebellar granule and basket neurons, over the locus containing SNCA. Red arrows indicate regions that are differentially accessible between granule and basket neurons. Highlighted are two regions that also contain SNPs that have been associated with human disease. Minimum and maximum data range values are indicated for each track.

Relationship between chromatin accessibility and gene expression in cerebellar granule and basket neurons, related to Figure 4.
(A) Stacked bar plot showing the proportion of peaks identified from ATAC-seq DNA accessibility assay from human and mouse cerebellar granule and basket neurons that map to various genomic regions. All: all peaks; DA: peaks that are differentially accessible between granule and basket neurons; n: total number of peaks for each category. Gene body contains peaks mapping to 5'UTR, exon, intron, 3'UTR, or TTS; Other contains peaks mapping to pseudogenes, miRNA, ncRNA, snoRNA, or rRNA. (B-C) Analysis of ATAC-seq DNA accessibility in for genes that are differentially expressed between granule and basket neurons. (B) Top: metagene analysis showing coverage in log2(FPKM) over the promoters of granule-enriched genes (1313 from mouse, 1100 from human) or basket-enriched genes (1124 from mouse, 1151 from human). Bottom: density plot showing enrichment of ATAC-seq reads over the promoters of each gene individually. (C) Same as (B) except coverage is shown over entire gene body. (D) Box plots showing normalized expression in granule (green) and basket (orange) neuron from human (left) and mouse (right), for genes associated with differentially accessible (DA) peaks between the two cell types. Expression is plotted separately for DA peaks located in promoter and transcription start site (TSS), 5' UTR, exon and intron, or 3' UTR and transcription terminal site (TTS) regions. T-test (two-sided, unequal variance) p-values comparing gene expression in granule cells versus basket cells are shown.
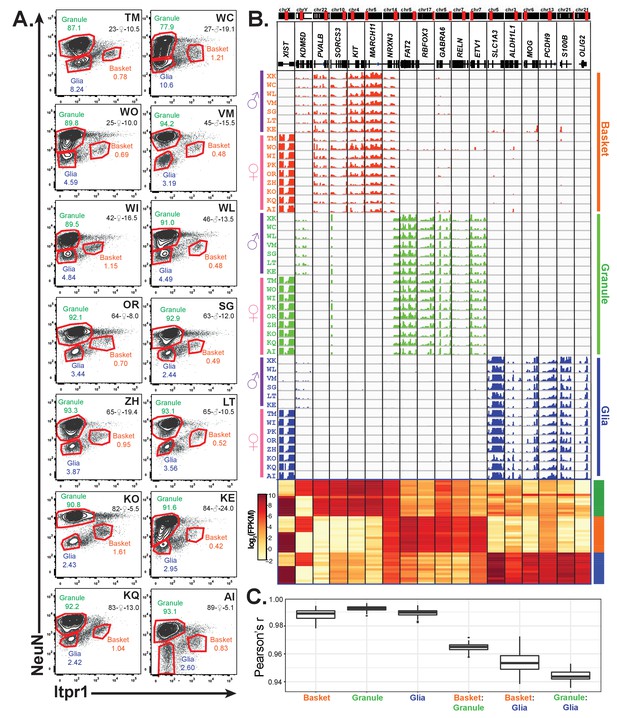
Profiling of three cell types in 16 human cerebellar samples.
(A) Fluorescence activated nuclear sorting of three cell types from the cerebella of 14 individuals. The percentage of each cell type in different individuals is shown. Each sample is identified by a two-letter code. Also shown are the age, gender, and post-mortem delay interval for each sample. (B) Browser view and heatmaps showing gene expression for gender and cell-type markers across 16 individuals (14 from A and two from Figure 4). For glia, samples from WC and WO were excluded due to granule cell contamination. (C) Boxplot showing pairwise Pearson’s correlation coefficient within cell types and between cell types for different individuals.
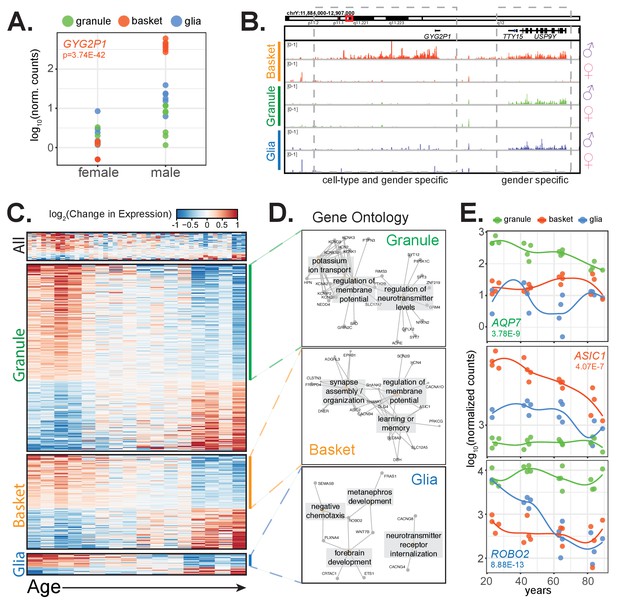
Clinical factors impact gene expression in a cell-type specific manner.
(A,B) GYG2P1: a basket-specific, male-specific gene. (A) quantification of GYG2P1 gene expression in granule, basket, and glial nuclei for female and male samples. (B) browser view showing expression of male-specific genes TTY15 and USP9Y and the male-specific and basket-specific gene GYG2P1. (C–E) Genes that significantly change with age. (C) Heatmap showing change in gene expression across all, granule, basket, or glia nuclei. Columns: samples are sorted by age. Rows: genes ranked by change in gene expression over age. (D) Enrichment map of significant gene ontology categories for aging down-regulated genes from granule cells, basket cells, and glia. (E) Scatterplot showing gene expression across age for three genes that are cell-type specific and down-regulated with age. See also Figure 6—figure supplement 1.
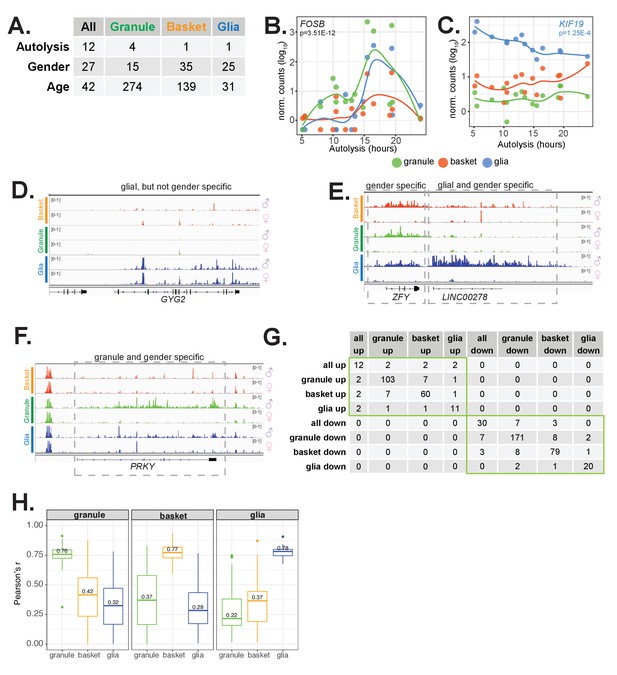
Analysis of clinical factors that contribute to gene expression variability across individuals, related to Figure 6.
(A) Table showing the number of differentially expressed genes (adjusted p<0.01, baseMean >50) for all cell types or each cell type individually when samples are stratified by clinical factors. For age and postmortem delay (PMD), numerical covariates were used for differential expression analysis. Gender indicates male versus female comparison. (B,C) Examples of genes that vary in expression depending on PMD. (B) FOSB is elevated in glia, granule, and basket cells in a few samples with intermediate PMDs. (C) KIF19 expression in glia declines as sample PMD increases. (D–F) Browser views showing expression of gene expression across males and females and in each cell type. Minimum and maximum scale values are indicated. Each track is a merged view of all samples corresponding to the indicated gender and cell type. (D) Expression of the autosomal gene GYG2. (E,F) Example of gender and cell-type specific genes in glia (E) and granule cells (F). Expression of PRKY in (F) is highest in granule cells, at intermediate levels in glia, and low in basket cells. (G) Table showing the number of genes that are in common between eight groups of differentially expressed aging genes. No overlaps were found between age up-regulated genes from any cell type and down-regulated genes from any other cell type. (H) Boxplots showing the distribution of Pearson’s correlation (r) values between gene expression and age. Correlations with age were computed using expression values from granule cells (green), basket cells (orange), and glia (blue) for 274 granule (left panel), 135 basket (middle panel), and 31 glial (right panel) age-associated genes. Median correlation values for each comparison are indicated.
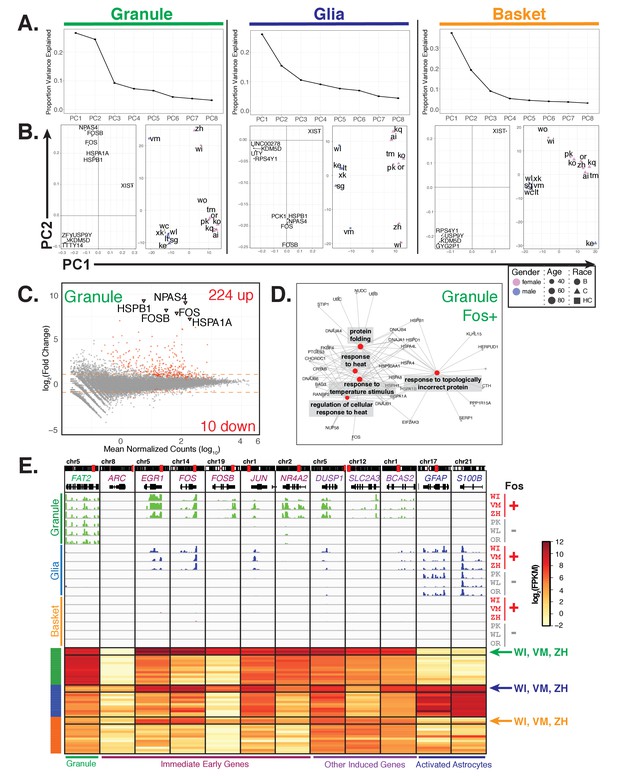
Additional sources of interindividual variability in gene expression.
(A, B) Principal component analysis of samples for each cell type identifies interindividual gene expression variability. (A) Plot showing the proportion variance explained by each of the first eight principal components for all granule, glia, and basket cell samples. (B) For each cell type, plots showing loadings (left) and scores (right) for PC1 and PC2. For loadings, the five genes with the highest absolute values from the loading vectors for PC1 or PC2 are shown. (C) MA-plot showing differential gene expression analysis of granule cells between three Fos+ samples versus the other 13 samples. (D) Network diagram of significant enriched GO categories for up-regulated genes in fos+ samples reveals enrichment for genes involved in stress response and protein folding. (E) Browser view showing expression of selected genes in three cell types from Fos+ (WI, VM, ZH) and Fos- (PK, WL, OR) human samples that are age and gender matched. Heatmap shows gene expression of selected genes in all human samples (granule – green, glia – blue, basket – orange). Samples from Fos+ donors WI, VM, and ZH are indicated. See also Figure 7—figure supplement 1.
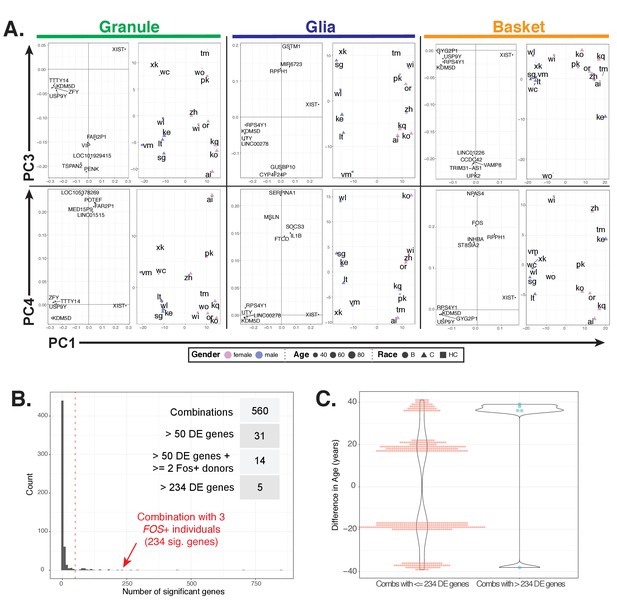
Additional analyses of interindividual variability in gene expression, related to Figure 7.
(A) For each cell type, plots showing loadings (left) and scores (right) for PC1 and PC3 (top row) or PC1 and PC4 (bottom row). For loadings, the five genes with the highest absolute values from the loading vectors for the indicated principle components are shown. (B) Resampling to determine interindividual variability in granule cell gene expression. 560 combinations partitioning the 16 individuals into two groups of three and thirteen individuals respectively were created and used for differential expression analysis. Histogram showing the distribution of the number of significant genes (padj <0.01, baseMean >50) from each analysis. 31 combinations resulted in the detection of over 50 significant genes (dashed red line), with 14 combinations including at least two out of three Fos+ donors (WI, VM, or ZH). The combination with all three Fos+ donors resulted in 234 DE genes and is indicated by the red arrow. Five combinations resulted in over 234 DE genes. (C) Median difference in age between the two groups for 555 combinations that results in 234 or fewer differentially expressed (DE) genes (left) or for the five combinations that result in more than 234 DE genes (right).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain (M. musculus) | NeuroD1 EGFP-L10a | (Doyle et al., 2008) | RRID:IMSR_JAX:030262 | |
| Strain (M. musculus) | Pcp2 EGFP-L10a | (Doyle et al., 2008) | RRID:IMSR_JAX:030267 | |
| Strain (M. musculus) | Sept4 EGFP-L10a | (Doyle et al., 2008) | RRID:IMSR_JAX:030271 | |
| Strain (M. musculus) | Glt25d2 EGFP-L10a | (Doyle et al., 2008) | RRID:IMSR_JAX:030257 | |
| Strain (M. musculus) | Ntsr1 EGFP-L10a | (Doyle et al., 2008) | RRID:IMSR_JAX:030264 | |
| Strain (M. musculus) | Wild type | Jackson labs | RRID:IMSR_JAX:000664 | |
| Antibody | anti-NeuN | Abcam | RRID:AB_2532109 | Rabbit monoclonal; 1:250 for nuclei staining; 1:250 for IF |
| Antibody | anti-NeuN | Millipore | RRID:AB_2298772 | Mouse monoclonal; 1:250 for nuclei staining; 1:250 for IF |
| Antibody | anti-Itpr1 | Abcam | ab190239 | Mouse monoclonal; 1:500 for nuclei staining; 1:500 for IF |
| Antibody | anti-Sorcs3 | Thermo | RRID:AB_2606387 | Goat polyclonal; 1:250 for nuclei staining; 1:250 for IF |
| Antibody | anti-EAAT1 | Abcam | RRID:AB_304334 | Rabbit polyclonal; 1:250 for nuclei staining |
| Antibody | anti-Olig2 | R and D | RRID:AB_2157554 | Goat polyclonal; 1:250 for nuclei staining |
| Antibody | anti-GFAP | Abcam | RRID:AB_880202 | Goat polyclonal; 1:500 for IF |
| Antibody | anti-Mog | Thermo | RRID:AB_2607363 | Goat polyclonal; 1:500 for IF |
| Antibody | anti-Mag | Cell Signaling | RRID:AB_2665480 | Rabbit polyclonal; 1:500 for IF |
| Antibody | anti-Pde1a | Acris | TA311317 | Goat polyclonal; 1:200 for IF |
| Antibody | anti-Pde1c | Santa Cruz | RRID:AB_11149544 | Rabbit polyclonal; 1:100 for IF |
| Sequence-based reagent | raw and processed sequencing data | this paper | GSE101918 | Details about all samples in this superseries are in Supplementary file 1 |
| Sequence-based reagent | raw and processed sequencing data | (Mo et al., 2015) | GSE63137 | Details about all samples in this superseries are in Supplementary file 1 |
| Sequence-based reagent | raw and processed sequencing data | (Habib et al., 2016) | GSE85721 | Details about all samples in this superseries are in Supplementary file 1 |
| Sequence-based reagent | raw and processed sequencing data | (Lake et al., 2018) | GSE97930 | Details about all samples in this superseries are in Supplementary file 1 |
| Sequence-based reagent | raw and processed sequencing data | (Saunders et al., 2018) | GSE116470/dropvis.org | Details about all samples in this superseries are in Supplementary file 1 |
| Commercial assay or kit | AllPrep FFPE | Qiagen | 80234 | |
| Commercial assay or kit | Rneasy Micro | Qiagen | 74004 | |
| Commercial assay or kit | MinElute Reaction Cleanup | Qiagen | 28206 | |
| Commercial assay or kit | Ovation RNAseq V2 | Nugen | 7102–32 | |
| Commercial assay or kit | Ultra II DNA Lirary Prep Kit for Illumina | NEB | E7645L | |
| Commercial assay or kit | Multiplex Adapters for Illumina | NEB | E7335L, E7500L | |
| Commercial assay or kit | Bioanalyzer Pico chips | Agilent | 5067–1513 | |
| Commercial assay or kit | TapeStation D1000 ScreenTape | Agilent | 5067–5583 | |
| Commercial assay or kit | TapeStation High Sensitivity D1000 ScreenTape | Agilent | 5067–5585 | |
| Software, algorithm | source code | (Xu, 2018) | https://github.com/xu-xiao/non_transgenic_cell_type_profiling | R scripts for analysis and generating figures; parameters for running command line tools. |
Additional files
-
Supplementary file 1
Summary of all RNA-seq datasets, including information about animals, sorts, and quality control metrics related to methods.
Also includes information on published RNA-seq datasets used in this manuscript.
- https://doi.org/10.7554/eLife.37551.019
-
Supplementary file 2
Clinical information for all human tissue donors, related to methods.
- https://doi.org/10.7554/eLife.37551.020
-
Supplementary file 3
Specificity index calculations for mouse, rat, and human cell types using either species-specific annotations or with mouse-rat-human orthologous gene annotations.
SIs for mouse and rat samples with orthologous gene annotations have been calculated using either all cell types (all) or without Purkinje samples.
- https://doi.org/10.7554/eLife.37551.021
-
Supplementary file 4
Differential expression analysis results for mouse and human-enriched genes.
Also mouse and human IDs for genes that are unchanged in gene expression between the two species.
- https://doi.org/10.7554/eLife.37551.022
-
Supplementary file 5
Annotation of ATAC peaks.
All: all MACS2 called peaks; DA: differentially accessible peaks between granule and basket neurons.
- https://doi.org/10.7554/eLife.37551.023
-
Supplementary file 6
Motif analysis of various ATAC-seq defined regions.
DA: regions that are differentially accessible between granule (gran) and basket (bsk) neurons in mouse (m) or human (h); HE: regions defined by peaks located in the promoter (p) or gene body (gb) of human (h) enriched genes for granule (gran) or basket (bsk) neurons; ME: regions defined by peaks located in the promoter (p) or gene body of mouse (m) enriched genes for granule (gran) or basket (bsk) neurons.
- https://doi.org/10.7554/eLife.37551.024
-
Supplementary file 7
Differentially accessible regions between human granule and basket neurons that contain single nucleotide polymorphisms (SNPs) associated with human disease.
The column Multiple specifies whether a SNP has been linked to a specific disease/trait in at least two publications.
- https://doi.org/10.7554/eLife.37551.025
-
Supplementary file 8
Differential expression analysis results for the influence of clinical factors on gene expression in human samples.
- https://doi.org/10.7554/eLife.37551.026
-
Supplementary file 9
Full results from all gene ontology (GO) analyses performed in the paper.
- https://doi.org/10.7554/eLife.37551.027
-
Transparent reporting form
- https://doi.org/10.7554/eLife.37551.028





