Intrinsic cooperativity potentiates parallel cis-regulatory evolution
Figures
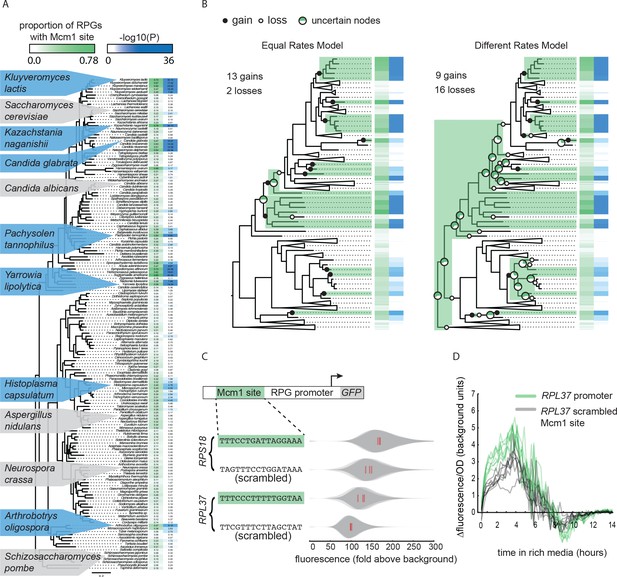
Repeated evolution of Mcm1 cis-regulatory sites at RPGs.
(A) The Mcm1 sites are found upstream of the ribosomal protein genes (RPGs) in several different clades in the Ascomycete fungi.The first column, colored in green, shows the proportion of ribosomal proteins in each species that contains at least one Mcm1 site at a cutoff of ~50% the maximum log likelihood score using a position weight matrix. The second column, colored in blue, shows the –log10(P) for the enrichment of these Mcm1 binding sites relative to upstream regulatory regions genome-wide, as expected under the hypergeometric distribution. Dis-enrichment values are not highlighted. The phylogeny is a maximum likelihood tree based on the protein sequences of 79 genes found in single copy in most species. Key species discussed in this paper and previous literature are highlighted with blue background indicating enrichment for Mcm1 binding sites or gray indicating no enrichment. (B) Results of two models estimating the numbers of gains and losses of Mcm1 cis-regulatory sites at the RPGs. Shown is one example tree out of 10,000 sampled for each of the two models. Gains are indicated in filled circles and losses are indicated in open circles. Major nodes with a high amount of uncertainty over all of the sampled trees (0.2 < proportion with Mcm1 sites < 0.8) are shown as pie graphs with the proportion of simulations with that ancestor having Mcm1 sites shown in green. Other nodes have a high proportion (>0.8) of trees matching the example tree. (C) Intergenic regions upstream of two ribosomal proteins in the species Kluyveromyces lactis were positioned upstream of a GFP reporter. The Mcm1 cis-regulatory sites were scrambled and the wild-type and mutant reporters were integrated into the Kl. lactis genome. Cells were grown for 6 hr in rich media and expression was measured by flow cytometry. Shown is the single-cell fluorescence distribution for three independent genetic isolates and the median (red bar), normalized by forward-scatter values. The values were divided by the average fluorescence for a cell lacking a GFP reporter (fold above background). (D) The RPL37 reporter strains were diluted into rich media and fluorescence and optical cell density (OD600) were measured every 15 min in a plate reader. Shown is the change in fluorescence between consecutive time points divided by the OD600 for eight technical replicates comprised of the three independent genetic isolates of each strain.
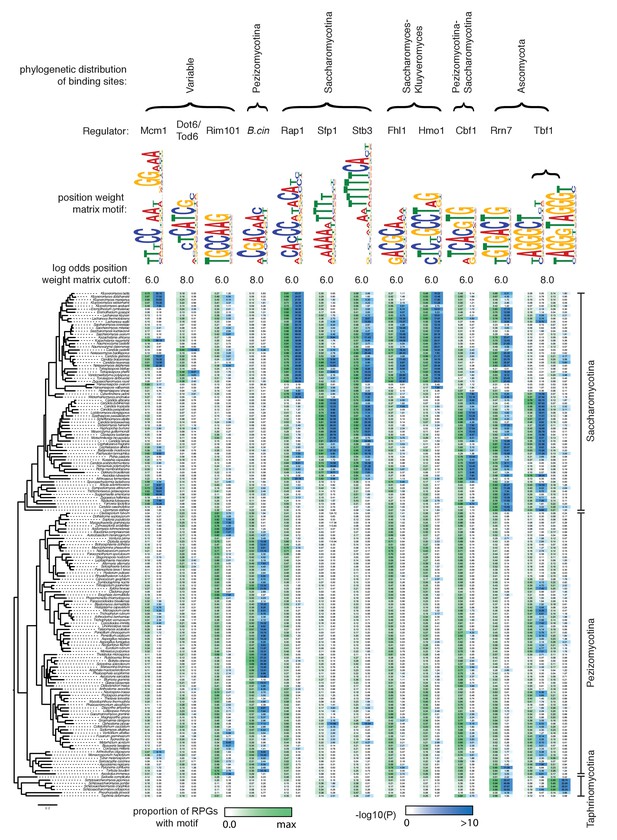
Evolution of RPG regulation.
The binding sites for 11 different known regulators of the ribosomal protein genes and one newly identified motif (‘B.cin’) found in many species was scored across 135 Ascomycete genomes.The S. cerevisiae ortholog gene name is shown above each column. For each regulator, the first column shows in green the proportion of ribosomal proteins in each species that contains at least one binding site at the log likelihood score cutoff indicated above. The second column shows the –log10(P) for the enrichment of these Mcm1 binding sites relative to upstream regulatory regions genome-wide, as expected under the hypergeometric distribution. The phylogeny is a maximum likelihood tree based on the protein sequences of 79 genes found in single copy in most species. At the top of the figure the broad distribution of the binding sites is indicated. Rrn7 corresponds to the Homol-D box, Tbf1 to the Homol-E box, and Hmo1 to the IFHL motif described in Tanay et al. (2005).
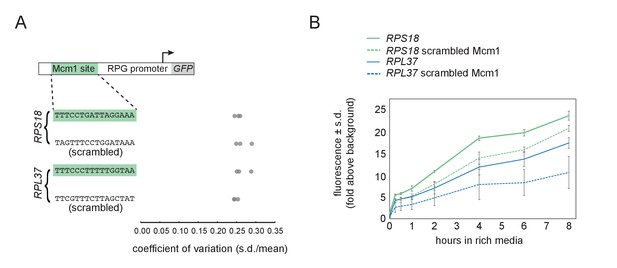
Mcm1 is an activator of the RPGs in Kl lactis.
(A) Reporter constructs from Figure 1C were analyzed for their cell-to-cell variation in expression.Shown is the coefficient of variation for each of three independent genetic isolates as in Figure 1C. (B) The reporter constructs from Figure 1C were measured over the course of 8 hr as they began to reach stationary phase. Shown is the mean fluorescence for the three independent genetic isolates for each construct.
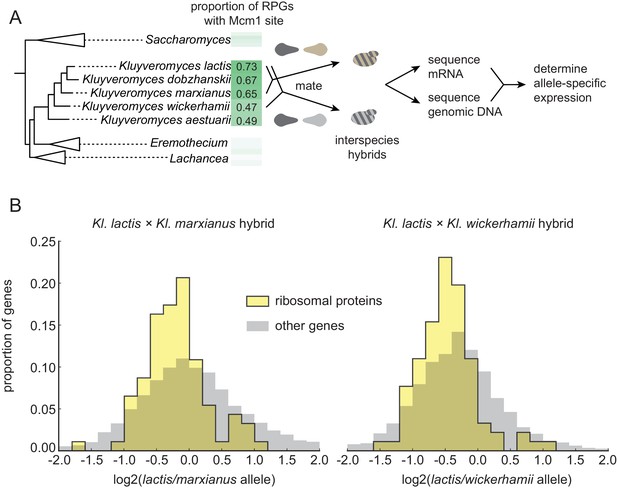
Selection on RPG expression level in Kluyveromyces.
(A) Schematic of the experimental approach to measure differences in gene expression between Kluyveromyces yeast species. Two interspecies hybrids were constructed through mating and mRNA and genomic DNA were sequenced. The differential allelic expression is the ratio of the number of reads mapping to the coding sequence of one gene vs. its ortholog in the genome of the other species. The mRNA reads for each gene were normalized to the total reads and to the genomic DNA reads mapping to the same region to control for biases introduced in the sequencing and analysis process. (B) The log2-ratio of allelic expression with the lactis allele in the numerator is shown for (left) the Kl. lactis ×Kl. marxianus hybrid (n = 3) and (right) the Kl. lactis ×Kl. wickerhamii hybrid (n = 7). Shown are histograms for ribosomal protein genes and the rest of the identified orthologs in the genome.
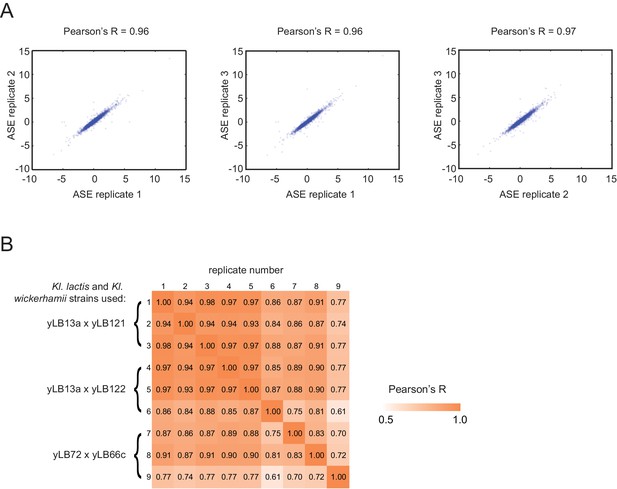
Technical validation of allele-specific experiments.
(A) Scatterplots for pairwise comparisons between replicates for the lactis-marxianus hybrid are shown. Shown is allele-specific expression (ASE) which includes the mRNA and gDNA read counts for each coding sequence in the genome. (B) Shown are Pearson’s R correlations for ASE between the nine replicates of the lactis-wickerhamii hybrid. Replicates 6 and 9 showed the lowest correlation due to chromosome loss and were not included in further analyses.
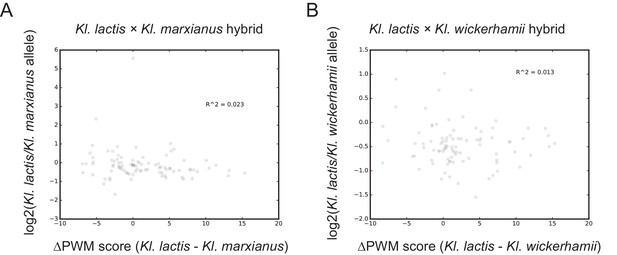
Mcm1 site evolution and allele-specific expression.
For each ribosomal protein gene, the difference in the Mcm1 position weight matrix score between the Kl. lactis ortholog and either (A) the Kl. marxianus or (B) the Kl. wickerhamii ortholog was calculated. This was plotted versus the allele-specific expression value for each gene.
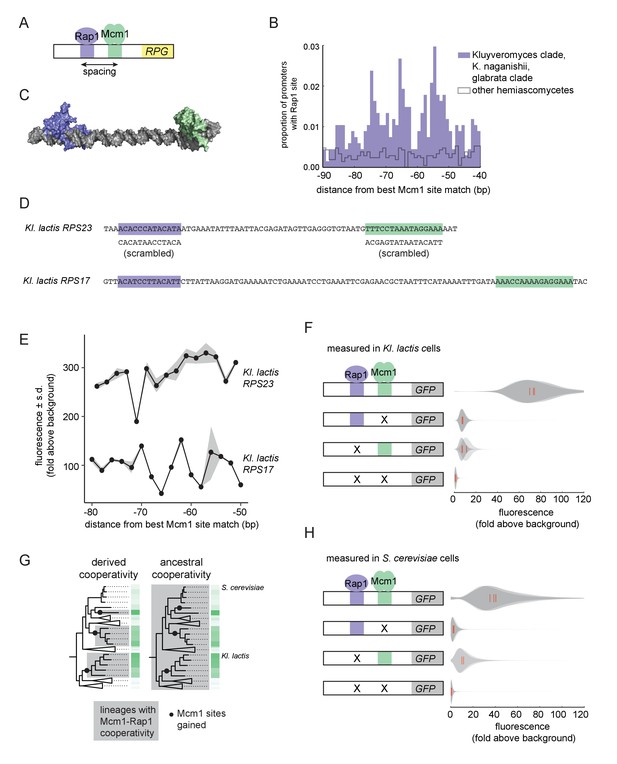
Ancestral cooperativity of Mcm1 with Rap1.
(A) Schematic of Mcm1 and Rap1 cis-regulatory sites at the RPGs. (B) RPG promoters were aligned at the strongest hit to the Mcm1 position weight matrix and the relative location of Rap1 cis-regulatory sites was plotted. Sites for Rap1 with log-likelihood >6.0 are shown at 1 bp resolution. Hemiascomycete yeast (the 29 species at the top of the tree in Figure 1A) are divided into those with large numbers of Mcm1 sites at the ribosomal protein genes (purple shading) and those without (gray line). (C) Schematic using published structures of Mcm1 and Rap1 DNA-binding domains (PDB IDs: 1MNM and 3UKG) bound to DNA connected by a DNA linker corresponding to 55 bp spacing between their cis-regulatory sites. Rap1 is shown in purple and Mcm1 is shown in green. (D–H) The ability of Mcm1 to work with the ribosomal protein gene regulator Rap1 was tested using a GFP reporter. (D) The Rap1 and Mcm1 cis-regulatory sites from Kl. lactis RPS23 and RPS17 were placed in a reporter containing the S. cerevisiae CYC1 basal promoter. Reporter variants were generated by altering the spacing between these sites and by mutating the sites individually and in combination. (E, F) Reporter variants were integrated into the genome of Kl. lactis. Cells were grown for 4 hr in rich media and expression was measured by flow cytometry. (E) Shown is the mean fluorescence for at least three independent genetic isolates. The values were divided by the average fluorescence for a cell lacking a GFP reporter (fold above background). The measurements for RPS23 and RPS17 were collected on separate days and are shown on the same axes for clarity. (F) Shown is the single-cell fluorescence distribution for three independent genetic isolates and the median (red bar), normalized by side-scatter values. (G) Diagrams showing the phylogenetic distribution of ancestral or derived cooperativity. (H) The RPS23 reporter variants were integrated into the genome of S. cerevisiae, a species that lacks Mcm1 cis-regulatory sequences at the ribosomal protein genes. Cells were grown and measured as described in (F). (For the third construct, one isolate had multiple reporter insertions and was not included.).
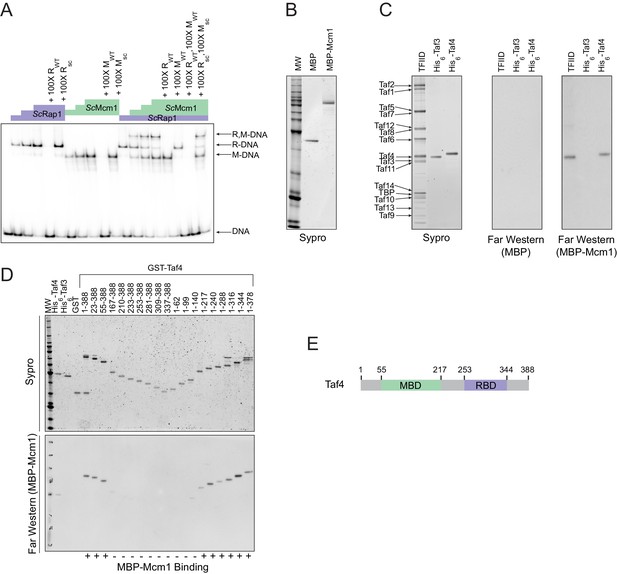
Mcm1 interacts with TFIID.
Experiments were performed in S.cerevisiae or using S. cerevisiae proteins to test the mechanism of Rap1-Mcm1 cooperativity. (A) Gel shift DNA binding assays were performed to test possible cooperative DNA binding between purified ScMcm1 and ScRap1. Gel shift reactions were performed by incubating 10 fmol (~7000 cpm) of a 79 bp 32P-labeled fragment of the Kl. lactis RPS23 promoter containing the Rap1 and Mcm1 binding sites (see Figure 3D) with either no protein, 2.5 fmol Rap1, 5 fmol Rap1, 10 fmol Rap1, 10 fmol Mcm1, 20 fmol Mcm1, 30 fmol Mcm1, or 2.5 fmol Rap1 with 10, 20, or 30 fmol Mcm1. Reactions also included either no cold competitor DNA or a 100-fold molar excess of cold ~20 bp DNA containing either the Rap1 WT (RWT) or Rap1 scrambled (Rsc) sequences and/or the Mcm1 WT (MWT) or Mcm1 scrambled (Msc) sequences in a final volume of 20 μl. Reactions were fractionated on non-denaturing polyacrylamide gels, vacuum dried, and imaged using a Bio-Rad Pharos FX imager. Radiolabeled species are indicated on the left (R,M-DNA = Rap1-Mcm1-DNA, R-DNA = Rap1 DNA, M-DNA = Mcm1 DNA) (B) Sypro Ruby stain of SDS-PAGE fractionated MBP (2.4 pmol) and MBP-Mcm1 (1.3 pmol) probe proteins used for Far Western protein-protein binding analyses. (C) Far Western protein-protein binding analysis of Mcm1 binding to TFIID. Purified TFIID, His6-Taf3, and His6-Taf4 were separated on two SDS-PAGE gels. One gel was stained with Sypro Ruby for total protein visualization (left panel). The other was electrotransferred to a membrane for protein-protein binding analysis (middle and right panels). Membranes were probed with either control MBP (middle panel) or MBP-Mcm1 (right panel). Binding of probe proteins to Tafs was detecting using an anti-MBP antibody. (D) Mapping the Mcm1 Binding Domain (MBD) of Taf4. Roughly equal molar amounts of His6-Taf4, His6-Taf3, GST, GST-Taf4, and GST-Taf4 deletion variants were fractionated on two SDS-PAGE gels. One gel was stained with Sypro Ruby for total protein visualization. The other gel was electrotransferred to a membrane and Mcm1-Taf protein-protein binding was assayed as described in (C) using the MBP-Mcm1 as the overlay protein. (E) Taf4 protein map indicating the location of the Taf4 Mcm1 Binding Domain mapped in this study (MBD, green) as well as the Rap1 Binding Domain (RBD, purple) mapped in a previous study (Layer et al., 2010).
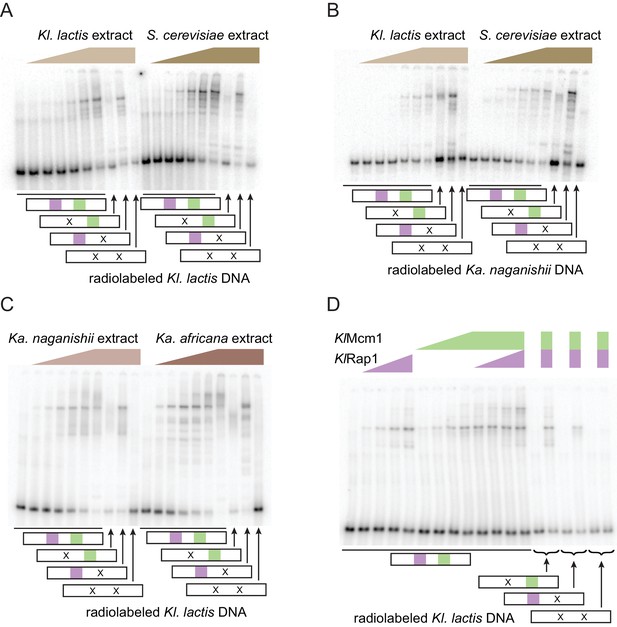
Rap1 and Mcm1 do not cooperatively bind DNA.
(A–C) Cleared cell extract from different species was incubated with radiolabeled probes containing Rap1 and Mcm1 binding sites, then run on polyacrylamide gels and imaged.A series of 4-fold dilutions is shown for each species’ extract. A fragment of the Kl. lactis RPS23 promoter containing the Rap1-Mcm1 binding sites spaced 53 bp apart (center-to-center) was used as the radiolabeled DNA oligonucleotide in (A, C and D). A fragment of the Ka. naganishii RPL1 promoter containing Rap1-Mcm1 binding sites 65 bp apart was used in (B). Rap1 sites are shown as purple squares, Mcm1 sites are shown as green squares, and scrambled sites are shown as ‘X’. (D) Gel shift using purified KlMcm1-HA and KlRap1-6-His. The highest concentration of KlMcm1-HA is approximately 100 nM, and the highest concentration of KlRap1-6-His is approximately 1 nM. A series of three-fold dilutions of the two proteins is shown as indicated above the gel image.
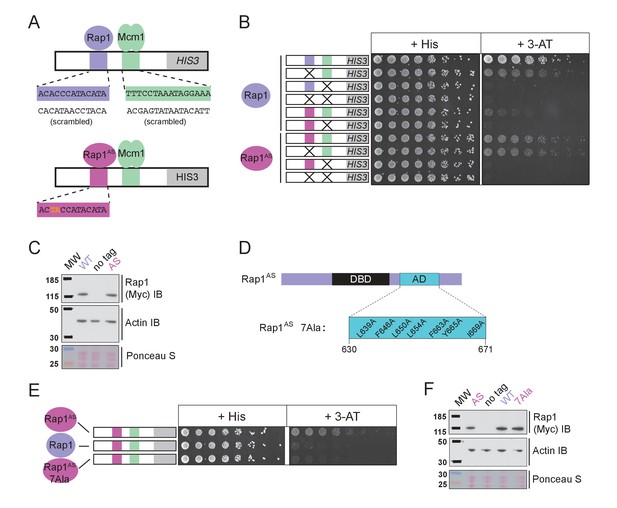
Mcm1-Rap1 cooperative activation requires Rap1-TFIID contacts.
(A) A series of reporter constructs were designed to test the mechanism of Rap1-Mcm1 cooperative activation. Experiments were performed in S. cerevisiae using S. cerevisiae proteins. (B) Growth analysis of yeast strains carrying the UASRap1-Mcm1 reporter (containing a fragment of the Kl. lactis RPS23 promoter) indicated in the diagram and either an altered DNA-binding specificity Rap1 variant (Rap1AS, magenta) or a second copy of Rap1WT (purple). To perform these analyses, yeast were grown overnight to saturation, serially diluted 1:4 and spotted using a pinning tool onto either non-selective media plates (+His) or plates containing 3-Aminotriazole (+3 AT), which selects for expression of the HIS3 reporter gene. Plates were incubated for 3 days at 30° C and imaged using a Bio-Rad ChemiDoc MP imager. (C) Immunoblot analyses of the expression levels of Myc-tagged Rap1WT and Myc-tagged Rap1AS (Myc IB) compared to actin (Actin IB) and total protein (Ponceau S) loading controls. (D) Rap1 protein map indicating the ScRap1 AD mapped to a location C-terminal of the Rap1 DBD and the seven key AD amino acids. These amino acids were mutated to alanine to inactivate the ScRap1 AD and create the Rap1AS 7Ala mutant variant. (E) Growth analyses performed using yeast carrying the UASRap1AS-Mcm1-HIS3 reporter and either Rap1AS, a second copy of Rap1WT, or Rap1AS7Ala performed as described in ‘B.’ (F) Immunoblot analysis of the Rap1 forms tested in (E) performed as described in (C).
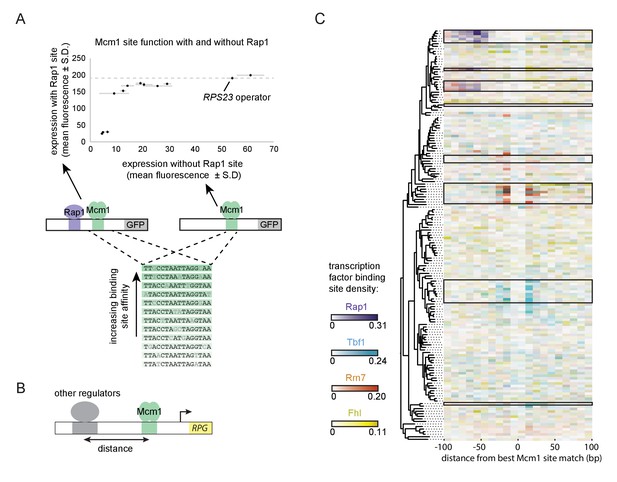
Evolutionary implications of intrinsic cooperative activation.
(A) A series of reporters were designed to test the transcriptional activation of a weak Mcm1 site in the presence and absence of a Rap1 site. A series of Mcm1 cis-regulatory sites were chosen with a range of affinities that correlate with transcription rate (Acton et al., 1997). The order of the sequences shown corresponds to their expression level on the x-axis. These sites were introduced to the S. cerevisiae CYC1 reporter and tested with (y-axis) and without (x-axis) an upstream Rap1 binding site. Cells were grown and measured as described in Figure 3E. The expression level of the WT RPS23 operator is shown as a dotted line. (B) A computational analysis was designed to detect evolution of Mcm1 sites at fixed distances from other ribosomal protein regulators. Ribosomal protein gene promoters were aligned at the strongest hit to the Mcm1 position weight matrix and the relative location of cis-regulatory sites for other transcription regulators was plotted. (C) The shading in each rectangle represents the proportion of ribosomal protein gene promoters in that species that have the given cis-regulatory site in that 10 bp interval. The clades with a large number of Mcm1 cis-regulatory sequences are shown with black boxes.
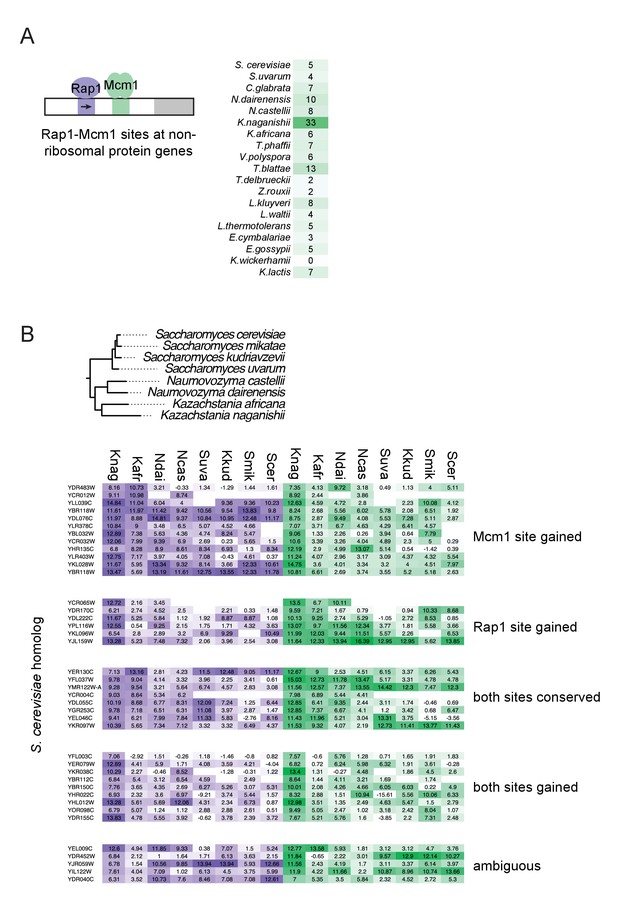
Evolution of Rap1-Mcm1 sites at additional genes.
(A) Genome-wide promoters for a subset of hemiascomycete yeasts were searched for Rap1 sites above ~%40 of the maximum score and that were oriented toward the activated gene (as seen in the ribosomal protein genes).Then, these genes were filtered by those that had an Mcm1 site between 52 and 78 bp downstream (where most of the Mcm1 sites are located in the ribosomal protein genes) and mapped to orthologs in S. cerevisiae. Shown are the number of orthologs that contained such a Rap1-Mcm1 site in the genome of each species. (B) Genes in Ka. naganishii with Rap1-Mcm1 sites as described in (A) were examined in closely related species. The scores reflect the maximum score for the Rap1 (left) or Mcm1 (right) position weight matrix in the intergenic region up to 1 kb. Based on the presence or absence in other species, they were categorized into how the sites arose in Ka. naganishii.
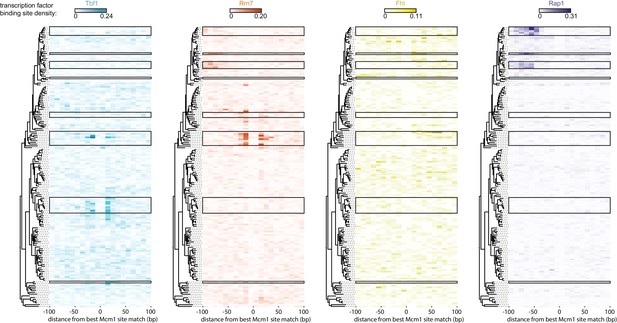
Evolution of Mcm1 cis-regulatory sites near sites for other regulators.
Shown are the locations of other cis-regulatory sites relative to the location of the best hit to the Mcm1 position weight matrix as in Figure 6C. Each of the regulators is shown separately for clarity.
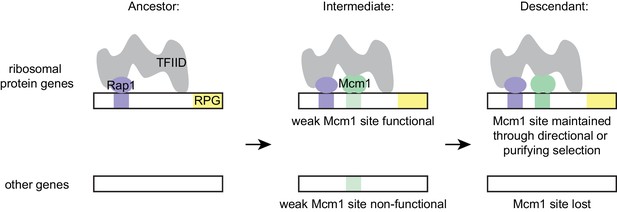
Model for evolution of cis-regulatory sites through intrinsic cooperativity.
Multiple gains of new Mcm1 sites occur in the ribosomal protein genes because Rap1 and Mcm1 both bind to TFIID, a general transcription factor. Due to the intrinsic cooperativity of Rap1 and Mcm1 (which is ancestral to the gains of Mcm1 cis-regulatory sequences) the evolution of even a weak Mcm1 site near an existing Rap1 site would produce an effect on transcription. Because they are more likely to be functional, weak Mcm1 cis-regulatory sequences are preferentially retained in the population if they arise at a specified distance (as determined by the shape of TFIID) from Rap1 cis-regulatory sequences. These sites would be preserved if there is direct selection to increase RPG expression, or if they are combined over time with the mutational degradation of other regulatory elements that bring the Mcm1 site under purifying selection.
Additional files
-
Supplementary file 1
A list of plasmids created for and used in this study.
The columns indicate the plasmid name, intended species for use, and purpose for creating the plasmid. A reference is given for plasmids from previous studies.
- https://doi.org/10.7554/eLife.37563.017
-
Supplementary file 2
A list of strains created for and used in this study.
The columns indicate the strain name, species, and genotype. A reference is given for strains from previous studies.
- https://doi.org/10.7554/eLife.37563.018
-
Supplementary file 3
A list of the genomes used for computational analyses.
The table indicates the genus and species, assembly version, and the publication or website from which the genome was obtained.
- https://doi.org/10.7554/eLife.37563.019
-
Supplementary file 4
Motifs used in the analysis of cis-regulatory sequences.
This file contains log odds matrices for each of the motifs used throughout the study. The columns indicate the scores for bases A, C, G, and T, respectively. The source of each motif is indicated in the methods section.
- https://doi.org/10.7554/eLife.37563.020
-
Supplementary file 5
Mcm1 sites at the RPGs in each species.
This file contains the sequence with the highest score matching the Mcm1 position weight matrix at each RPG in each species. The first column is the species identifier, followed by the gene name, position weight matrix score, location of the match, sequence of the match, and the strand of the match. For the purposes of the computational analysis, an Mcm1 site was considered present if the score was above 6.0.
- https://doi.org/10.7554/eLife.37563.021
-
Transparent reporting form
- https://doi.org/10.7554/eLife.37563.022





