Prolonged cross-bridge binding triggers muscle dysfunction in a Drosophila model of myosin-based hypertrophic cardiomyopathy
Figures
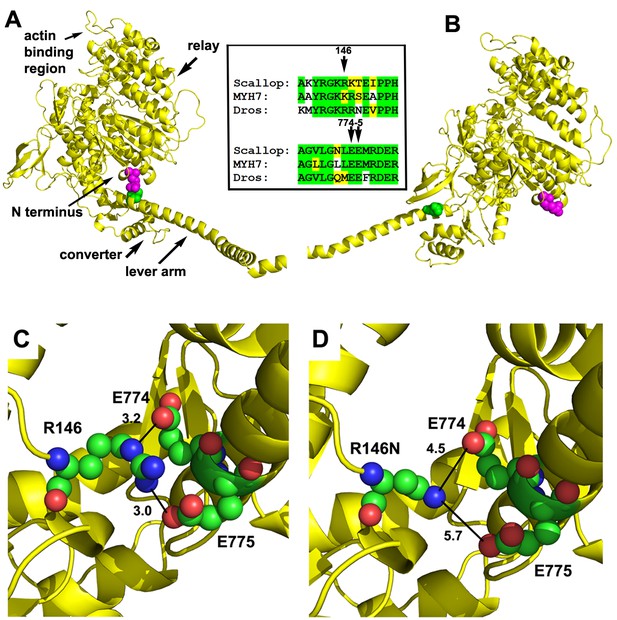
Location and interactions of R146 within the myosin molecule during pre- and post-power stroke states.
(A) Scallop myosin S1 in the pre-power stroke state (1QVI) was used as a template to model Drosophila IFM S1. R146, the positively charged residue involved in HCM is depicted with magenta spheres. In this state, the lever arm is in close proximity to R146, with the site of interaction colored green. The inset shows the amino acid context for R146 (top) and its interaction sites on the lever arm (bottom) in scallop, Drosophila and human β-cardiac myosin. Identical (green) and conserved (yellow) residues demonstrate a high degree of sequence similarity. (B) Scallop myosin S1 in the post-power stroke state (1KK8) was used as a template to model Drosophila IFM S1. In this state, R146 is distant from the lever arm interaction site (green). (C) Close-up view of potential interactions of R146 in modeled Drosophila IFM S1 in the pre-power stroke state. R146 displays charge interaction with E774 with a contact distance of 3.2 Å. A second charge interaction occurs between R146 and E775 with a contact distance of 3.0 Å. The close contact distances permit the formation of salt bridges between the positively charged R146 and the negatively charged E774 and E775 residues. (D) The R146N mutation eliminates the close contacts with E774 and E775 during the pre-power stroke state. R146N to E774 distance is 4.5 Å and R146N to E775 distance is 5.7 Å.
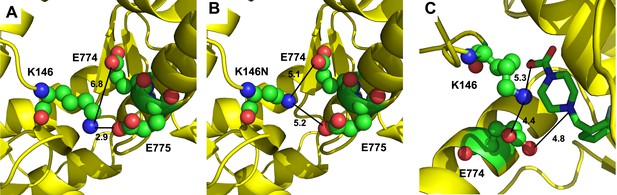
Analysis of the K146 residue in mammalian cardiac myosin in the pre-power stroke state.
(A) Scallop myosin S1 in the pre-power stroke state (PDB 1QVI) was used as a template to model the human β-cardiac myosin sequence. K146, the positively charged residue involved in HCM is in close proximity to E775 with a contact distance of 2.9 Å. (B) By modeling hypertrophic cardiomyopathy mutation K146N of β-cardiac myosin during the pre-power stroke state, it is observed that the close contact with E775 is eliminated and the mutant residue remains distant from E774. (C) Bovine β-cardiac myosin structure in the presence of omecamtiv mecarbil (PDB 5N69) shows that the drug, which stabilizes the pre-power stroke state, interacts with both K146 and E774. There is a 4.4 Å distance between these two β-cardiac myosin residues in the presence of the drug.
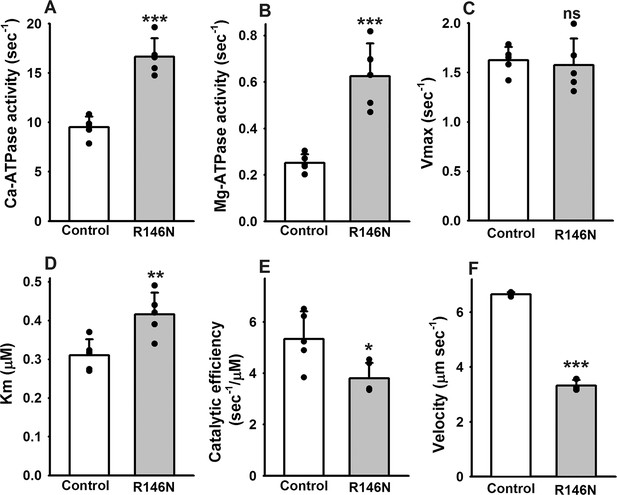
Effects of R146N on myosin enzymatic activity and in vitro actin sliding velocity.
Myosin isolated from IFMs of wild-type transgenic control (PwMhc2) and R146N lines was assessed for ATPase parameters (N = 5): (A) Ca-ATPase activity, (B) basal Mg-ATPase activity (C) actin-activated Mg-ATPase activity (Vmax), (D) actin affinity relative to ATPase (Km) and (E) catalytic efficiency (Vmax/Km). Actin sliding velocity (F) was determined using the in vitro motility assay, with at least 30 filaments tracked per myosin preparation (N = 3). For all parameters, mean ± S.D. values are given. These were assessed for statistically significant differences by Student’s t-tests. *p<0.05, **p<0.01, ***p<0.001, ns, not significant.
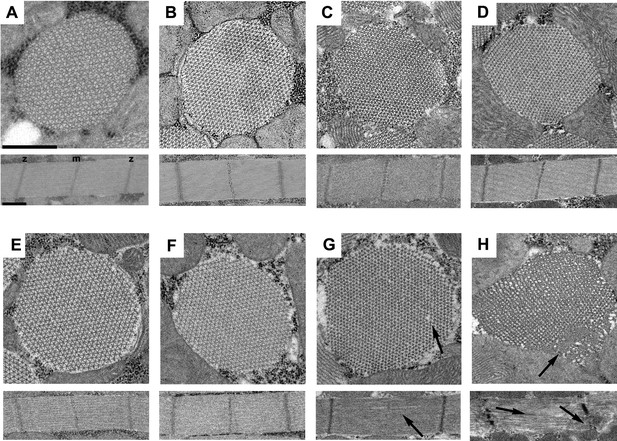
The R146N myosin mutation disrupts IFM myofibril stability.
Transverse and longitudinal sections are shown from the homozygous control line, PwMhc2 (A–D), and R146N homozygotes (E–H), for four different developmental stages: (A, E) late-stage pupae, (B, F) 2-hr-old adults, (C, G) 2-day-old adults, (D, H) 7-day-old adults. Each panel shows one myofibril that is representative of the population for that stage of development. Thick and thin filaments are arrayed in a normal double hexagonal pattern and well formed Z- and M-lines are observed in the sarcomeres at all stages in control organisms and at the late pupal and 2-hr-old adult stages for R146N. Mild disorder in the hexagonal packing of thick and thin filaments and some M-line disruption of sarcomeres (arrows) is observed in 2-day-old mutant adults (G). The structure of the mutant further deteriorates (arrows) in seven-day-old adults (H), with myofilament packing defects and strong disruption of both M- and Z-lines (m, m-line, z, z-line). All three R146N lines yielded similar phenotypes. Scale bars, 0.5 µm. Full genotypes are shown in parentheses: PwMhc2 homozygous control (P{PwMhc2}/P{PwMhc2}; Mhc10/Mhc10); R146N homozygote (Mhc10/Mhc10; P{R146N}/P{R146N}).
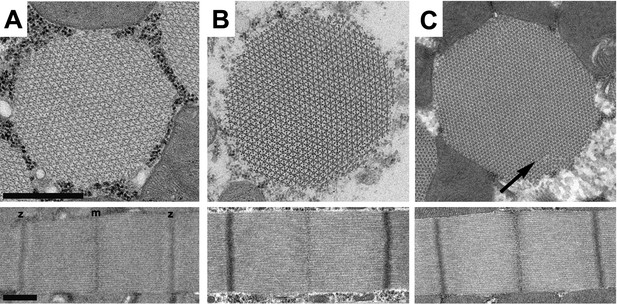
The R146N myosin mutation acts in a dominant fashion to cause minor disruption in IFM myofibril stability.
Transverse and longitudinal sections of IFM are shown from R146N/+ (full genotype: Mhc10/+; P{R146N}) (A–C), for three different developmental stages: (A) late-stage pupa, (B) 2-hr-old adult, (C) 2-day-old adult. Each panel shows one myofibril that is representative of the population for that stage of development. Thick and thin filaments are arrayed in a normal double hexagonal pattern and well formed Z- and M-lines are observed in the sarcomeres at the late pupal and 2-h-old adult stages. Mild disorder in the hexagonal packing of thick and thin filaments (arrow) is observed in 2-day-old R146N/+ heterozygotes (C). m, m-line, z, z-line. All three R146N lines yielded similar phenotypes. Scale bars, 0.5 μm.
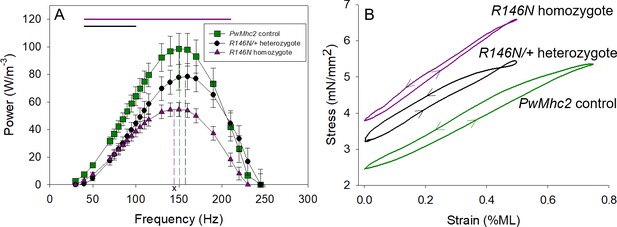
Power and work are decreased by the R146N mutation.
(A) Muscle power was measured by small amplitude, 0.125% muscle length (ML), sinusoidal analysis over a range of oscillation frequencies. Control fibers (PwMhc2 homozygotes) are represented by green squares, heterozygous fibers (R146N-15/+) by black circles and homozygous fibers (R146N-15/R146N-15) by purple triangles. Dashed lines in green, black and purple represent the respective fibers’ fmax. x Significant difference from R146N-15/+ heterozygotes at p<0.05. Bars indicate statistical significance from control, p<0.05, Student’s t-test. N = 12 for control and heterozygous fibers, 16 for homozygous fibers. (B) Work and power were measured using the work loop technique, which allows for variation in ML oscillation. In this representative example, the fibers were subjected to different ML amplitudes and oscillation frequencies until conditions were found that generated maximum power. The area within the loop is the amount of net work generated per cycle, and power is calculated by multiplying net-work times oscillation frequency. The control fiber (green) has a larger loop area than those of the heterozygous (black) or homozygous (purple) mutant fibers. Arrows show the counter-clockwise direction of the work loops indicating net work production rather than net work absorption. The mutant fibers perform best at 0.5% ML, 125 Hz and 150 Hz, for the R146N-15 homozygous and R146N-15/+ heterozygous fibers, respectively, while the control fiber produced maximum power at 0.75% ML, 125 Hz. Full genotypes are shown in parentheses: PwMhc2 homozygote control (P{PwMhc2}/P{PwMhc2}; Mhc10/Mhc10); R146N-15/+ heterozygote (Mhc10/+; P{R146N-15}); R146N-15 homozygote (Mhc10/Mhc10; P{R146N-15}/P{R146N-15}).
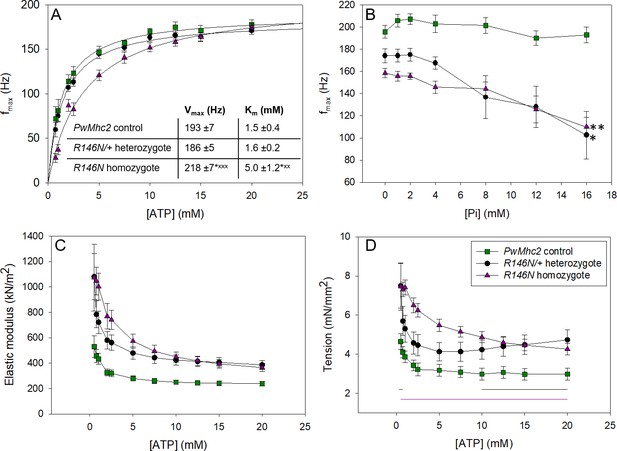
Varying [ATP] and [Pi] reveals information about cross-bridge kinetics.
(A) The frequency at which small amplitude sinusoidal power was maximal, fmax, was measured for a range of ATP concentrations for control (PwMhc2 homozygotes), heterozygous (R146N-15/+) and homozygous (R146N-15/R146N-15) fibers. The curves were fit with the Michaelis-Menten equation and Vmax and Km values determined (see inset). Student’s t-test with p<0.05 significantly different from control (*p<0.05, **p<0.01, ***p<0.001) or heterozygote (xp<0.05, xxp<0.01, xxxp<0.001). (B) fmax relative to inorganic phosphate concentration. fmax decreased with increasing [Pi], suggesting phosphate is competing with ATP for the rigor state. * indicates statistically significant difference from 0 mM Pi (Student’s t-test with p<0.05 significantly different (*p<0.05, **p<0.01). Linear regression analysis showed mutants had a steeper decline of fmax and larger negative slopes than the control. Seven control, 12 heterozygous, and seven homozygous fibers were tested. (C) Change in active stiffness with increasing ATP concentration. In-phase stiffness (elastic modulus) was measured at pCa 5.0 at 500 Hz ML oscillation using small amplitude sinusoidal analysis. All R146N stiffness values are statistically greater than the control’s by a Student’s t-test at p<0.05. (D) Active isometric tension generation as a function of ATP concentration at pCa 5.0. For the ATP study, N = 12 control, 12 heterozygous and 14 homozygous fibers. Black and purple horizontal lines indicate [ATP] where heterozygous and homozygous tension values are statistically greater than control’s (t-test, p<0.05). Full genotypes are shown in parentheses: PwMhc2 homozygote control (P{PwMhc2}/P{PwMhc2}; Mhc10/Mhc10); R146N-15/+ heterozygote (Mhc10/+; P{R146N-15}); R146N-15 homozygote (Mhc10/Mhc10; P{R146N-15}/P{R146N-15}).
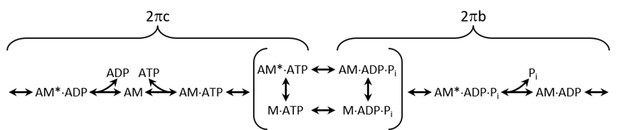
Acto-myosin cross-bridge cycle scheme.
A is actin, M is myosin and Pi is inorganic phosphate. Asterisks indicate a second conformational state with the same nucleotide. Muscle apparent rate constants 2πb and 2πc are derived from sinusoidal analysis of Drosophila IFM fibers (Swank, 2012). The primary cross-bridge transitions that influence 2πb and 2πc are bracketed. Cross-bridge transitions on the right hand side generally produce work (Pi release and the power stroke) in the muscle and hence primarily influence 2πb, while those on the left side of the diagram generally absorb work (ADP release, ATP induced dissociation of actin and myosin) and primarily influence 2πc.
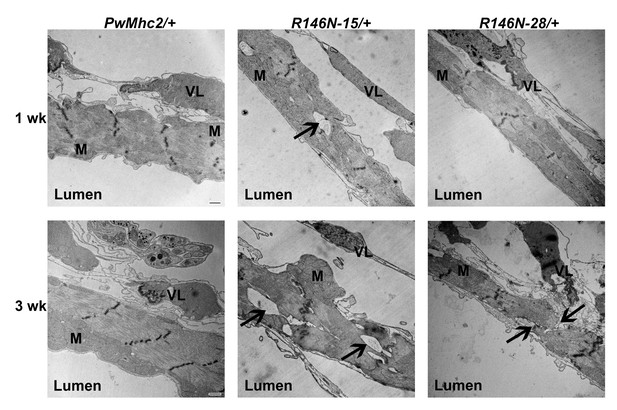
Transmission electron microscopy images of hearts of 1- and 3-week-old PwMhc2/+ control and R146N/+ mutant heterozygote lines.
Micrographs show transverse images through the cardiac tube between the second and third sets of ostia. M, mitochondria. VL, supportive ventral-longitudinal fibers. ‘Lumen’ refers to the hemolymph-containing center of the heart tube. Arrows indicate areas of myofibrillar discontinuities, which are only observed in mutant hearts. Scale bars, 500 nm. Full genotypes are shown in parentheses: PwMhc2/+ (P{PwMhc2}; Mhc1/+); R146N/+ (Mhc1/+; P{R146N}).
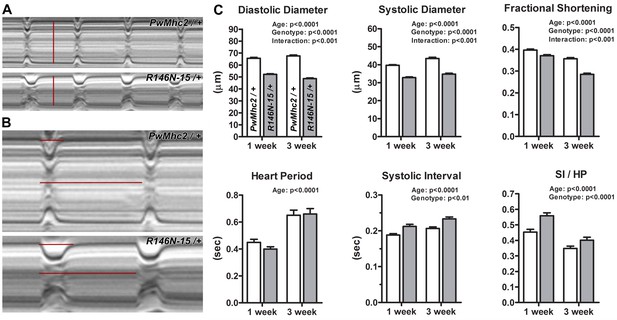
Expression of R146N myosin engenders a restricted cardiac physiology with diastolic dysfunction.
M-mode kymograms generated from high-speed videos of beating 3-week-old PwMhc2/+ and R146N-15/+ hearts. (A) Vertical red lines delineate diastolic diameters. Mutant hearts display cardiac restriction. (B) Horizontal lines demarcate systolic intervals (SI, top) and heart periods (HP, bottom). Mutant hearts display prolonged systolic phases. (C) R146N-15/+ Drosophila exhibit highly significant alterations in several cardiac functional parameters relative to PwMhc2/+ control flies. Both lines were examined at 1- and 3-weeks of age to track potential physiological defects and cardiac remodeling over time. Decreased cardiac dimensions, fractional shortening, and extended periods of systole are observed in 1- and 3-week-old mutants relative to controls. Data are presented as mean ±S.E.M. (N = 40–44 for each genotype and age group) and were evaluated using two-way ANOVAs with Bonferroni multiple comparisons tests. Significance was assessed at p<0.05. Full genotypes are shown in parentheses: PwMhc2/+ (P{PwMhc2}; Mhc1/+); R146N/+ (Mhc1/+; P{R146N}).
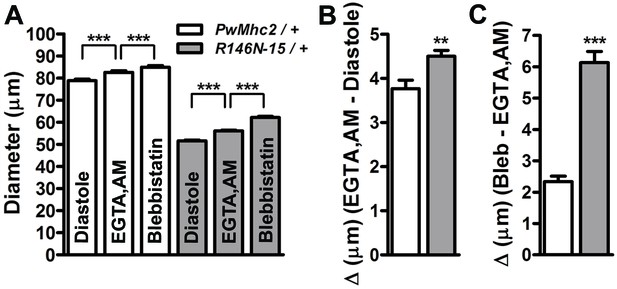
Excessive Ca2+-dependent and Ca2+-independent actomyosin associations during diastole promote incomplete relaxation of R146N-15/+ cardiomyocytes.
(A) Significant, incremental increases in cardiac diameters (repeated measures ANOVA with Bonferroni’s post-hoc tests) occur for PwMhc2/+ and R146N-15/+ hearts following extra- and intra-cellular Ca2+ chelation (EGTA/EGTA,AM) and again, upon blebbistatin incubation (***p≤0.0001). (B) The change in diameter across the heart wall in response to EGTA/EGTA,AM is higher in R146N-15/+ cardiac tubes, indicative of impaired Ca2+ homeostasis and a Ca2+-dependent increase in resting myocardial tension contributing to the restricted cardiac diameter (**p≤0.01). (C) Addition of blebbistatin after chelation of extra- and intracellular Ca2+ also prompted a significantly greater increase in cardiac dimensions of R146N-15/+ hearts relative to that observed for PwMhc2/+ hearts (***p≤0.0001). This implies that excessive Ca2+-independent cross-bridge cycling occurs in R146N-15/+ hearts, resulting in further enhanced resting basal tension and incomplete relaxation. Data are presented as mean ± S.E.M. (N = 20). The effects of EGTA, EGTA-AM and blebbistatin treatment on diameters were evaluated using paired t-tests. Unpaired two-tailed t-tests were used to distinguish significant differences in cardiac responses to EGTA, EGTA.AM and blebbistatin between PwMhc2/+ and R146N-15/+ hearts. Significance was assessed at p<0.05. Full genotypes are shown in parentheses: PwMhc2/+ (P{PwMhc2}; Mhc1/+); R146N/+ (Mhc1/+; P{R146N}).
Tables
Small amplitude power, work, and isometric tension.
Means ± S.E.M are reported. Student’s t-test with p<0.05 significantly different from control (*p<0.05, **p<0.01, ***p<0.001) or heterozygote (xp<0.05, xxp<0.01, xxxp<0.001). fmax: the frequency at which maximum power was generated, fWmax: frequency at which maximum work was generated as determined using small amplitude sinusoidal analysis. Passive tension was measured at pCa 8.0 and active at pCa 5.0. N = 12 fibers for control and mutant heterozygotes; N = 16 for mutant homozygotes. Full genotypes are shown in parentheses: PwMhc2 control (P{PwMhc2}/P{PwMhc2}; Mhc10/Mhc10); R146N-15/+ heterozygote (Mhc10/+; P{R146N-15}); R146N-15 homozygote (Mhc10/Mhc10; P{R146N-15}/P{R146N-15}).
| Line | Power (W/m3) | fmax (Hz) | Work (J/m3) | fWmax (Hz) | Passive Tension | Active Tension |
|---|---|---|---|---|---|---|
| PwMhc2 control | 100 ± 11 | 151 ± 3 | 0.71 ± 0.08 | 128 ± 2 | 1.94 ± 0.30 | 3.07 ± 0.32 |
| R146N-15/+ heterozygote | 81 ± 10 | 158 ± 4 | 0.54 ± 0.07 | 136 ± 4 | 3.16 ± 0.40* | 4.40 ± 0.51* |
| R146N-15 homozygote | 57 ± 5***x | 145 ± 4x | 0.41 ± 0.04** | 122 ± 4x | 2.73 ± 0.32 | 4.59 ± 0.29** |
Optimized work loops.
Muscle length (ML) change amplitude and frequency of large amplitude sinudoidal oscillations were varied until maximum power generating parameters were obtained. fPmax: the frequency at which maximum power was generated as determined using the work loop technique. Student’s t-test with p<0.05 significantly different from control (*p<0.05, **p<0.01, ***p<0.001) or heterozygote (xp<0.05, xxp<0.01, xxxp<0.001). N = 12 for control and mutant heterozygotes; N = 16 for mutant homozygotes. Full genotypes given in legend to Table 1.
| Line | Power (W/m3) | Net work (J/m3) | Work Gen. (J/m3) | Work abs. (J/m3) | % ML Amplitude | fPmax (Hz) |
|---|---|---|---|---|---|---|
| PwMhc2 control | 240 ± 34 | 2.18 ± 0.3 | 36.16 ± 4.19 | 33.98 ± 3.95 | 0.79 ± 0.06 | 115 ± 8 |
| R146N-15/+ heterozygote | 137 ± 20* | 0.93 ± 0.1*** | 21.92 ± 2.70** | 20.99 ± 2.59** | 0.5 ± 0.03*** | 150 ± 5** |
| R146N-15 homozygote | 85 ± 7***x | 0.65 ± 0.06*** | 22.50 ± 1.51** | 21.84 ± 1.46** | 0.52 ± 0.03*** | 130 ± 3xx |
Muscle apparent rate constants derived from sinusoidal analysis (Kawai and Brandt, 1980).
Means ± S.E.M are reported. Student’s t-test with p<0.05 significantly different from control (*p<0.05, **p<0.01, ***p<0.001). N = 12 for control and mutant heterozygotes; N = 16 for mutant homozygotes (see Figure 5—figure supplement 1 for an explanation of how the apparent rate constants relate to the cross-bridge cycle). Full genotypes given in legend to Table 1.
| Line | A | B | 2πb (s -1) | C | 2πc (s -1) |
|---|---|---|---|---|---|
| PwMhc2 control | 339 ± 23 | 741 ± 73 | 1036 ± 35 | 923 ± 76 | 5376 ± 169 |
| R146N-15/+ heterozygote | 305 ± 33 | 832 ± 75 | 1343 ± 52*** | 1009 ± 85 | 4928 ± 262 |
| R146N-15 homozygote | 304 ± 33 | 751 ± 87 | 1224 ± 80 | 934 ± 93 | 4671 ± 232* |
Flight ability and wing beat frequency (WBF) of R146N mutants.
Adult female flies were aged for 2 days prior to flight testing. Transgenic flies were then assayed for the ability to fly up (U), horizontal (H), down (D) or not at all (N). Flight index = 6 U/T+4 H/T+2 D/T+0 N/T; T is the total number of flies tested (given in parentheses). Flight index and WBF values are mean ±S.E.M. Student’s t-test with p<0.05 significantly different from 2-day-old PwMhc2 (*p<0.05, **p<0.01, ***p<0.001) or statistically different from heterozygous mutant (xp<0.05, xxp<0.01, xxxp<0.001). Full genotypes given in legend to Table 1.
| Line | Age (days) | Flight index 22°C | Flight index 15°C | WBF 22°C (Hz) | WBF 15°C (Hz) |
|---|---|---|---|---|---|
| PwMhc2 control | 2 | 4.1 ± 0.16 (111) | 2.3 ± 0.12 (111) | 196 ± 3 (10) | 156 ± 3 (10) |
| R146N-15/+ heterozygote | 2 | 3.5 ± 0.16 (111)*** | 2.0 ± 0.10 (111) | 194 ± 3 (10) | 150 ± 2 (10) |
| R146N-15 homozygote | 2 | 1.1 ± 0.11 (111)*** xxx | 1.1 ± 0.10 (111)***xxx | 182 ± 3 (10)*** xx | 141 ± 4 (10)** x |
Additional files
-
Supplementary file 1
Power generation measured using the work loop technique under the same muscle length (ML) and frequency parameters.
All three fiber types were oscillated through ten 0.75% ML amplitude and 125 Hz contraction cycles. These parameters are the optimal power producing parameters for most of the control fibers (average results are shown in Table 2). Means ± S.E.M are reported. N = 12 for the control and homozygous fibers, 13 for heterozygous fibers. Student’s t-test with p<0.05 significantly different from control (*p<0.05, **p<0.01, ***p<0.001) or heterozygote (xp<0.05, xxp<0.01, xxxp<0.001). Full genotypes are shown in parentheses: PwMhc2 control (P{PwMhc2}/P{PwMhc2}; Mhc10/Mhc10); R146N-15/+ heterozygote (Mhc10/+; P{R146N-15}); R146N-15 homozygote (Mhc10/Mhc10; P{R146N-15}/P{R146N-15}).
- https://doi.org/10.7554/eLife.38064.018
-
Supplementary file 2
Flight ability of R146N homozygous adults at two and seven days.
Adult female flies were aged for two or seven days prior to flight testing. Transgenic flies were then assayed for the ability to fly up (U), horizontal (H), down (D) or not at all (N). Flight index = 6 U/T+ 4 H/T+ 2 D/T+ 0 N/T; T is the total number of flies tested, listed in parentheses. Flight index is mean ±S.E.M. Student’s t-test with p<0.05 significantly different from same age PwMhc2 (*p<0.05, **p<0.01, ***p<0.001) or from same fly line at 2 days of age (xp<0.05, xxp<0.01, xxxp<0.001). Full genotypes are shown in parentheses: PwMhc2 (P{PwMhc2}/P{PwMhc2}; Mhc10/Mhc10); R146N (Mhc10/Mhc10; P{R146N}/P{R146N}).
- https://doi.org/10.7554/eLife.38064.019
-
Supplementary file 3
Cardiomyocyte thickness measurements.
At least three hearts were analyzed per line and genotype, including three sections along the anterior-poster axis at least 10 microns apart per sample, and ≥3 images each for dorsal or ventral areas per section analyzed. Means ± S.E.M are reported. A one-way ANOVA with the Bonferroni correction determined that no statistically significant differences (p<0.05) exist for any of the comparisons within or between samples at the same or different ages. Full genotypes are shown in parentheses: PwMhc2/+ (P{PwMhc2}; Mhc1/+); R146N/+ (Mhc1/+; P{R146N}).
- https://doi.org/10.7554/eLife.38064.020
-
Supplementary file 4
Cardiac parameters for R146N-15/+ and R146N-28/+ mutant lines relative to PwMhc2/+ control.
Means ± S.E.M are reported. Two-way ANOVA results summarizing the statistical differences between the control and mutant flies can be found in Figure 7. No significant differences (p<0.05) in any cardiac physiological indices reported were noted between the two mutant lines via two-way ANOVA. Full genotypes are shown in parentheses: PwMhc2/+ (P{PwMhc2}; Mhc1/+); R146N/+ (Mhc1/+; P{R146N}).
- https://doi.org/10.7554/eLife.38064.021
-
Transparent reporting form
- https://doi.org/10.7554/eLife.38064.022





