An assay for de novo kinetochore assembly reveals a key role for the CENP-T pathway in budding yeast
Figures
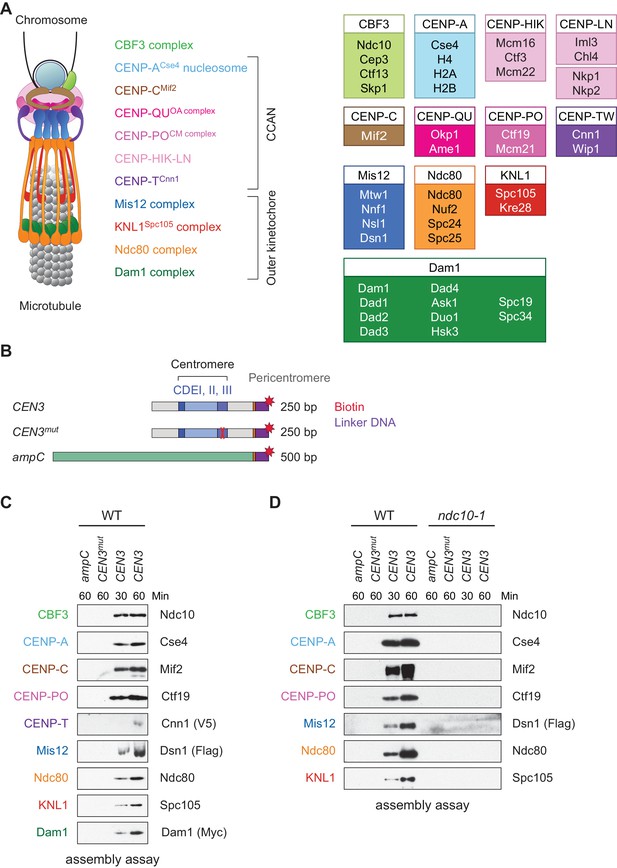
Assembly of kinetochores de novo on a centromere DNA template.
(A) A schematic of the budding yeast kinetochore. Inner kinetochore subcomplexes assemble onto centromeres, serving as the platform for outer kinetochore recruitment. The listed subcomplexes are ordered based on physical interactions, and the yeast proteins in each kinetochore subcomplex are shown on the right. (B) DNA templates for the assembly assay. The templates include 500 bp from the E. coli ampC gene that encodes for β-lactamase (green) as a negative control, the 117 bp chromosome III centromere (CEN3), or a mutant CEN3 (CEN3mut) containing three point mutations in the CBF3 binding site (red ‘X’). The three Centromere-Determining Elements (CDEs) are indicated and ~70 bp of flanking pericentromeric DNA on either side is shown (grey). The DNA templates also contain linker DNA (purple) before the biotinylation (red star) at the 3’ end of the centromere. (C) Kinetochores assembled in vitro are centromere-specific and span the entire kinetochore. The indicated DNA templates were incubated in WT whole cell extracts prepared from a CNN1-3V5 DSN1-3Flag DAM1-9myc strain (SBY17228) for the indicated time (min). DNA-bound proteins were analyzed by immunoblotting with the indicated antibodies. Extracts are shown in Figure 1—figure supplement 1. (D) Kinetochore assembly is inhibited in an ndc10-1 temperature sensitive mutant. Extracts from a DSN1-6His-3Flag strain (SBY8253) or a DSN1-6His-3Flag ndc10-1 strain (SBY8361) shifted to the non-permissive temperature were used for assembly assays. DNA-bound proteins were analyzed by immunoblotting with the indicated antibodies. Extracts in Figure 1—figure supplement 2.
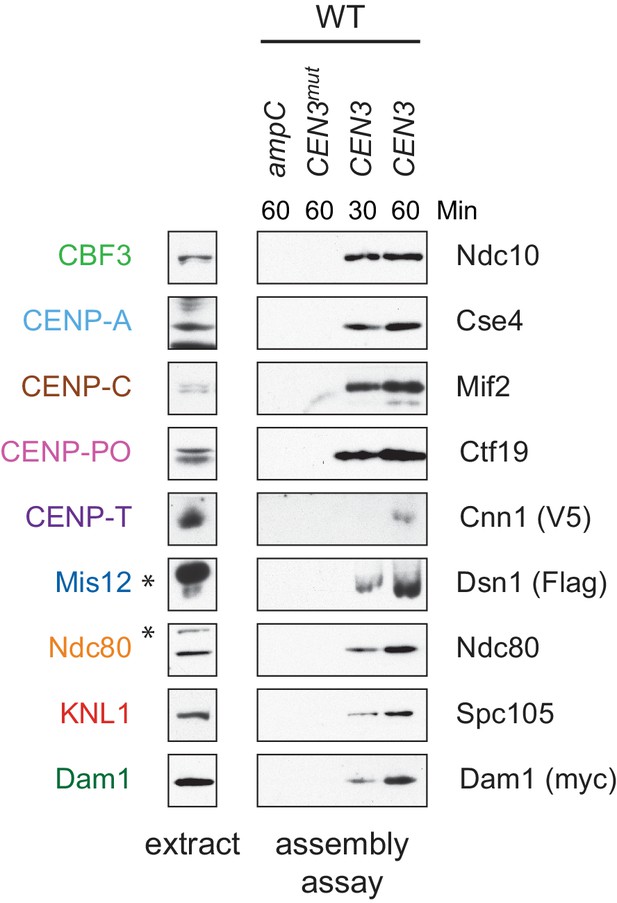
Whole cell extracts (left) and assembled kinetochores (right) from Figure 1C, immunoblotted with the indicated antibodies. * indicates a background band in the extract in all figures.
https://doi.org/10.7554/eLife.37819.003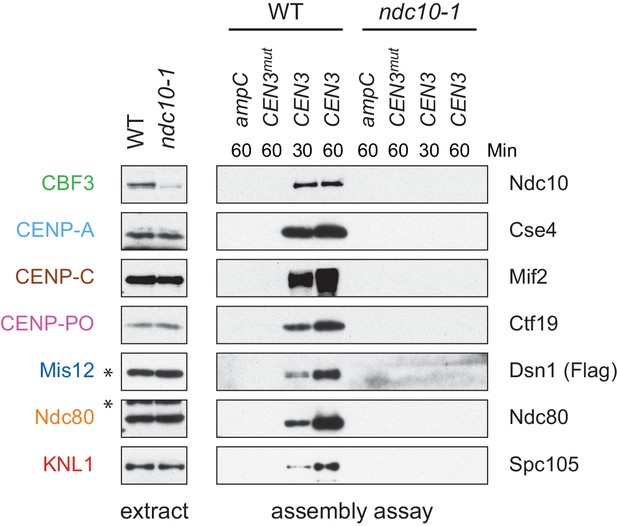
Whole cell extracts (left) and assembled kinetochores (right) from Figure 1D, immunoblotted with the indicated antibodies.
https://doi.org/10.7554/eLife.37819.004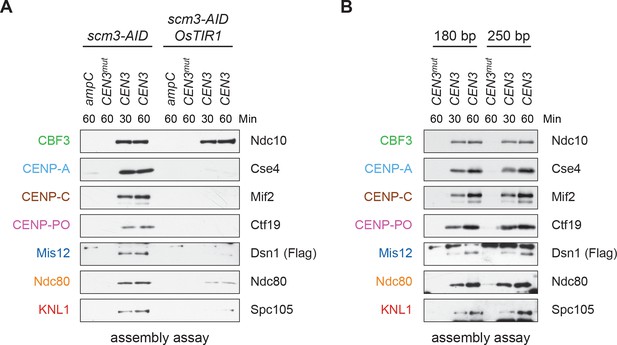
Assembled kinetochore particles contain a single, chaperone-dependent CENP-ACse4 nucleosome.
(A) Degradation of HJURPScm3 blocks assembly of the kinetochore beginning with CENP-ACse4. A DSN1-3Flag scm3-EGFP-AID strain (SBY16440) and a DSN1-3Flag scm3-EGFP-AID OsTIR1-myc strain (SBY16438) were treated with auxin and the extracts were used for assembly assays. DNA-bound proteins were analyzed by immunoblotting for the indicated proteins. Extracts in Figure 2—figure supplement 2. (B) Assembly on a centromeric DNA template of only 180 bp is similar to a 250 bp template. Extract from a DSN1-3Flag CNN1-3V5 DAM1-9myc (SBY17228) strain was used for assembly assays with the indicated DNA templates. DNA-bound proteins were analyzed by immunoblotting with the indicated antibodies. Extracts in Figure 2—figure supplement 3.
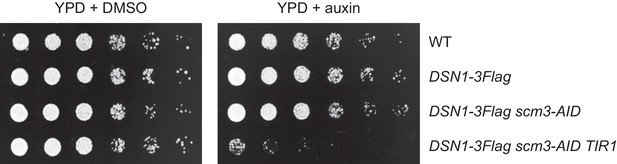
Degradation of HJURPScm3 is lethal to cells.
The Scm3-AID protein is degraded in the presence of both auxin and OsTIR1, resulting in lethality. Saturated cultures were serial diluted and plated on the indicated media. The strains used are WT (SBY4), DSN1-3Flag (SBY14441), DSN1-3Flag scm3-EGFP-AID (SBY16440), and DSN1-3Flag scm3-EGFP-AID OsTIR1-myc (SBY16438).
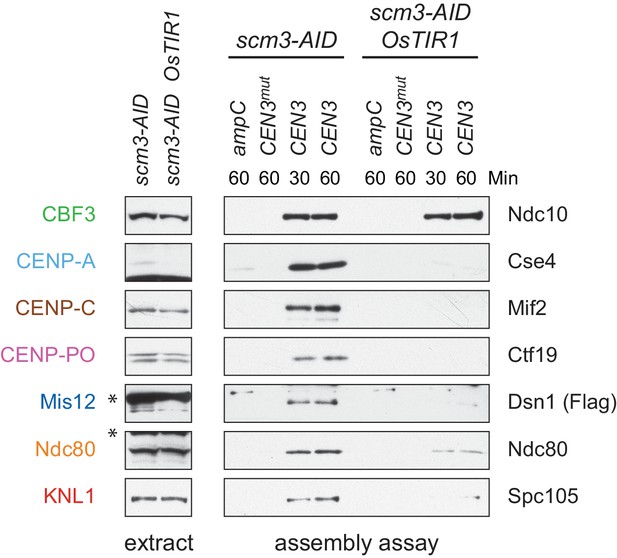
Whole cell extracts (left) and assembled kinetochores (right) from Figure 2A, immunoblotted with the indicated antibodies.
https://doi.org/10.7554/eLife.37819.008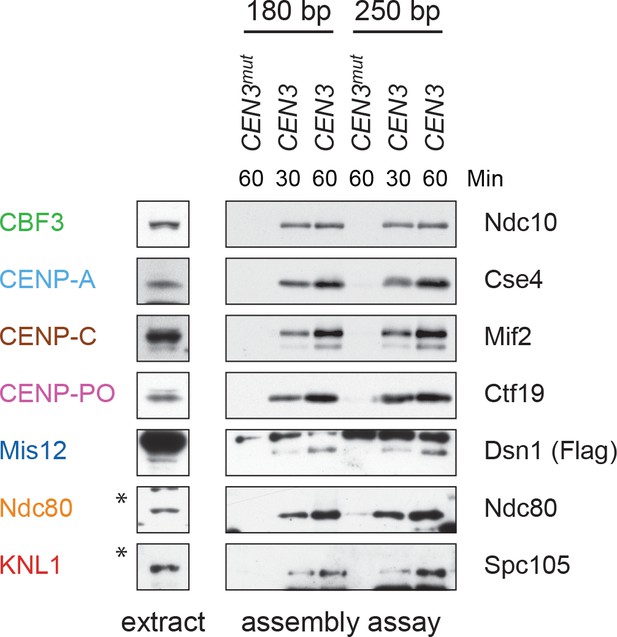
Whole cell extracts (left) and assembled kinetochores (right) from Figure 2B, immunoblotted with the indicated antibodies.
https://doi.org/10.7554/eLife.37819.009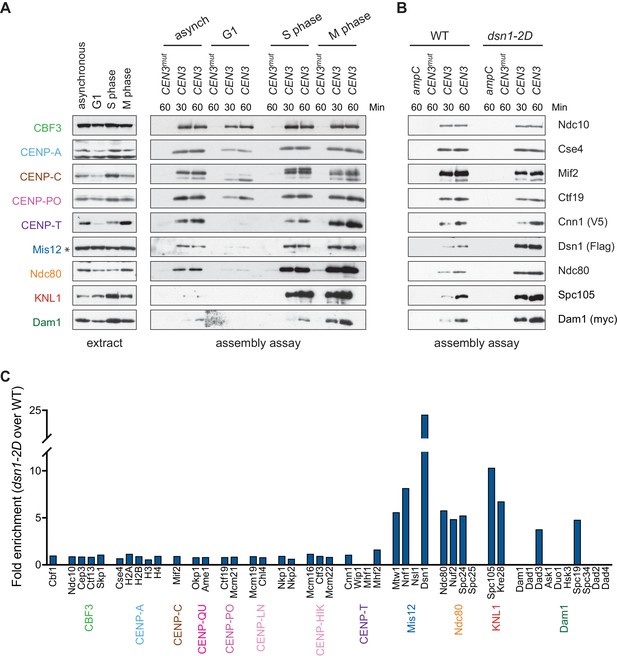
Kinetochore assembly in vitro is regulated by the cell cycle and phosphorylation.
(A) Assembly in vitro is most efficient in extracts made from mitotically arrested cells. Kinetochores were assembled using extract from WT cells (DSN1-3Flag CNN1-3V5 DAM1-9myc (SBY17227)) that were either asynchronously growing or arrested in G1 (with alpha factor), S phase (with hydroxyurea), or early mitosis (with benomyl). Diluted whole cell extracts (left) and DNA-bound proteins (right) were analyzed by immunoblotting with the indicated antibodies. (B) Outer kinetochore assembly is enhanced in dsn1-2D extracts. Assembly assays were performed using extracts prepared from benomyl-arrested DSN1-3Flag CNN1-3V5 DAM1-9myc (SBY17228) and dsn1-2D-3Flag CNN1-3V5 DAM1-9myc (SBY17234) strains on the indicated DNA templates. DNA-bound proteins were analyzed by immunoblotting with the indicated antibodies. Extracts in Figure 3—figure supplement 1. (C) dsn1-2D enhances the assembly of most outer kinetochore proteins by at least 5-fold. Assembly assays were performed using extracts from DSN1-3Flag (SBY14441) and dsn1-2D-3Flag (SBY14151) on CEN3 DNA. Assembled proteins were labeled with tandem mass tags and analyzed by quantitative mass spectrometry. For each protein, the relative abundance in dsn1-2D assembled kinetochores was divided by the relative abundance in WT to calculate the fold enrichment in the dsn1-2D assembled kinetochores.
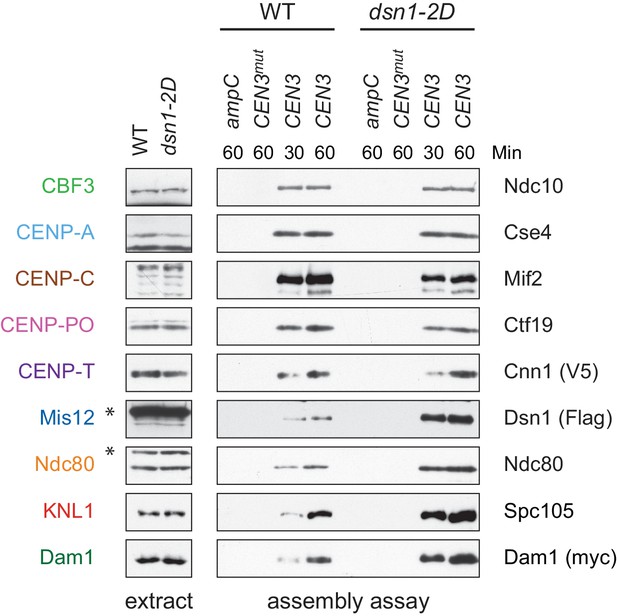
Whole cell extracts (left) and assembled kinetochores (right) from Figure 3B, immunoblotted with the indicated antibodies.
https://doi.org/10.7554/eLife.37819.011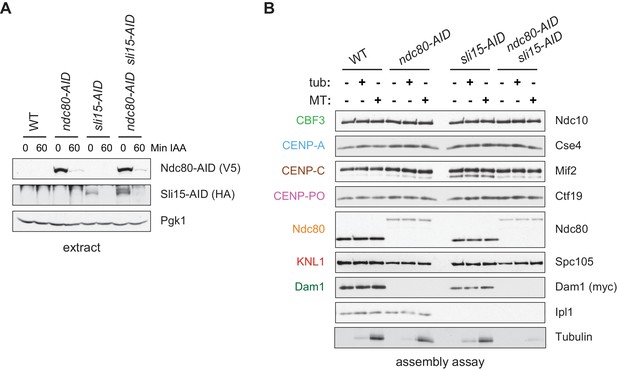
Assembled kinetochore particles bind to microtubules.
(A) The Ndc80-3V5-AID and Sli15-3HA-AID proteins are degraded after one hour of auxin treatment as determined by immunoblotting of whole cell extracts. (B) Assembled kinetochores bind microtubules but not free tubulin. Assembly assays were performed using extracts from the following strains: dsn1-2D-3Flag DAM1-9myc OsTIR1 (SBY14343), dsn1-2D-3Flag DAM1-9myc OsTIR1 ndc80-3V5-AID (SBY14336), dsn1-2D-3Flag DAM1-9myc OsTIR1 sli15-3HA-AID (SBY14890), and dsn1-2D-3Flag DAM1-9myc OsTIR1 ndc80-3V5-AID sli15-3HA-AID (SBY17238). All strains were arrested in benomyl and treated with auxin before harvesting. The assembled kinetochores were then incubated with buffer, free tubulin, or taxol-stabilized microtubules. The free tubulin and the microtubules contained alexa-647-labeled tubulin. DNA-bound proteins were analyzed by immunoblotting with the indicated antibodies, and the tubulin and microtubules were analyzed by fluorescence imaging. The Ndc80-3V5-AID protein migrates slower than untagged Ndc80. Extracts and tubulin input in Figure 4—figure supplement 1.
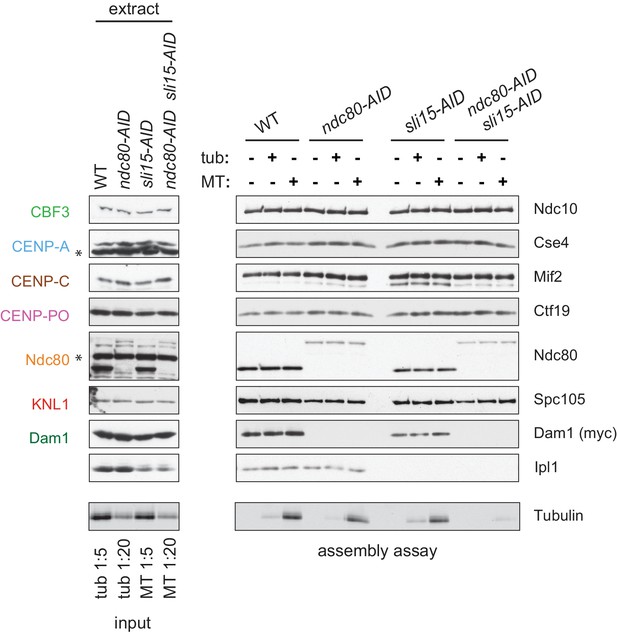
Whole cell extracts (left) and assembled kinetochores (right) from Figure 4B, immunoblotted with the indicated antibodies.
Free tubulin and microtubule inputs are loaded at 1:5 and 1:20 of the amount introduced to assembled kinetochores.
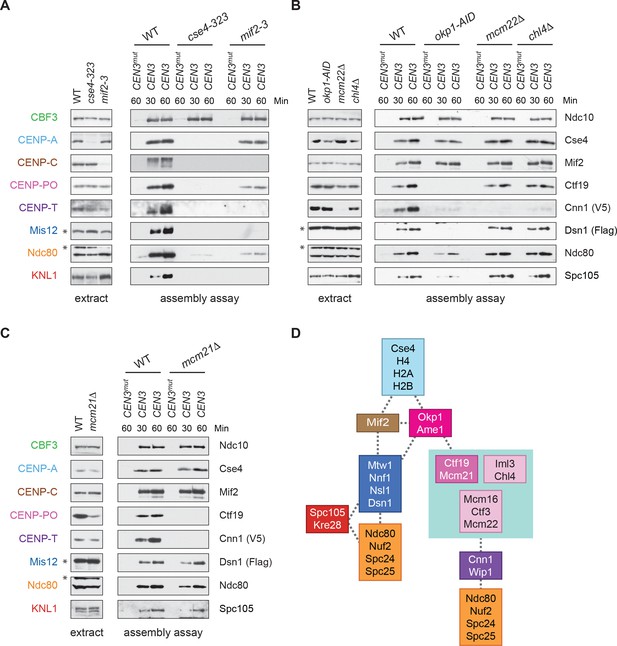
CENP-TCnn1 localization to the kinetochore requires the CCAN.
(A–C) CENP-TCnn1 assembly occurs downstream of all other inner kinetochore components. Assembly assays were performed on the indicated DNA templates using extracts prepared from cells arrested in benomyl. The strains used in (A) were also shifted to the non-permissive temperature for three hours before harvesting: DSN1-3Flag CNN1-3V5 (SBY17230), DSN1-3Flag CNN1-3V5 cse4-323 (SBY17770), and DSN1-3Flag CNN1-3V5 mif2-3 (SBY17603). The strains used in (B) were treated with auxin for three hours before harvesting: DSN1-3Flag CNN1-3V5 (SBY17230), DSN1-3Flag CNN1-3V5 okp1-3V5-AID OsTIR1 (SBY17764), DSN1-3Flag CNN1-3V5 mcm22Δ (SBY17460), and DSN1-3Flag CNN1-3V5 chl4Δ (SBY17607). The strains used in (C) were benomyl treated only: DSN1-3Flag CNN1-3V5 (SBY17230) and DSN1-3Flag CNN1-3V5 mcm21Δ (SBY18304). (D) A schematic distinguishing the proteins involved in the CENP-TCnn1 and Mis12 recruitment pathways.
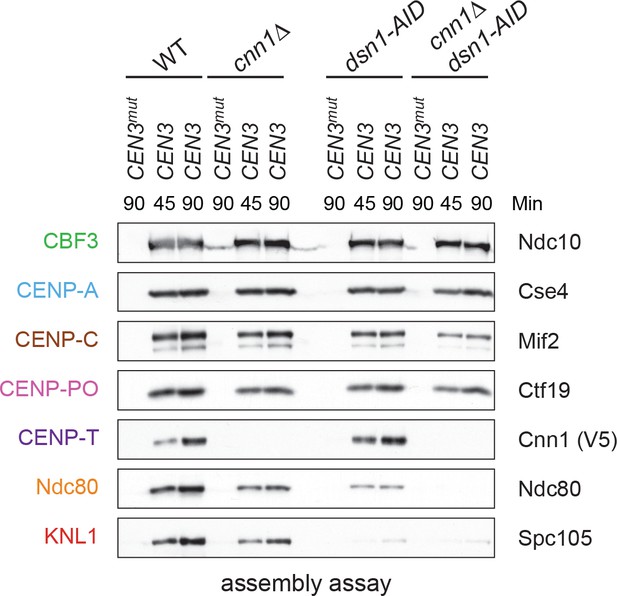
Kinetochore assembly utilizes two pathways for Ndc80 recruitment.
Dsn1 and CENP-TCnn1 both contribute to Ndc80 recruitment. Kinetochores were assembled using extract from WT, dsn1-AID, cnn1Δ, or dsn1-AID cnn1Δ double mutant cells that were arrested in benomyl and treated with auxin: DSN1-3Flag Cnn1-3V5 OsTIR1 (SBY17548), DSN1-3Flag cnn1Δ OsTIR1 (SBY17546), dsn1-3HA-AID Cnn1-3V5 OsTIR1 (SBY17544), and dsn1-3HA-AID cnn1Δ OsTIR1 (SBY17380). Extracts in Figure 6—figure supplement 1.
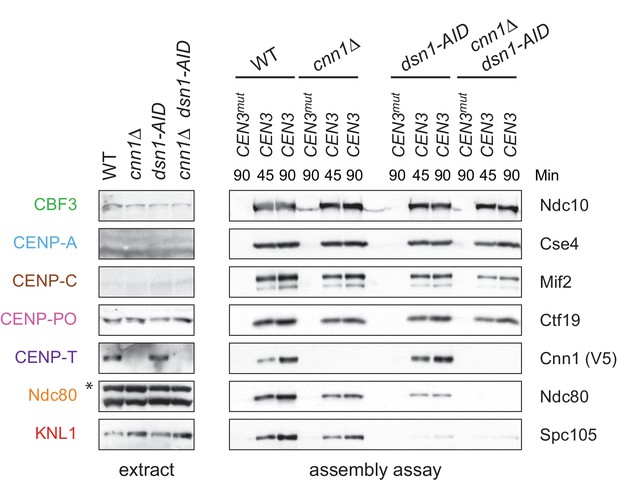
Whole cell extracts (left) and assembled kinetochores (right) from Figure 6, immunoblotted with the indicated antibodies.
https://doi.org/10.7554/eLife.37819.017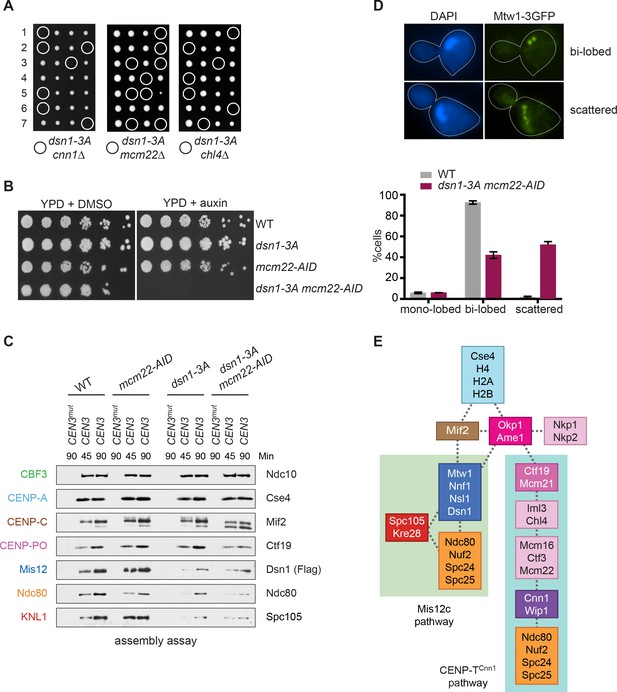
Cells require CENP-TCnn1 when the Mis12 pathway is impaired.
(A) The CENP-TCnn1 pathway is required for viability when the Mis12c assembly pathway is compromised. Dsn1-3A is synthetic lethal with cnn1Δ and deletions of other genes (MCM22 and CHL4) in the CENP-TCnn1 recruitment pathway. A dsn1-3A strain (SBY14170) was crossed to cnn1Δ (SBY13386), mcm22Δ (SBY6997), and chl4Δ (SBY8788). The meiotic products (tetrads) of the resulting diploids are oriented left to right, haploid spores were genotyped, and double mutants are indicated with circles. (B) A dsn1-3A mcm22-AID double mutant is lethal when treated with auxin. Serial dilutions of the following yeast strains were plated on the indicated media: WT (SBY3), dsn1-3A-3Flag (SBY14170), mcm22-3HA-AID OsTIR1 (SBY17982), and dsn1-3A-3Flag mcm22-3HA-AID OsTIR1 (SBY18171). (C) The CENP-TCnn1 pathway recruits Ndc80 when Mis12 complex assembly is compromised. Assembly was performed with extracts from HU-arrested strains that were treated with auxin: DSN1-3Flag OsTIR1 (SBY14131), DSN1-3Flag mcm22-3HA-AID OsTIR1 (SBY18044), dsn1-3A-3Flag OsTIR1 (SBY14169), and dsn1-3A-3Flag mcm22-3HA-AID OsTIR1 (SBY18034). Extracts in Figure 7—figure supplement 2. (D) WT (SBY18498) and dsn1-3A mcm22-3HA osTIR1 (SBY18324) cells containing MTW1-3GFP were released from G1 and kinetochores were analyzed by fluorescence microscopy during metaphase. The percentage of cells containing mono-lobed, bi-lobed or scattered kinetochores was quantified and a representative picture of the bi-lobed and scattered categories is shown above the graph. The p value for the difference between WT and the double mutant for bi-lobed kinetochores is 0.04 and for scattered kinetochores is 0.036. (E) The sequential order of kinetochore subcomplex recruitment to the DNA, as determined from our data and from (Pekgöz Altunkaya et al., 2016). Dotted lines indicate physical interactions.
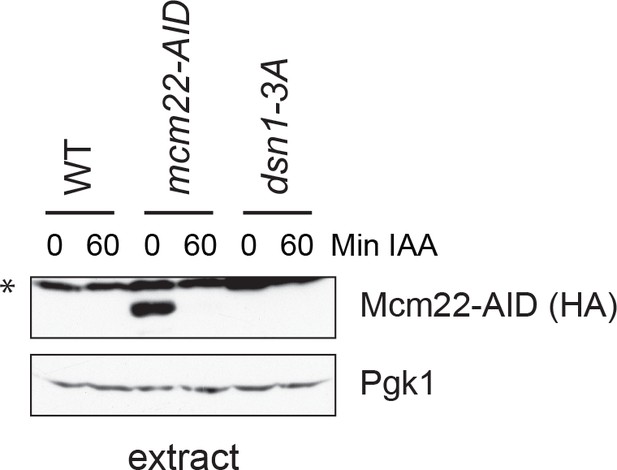
Mcm22-3HA-AID is efficiently degraded after 60 min of auxin treatment.
The strains are DSN1-3Flag CNN1-3V5 OsTIR1 (SBY18040), DSN1-3Flag CNN1-3V5 mcm22-3HA-AID OsTIR1 (SBY18042), and dsn1-3A-3Flag CNN1-3V5 OsTIR1 (SBY18028).
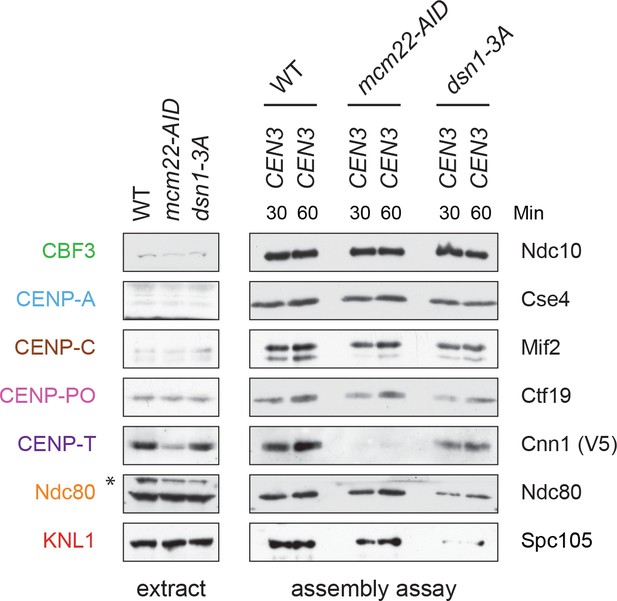
Cnn1 recruitment is blocked in Mcm22-3HA-AID strains.
Whole cell extracts and assembly assays for strains DSN1-3Flag CNN1-3V5 OsTIR1 (SBY18040), DSN1-3Flag CNN1-3V5 mcm22-3HA-AID OsTIR1 (SBY18042), and dsn1-3A-3Flag CNN1-3V5 OsTIR1 (SBY18028). (C) Mcm22-3HA-AID degradation for the experiment in Figure 7C.
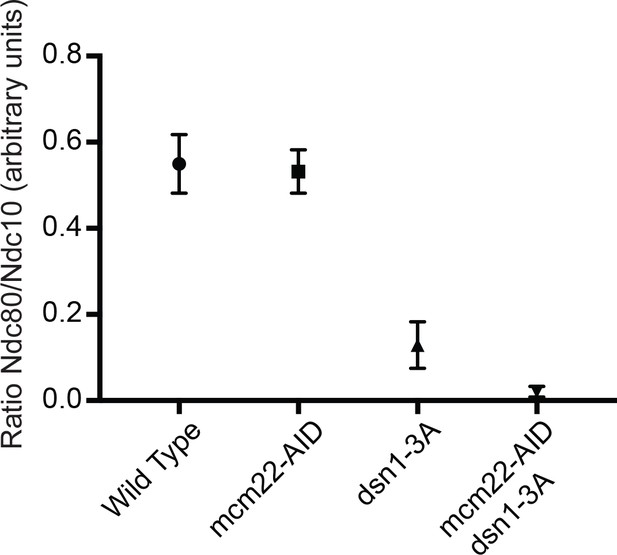
Assembly assays were performed using the following strains that were arrested in hydroxyurea for two hours and then treated with IAA for one hour: WT (SBY14131), mcm22-3HA-AID OsTIR1 (SBY18453), dsn1-3A (SBY14169) and dsn1-3A mcm22-3HA-AID OsTIR1 (SBY18172).
The levels of Ndc10 and Ndc80 were quantified and the relative ratio was graphed. The standard deviation is indicated.
Tables
Components from each of the core subcomplexes are detected on assembled kinetochores.
Kinetochores were assembled on ampC, CEN3mut, or CEN3 DNA from an asynchronous WT DSN1-3Flag (SBY14441) extract and analyzed by LC/MS/MS mass spectrometry. The table indicates the human ortholog (if applicable) of each yeast protein, the percent coverage, and the number of unique and total peptides detected from each assembly. We included the only detected microtubule-associated protein.
| Table 1. WT assembled kinetochores | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ampC | ampC | ampC | CEN3mut | CEN3mut | CEN3mut | CEN3 | CEN3 | CEN3 | ||||
| Subcomplex | Yeast Protein | Human Protein | % Coverage | Unique Peptides | Total Peptides | % Coverage | Unique Peptides | Total Peptides | % Coverage | Unique Peptides | Total Peptides | |
| CPC | Ipl1 | Aurora B | Not present | Not present | 23.7 | 8 | 10 | |||||
| Sli15 | INCENP | 7 | 2 | 2 | 14.8 | 6 | 7 | 64.3 | 54 | 113 | ||
| Bir1 | Survivin | 15.1 | 10 | 11 | 22.6 | 14 | 17 | 59.2 | 70 | 177 | ||
| Nbl1 | Borealin | Not present | Not present | 76.7 | 6 | 11 | ||||||
| CCAN | Cbf1 | Cbf1 | Not present | 62.7 | 30 | 77 | 59.3 | 29 | 60 | |||
| Cbf3 | Ndc10 | 11.4 | 7 | 8 | 32.9 | 23 | 26 | 63.2 | 78 | 194 | ||
| Cep3 | 3.5 | 1 | 2 | 15.3 | 8 | 9 | 34.2 | 25 | 76 | |||
| Ctf13 | 2.7 | 1 | 1 | 18.4 | 5 | 5 | 46 | 22 | 38 | |||
| Skp1 | 28.4 | 3 | 3 | 19.6 | 2 | 2 | 41.8 | 12 | 24 | |||
| Nucleosome | Cse4 | CENP-A | 13.1 | 3 | 3 | 24.5 | 6 | 8 | 49.8 | 10 | 31 | |
| Hta2 | H2A | 35.6 | 7 | 20 | 35.6 | 5 | 13 | 35.6 | 6 | 19 | ||
| Htb2 | H2B | 45 | 8 | 18 | 39.7 | 7 | 29 | 39.7 | 7 | 29 | ||
| Hht1 | H3 | 5.1 | 1 | 1 | 5.1 | 1 | 1 | Not present | ||||
| Hhf1 | H4 | 45.6 | 7 | 11 | 56.3 | 8 | 19 | 46.6 | 8 | 13 | ||
| Nucleosome | Psh1 | 3.9 | 1 | 1 | Not present | Not present | ||||||
| Associated | Scm3 | HJURP | Not present | 6.3 | 1 | 1 | 28.3 | 8 | 10 | |||
| Mif2 | Mif2 | CENP-C | Not present | 9.7 | 4 | 4 | 58.7 | 29 | 39 | |||
| OA | Okp1 | CENP-Q | Not present | 20.9 | 7 | 9 | 42.6 | 21 | 34 | |||
| Ame1 | CENP-U | Not present | 20.4 | 5 | 6 | 61.4 | 22 | 41 | ||||
| CM | Ctf19 | CENP-P | Not present | 6.8 | 2 | 2 | 44.7 | 21 | 31 | |||
| Mcm21 | CENP-O | Not present | 10.3 | 3 | 4 | 65.8 | 26 | 39 | ||||
| Iml3 | Iml3 | CENP-L | Not present | Not present | 60.8 | 13 | 19 | |||||
| Chl4 | CENP-N | Not present | 7.9 | 3 | 3 | 37.1 | 16 | 19 | ||||
| Nkp1 | Not present | 26.9 | 4 | 6 | 57.6 | 17 | 28 | |||||
| Nkp2 | Not present | 15 | 2 | 4 | 55.6 | 7 | 10 | |||||
| Ctf3 | Mcm16 | CENP-H | Not present | 22.7 | 2 | 3 | 48.6 | 7 | 11 | |||
| Ctf3 | CENP-I | Not present | 5.3 | 3 | 3 | 23.7 | 17 | 27 | ||||
| Mcm22 | CENP-K | Not present | 25.5 | 3 | 4 | 81.6 | 18 | 27 | ||||
| Cnn1 | Cnn1 | CENP-T | Not present | Not present | 45.7 | 13 | 18 | |||||
| Wip1 | CENP-W | Not present | Not present | 39.3 | 2 | 2 | ||||||
| Mhf1 | CENP-S | 48.9 | 4 | 4 | 48.9 | 3 | 7 | 40 | 2 | 5 | ||
| Mhf2 | CENP-X | 43.8 | 4 | 8 | 47.5 | 4 | 6 | 28.8 | 3 | 4 | ||
| Outer Kt | Mtw1 | Mtw1 | Mis12 | Not present | Not present | 22.8 | 4 | 4 | ||||
| Nnf1 | PMF1 | Not present | Not present | 13.9 | 2 | 2 | ||||||
| Nsl1 | Nsl1 | Not present | Not present | 24.1 | 3 | 3 | ||||||
| Dsn1 | Dsn1 | Not present | Not present | 7.3 | 2 | 2 | ||||||
| Ndc80 | Ndc80 | HEC1 | Not present | Not present | 28.4 | 15 | 16 | |||||
| Nuf2 | NUF2 | Not present | Not present | 32.8 | 12 | 13 | ||||||
| Spc24 | SPC24 | Not present | Not present | 57.3 | 8 | 8 | ||||||
| Spc25 | SPC25 | Not present | Not present | 27.1 | 5 | 5 | ||||||
| Spc105 | Spc105 | KNL1 | Not present | Not present | 5 | 3 | 3 | |||||
| Kre28 | Zwint1 | Not present | Not present | Not present | ||||||||
| Dam1 | Dam1 | Not present | Not present | 10.8 | 2 | 2 | ||||||
| Dad1 | 26.6 | 1 | 1 | 26.6 | 1 | 2 | 26.6 | 1 | 1 | |||
| Dad3 | Not present | Not present | Not present | |||||||||
| Ask1 | Not present | Not present | 8.2 | 1 | 1 | |||||||
| Duo1 | Not present | Not present | 7.3 | 1 | 1 | |||||||
| Hsk3 | 15.9 | 1 | 1 | 15.9 | 1 | 1 | 15.9 | 1 | 1 | |||
| Spc19 | Not present | Not present | 8.5 | 1 | 1 | |||||||
| Spc34 | Not present | Not present | 4.7 | 1 | 2 | |||||||
| Dad2 | Not present | Not present | Not present | |||||||||
| Dad4 | Not present | Not present | Not present | |||||||||
| MAPs | Stu2 | CHTOG | 3.5 | 2 | 2 | Not present | Not present | |||||
Outer kinetochore assembly is enhanced by Dsn1 phosphorylation.
Kinetochores were assembled on the indicated DNA templates from an asynchronous dsn1-2D-3Flag (SBY14151) extract and analyzed by mass spectrometry as in Table 1. We included the detected microtubule-associated proteins.
| Table 2. dsn1-2D assembled kinetochores | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ampC | ampC | ampC | CEN3mut | CEN3mut | CEN3mut | CEN3 | CEN3 | CEN3 | ||||
| Subcomplex | Yeast Protein | Human Protein | % Coverage | Unique Peptides | Total Peptides | % Coverage | Unique Peptides | Total Peptides | % Coverage | Unique Peptides | Total Peptides | |
| CPC | Ipl1 | Aurora B | Not present | Not present | 24.5 | 9 | 15 | |||||
| Sli15 | INCENP | 11 | 4 | 5 | 21.9 | 10 | 11 | 62.6 | 61 | 221 | ||
| Bir1 | Survivin | 20.2 | 13 | 15 | 25.5 | 17 | 20 | 64.3 | 71 | 257 | ||
| Nbl1 | Borealin | 23.3 | 1 | 1 | 21.9 | 1 | 1 | 61.6 | 7 | 19 | ||
| CCAN | Cbf1 | Cbf1 | Not present | 65 | 27 | 53 | 59.3 | 30 | 92 | |||
| Cbf3 | Ndc10 | 21.2 | 2 | 14 | 24.5 | 19 | 22 | 58.9 | 68 | 563 | ||
| Cep3 | 20.6 | 8 | 9 | 15.3 | 8 | 10 | 34.5 | 21 | 111 | |||
| Ctf13 | 2.7 | 1 | 1 | 12.3 | 5 | 5 | 40.6 | 21 | 71 | |||
| Skp1 | 8.8 | 1 | 2 | 22.2 | 3 | 3 | 44.3 | 11 | 32 | |||
| Nucleosome | Cse4 | CENP-A | 20.5 | 5 | 5 | 14 | 4 | 6 | 49.3 | 10 | 34 | |
| Hta2 | H2A | 35.6 | 5 | 13 | 35.6 | 7 | 20 | 35.6 | 5 | 34 | ||
| Htb2 | H2B | 39.7 | 7 | 18 | 39.7 | 7 | 31 | 31.3 | 6 | 29 | ||
| Hht1 | H3 | Not present | 5.1 | 1 | 1 | Not present | ||||||
| Hhf1 | H4 | 56.3 | 9 | 11 | 56.3 | 8 | 16 | 55.3 | 8 | 22 | ||
| Nucleosome Associated | Psh1 | 7.4 | 2 | 2 | Not present | Not present | ||||||
| Scm3 | HJURP | Not present | Not present | 30.5 | 8 | 17 | ||||||
| Mif2 | Mif2 | CENP-C | Not present | 13.7 | 5 | 5 | 55.7 | 27 | 54 | |||
| OA | Okp1 | CENP-Q | Not present | 22.7 | 8 | 9 | 43.6 | 21 | 50 | |||
| Ame1 | CENP-U | Not present | 28.7 | 5 | 5 | 54.9 | 19 | 43 | ||||
| CM | Ctf19 | CENP-P | Not present | 9.8 | 3 | 3 | 42.3 | 17 | 41 | |||
| Mcm21 | CENP-O | Not present | 25.8 | 7 | 8 | 48.6 | 23 | 42 | ||||
| Iml3 | Iml3 | CENP-L | Not present | 15.5 | 3 | 3 | 60.8 | 13 | 28 | |||
| Chl4 | CENP-N | Not present | 7.2 | 3 | 3 | 29 | 12 | 21 | ||||
| Nkp1 | Not present | 26.5 | 4 | 6 | 58 | 18 | 35 | |||||
| Nkp2 | Not present | 15 | 2 | 3 | 35.9 | 5 | 14 | |||||
| Ctf3 | Mcm16 | CENP-H | 14.9 | 1 | 1 | 19.9 | 2 | 2 | 44.8 | 6 | 12 | |
| Ctf3 | CENP-I | Not present | 4 | 2 | 2 | 13.9 | 12 | 19 | ||||
| Mcm22 | CENP-K | Not present | 14.6 | 2 | 2 | 74.9 | 17 | 34 | ||||
| Cnn1 | Cnn1 | CENP-T | Not present | Not present | 27.1 | 8 | 11 | |||||
| Wip1 | CENP-W | Not present | Not present | 21.1 | 2 | 2 | ||||||
| Mhf1 | CENP-S | 48.9 | 3 | 4 | 48.9 | 4 | 9 | 21.1 | 2 | 2 | ||
| Mhf2 | CENP-X | 62.5 | 7 | 8 | 47.5 | 4 | 4 | 28.8 | 2 | 3 | ||
| Outer KT | Mtw1 | Mtw1 | Mis12 | Not present | 18.7 | 4 | 4 | 48.1 | 13 | 21 | ||
| Nnf1 | PMF1 | Not present | 16.9 | 2 | 2 | 30.3 | 10 | 13 | ||||
| Nsl1 | Nsl1 | Not present | 15.3 | 2 | 2 | 69 | 15 | 21 | ||||
| Dsn1 | Dsn1 | Not present | 13.4 | 4 | 4 | 39.2 | 21 | 31 | ||||
| Ndc80 | Ndc80 | HEC1 | Not present | 18.5 | 8 | 9 | 56.4 | 37 | 63 | |||
| Nuf2 | NUF2 | Not present | 15.7 | 6 | 6 | 51 | 27 | 42 | ||||
| Spc24 | SPC24 | Not present | 38.5 | 4 | 5 | 63.4 | 12 | 26 | ||||
| Spc25 | SPC25 | Not present | 8.1 | 1 | 1 | 37.6 | 8 | 10 | ||||
| Spc105 | Spc105 | KNL1 | Not present | 3.1 | 2 | 2 | 45.9 | 41 | 60 | |||
| Kre28 | Zwint1 | Not present | 3.6 | 1 | 1 | 7 | 2 | 2 | ||||
| Dam1 | Dam1 | Not present | Not present | 21.6 | 5 | 6 | ||||||
| Dad1 | 26.6 | 1 | 1 | 26.6 | 1 | 1 | 37.2 | 2 | 4 | |||
| Dad3 | Not present | Not present | 29.8 | 2 | 3 | |||||||
| Ask1 | Not present | Not present | 21.2 | 3 | 4 | |||||||
| Duo1 | Not present | Not present | 18.2 | 4 | 4 | |||||||
| Hsk3 | 15.9 | 1 | 1 | 15.9 | 1 | 1 | 15.9 | 1 | 1 | |||
| Spc19 | Not present | Not present | 30.3 | 4 | 6 | |||||||
| Spc34 | Not present | Not present | 31.5 | 6 | 11 | |||||||
| Dad2 | Not present | Not present | Not present | |||||||||
| Dad4 | Not present | Not present | Not present | |||||||||
| MAPs | Stu2 | CHTOG | Not present | Not present | 33.6 | 23 | 31 | |||||
| Bim1 | Not present | Not present | 17.7 | 4 | 5 | |||||||
| Slk19 | Not present | Not present | 1.6 | 1 | 1 | |||||||
| Bik1 | Not present | Not present | 10 | 3 | 4 | |||||||
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (S. cerevisiae) | See supplementary file 1 | |||
| Strain, strain background (Saccharomyces cerevisiae) | W303 | |||
| Genetic reagent (S. cerevisiae) | See supplementary file 1 | |||
| Antibody | anti-Ndc10 (rabbit polyclonal) | Desai lab | OD1 | (1:5,000) |
| Antibody | anti-Cse4 (rabbit polyclonal) | Biggins lab | 9536 | (1:500) |
| Antibody | anti-Mif2 (rabbit polyclonal) | Desai lab | OD2 | (1:6,000) |
| Antibody | anti-Ctf19 (rabbit polyclonal) | Desai lab | OD10 | (1:15,000) |
| Antibody | anti-Ndc80 (rabbit polyclonal) | Desai lab | OD4 | (1:10,000) |
| Antibody | anti-Spc105 (rabbit polyclonal) | Biggins lab | PAC4065 | (1:10,000) |
| Antibody | anti-Ipl1 (rabbit polyclonal) | Desai lab | OD121 | (1:300) |
| Antibody | anti-HA (mouse monoclonal) | Roche | 12AC5, Catalog #1-583-816 | (1:10,000) |
| Antibody | anti-V5 (mouse monoclonal) | Invitrogen | Catalog #R960-25 | (1:5,000) |
| Antibody | anti-Flag (mouse monoclonal) | Sigma-Aldrich | Catalog #F3165 | (1:3,000) |
| Antibody | anti-Myc (mouse monoclonal) | Covance | 9E10, Catalog #MMS-150R | (1:10,000) |
| Antibody | anti-mouse secondary (goat monoclonal) | GE Healthcare BioSciences | NA931 | (1:10,000) |
| Antibody | anti-rabbit secondary (goat monoclonal) | GE Healthcare BioSciences | NA934 | (1:10,000) |
| Recombinant DNA reagent | See supplementary file 2 | |||
| Sequence-based reagent | See supplementary file 3 | |||
| Chemical compound, drug | α-factor | United Biochemical Research Inc. | 10 mg/mL | |
| Chemical compound, drug | hydroxyurea | Sigma | H8627 | 0.2M |
| Chemical compound, drug | benomyl | Sigma | 381586–25G | 60 mg/mL |
| Chemical compound, drug | indole-3-acetic acid (IAA) | Sigma | I3750-5G-A | 500 mM |
| Other | Dynabeads M-280 Streptadivin | Invitrogen | 112-05D |
Additional files
-
Supplementary file 1
Yeast strains used in this study.
Complete genotypes of the Saccharomyces cerevisiae strains used are listed along with the strain number to reference. Replicating plasmids are indicated in brackets. All strains are isogenic with W303.
- https://doi.org/10.7554/eLife.37819.022
-
Supplementary file 2
Plasmids used in this study.
The relevant genes and markers on each plasmid used are listed.
- https://doi.org/10.7554/eLife.37819.023
-
Supplementary file 3
DNA primers used in this study.
The sequence and purpose of each primer used is listed.
- https://doi.org/10.7554/eLife.37819.024
-
Transparent reporting form
- https://doi.org/10.7554/eLife.37819.025





