Structural basis for Scc3-dependent cohesin recruitment to chromatin
Figures
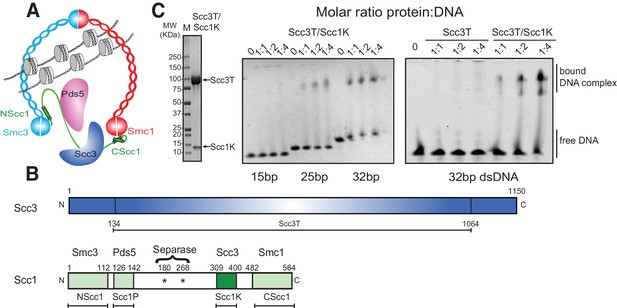
DNA binding by the Scc3-Scc1 subcomplex.
(A) Cartoon of the cohesin complex. (B) Domain structure of Scc3 and Scc1. Construct boundaries used and their acronyms are shown below. (C) SDS-PAGE analysis of purified Scc3T-Scc1K and DNA binding analysis by EMSA. Scc3 binds to longer DNA more efficiently compared to shorter DNA. The DNA binding capacity of Scc3T is enhanced by Scc1K.
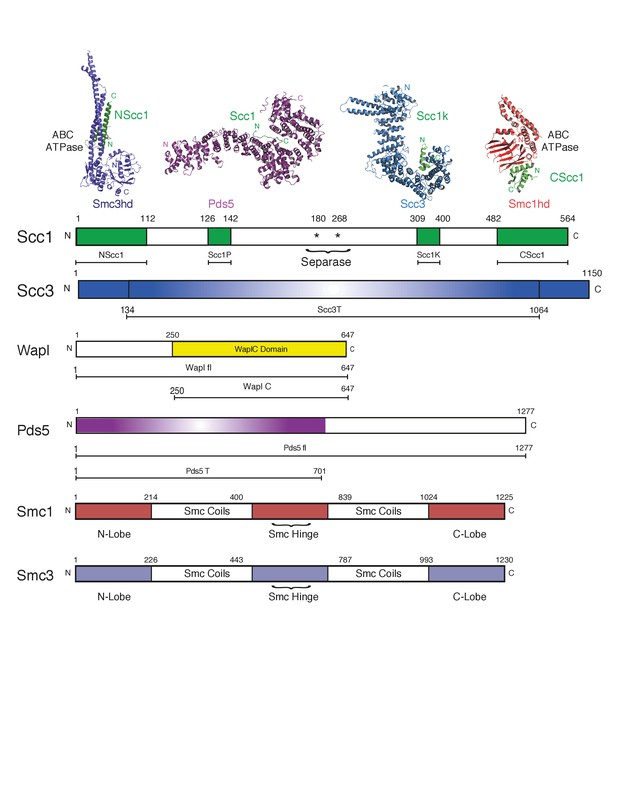
Cartoon of domain boundaries and constructs used.
The domain boundaries of different cohesin components and subcomplexes are indicated. Expression constructs and acronyms used in the text are indicated below each cohesin component. NScc1 comprises an expression construct spanning amino acids 1–112 of Scc1. Scc1P (amino acids 126–142). Scc1K (amino acids 309–400). CScc1 (amino acids 482–564). Scc3T (amino acids 134–1064). Wapl fl (amino acids 1–647). WaplC (amino acids 250–647). Pds5 fl (amino acids 1–1277). Pds5T (amino acids 1–701).
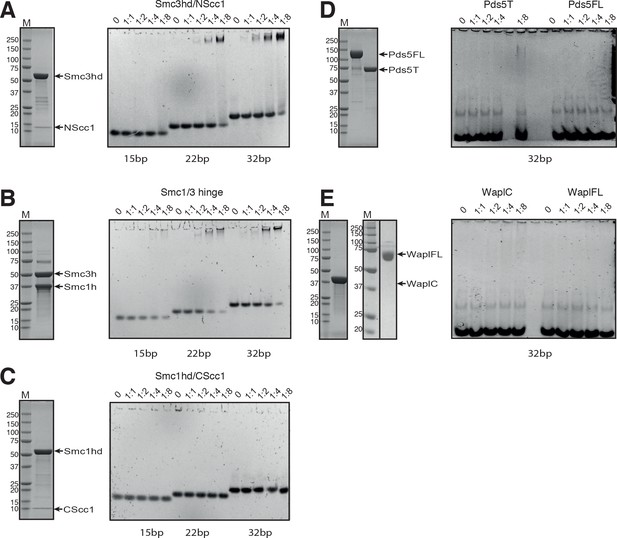
SDS-PAGE analysis of the purified cohesin components and DNA binding analysis by EMSA.
(A) SDS-PAGE and DNA binding analysis of the Smc3hd/NScc1 complex. (B) SDS-PAGE and DNA binding analysis of the Smc3/Smc1 hinge heterodimer. (C) SDS-PAGE of the purified Smc1hd/CScc1 complex and DNA binding analysis. (D) SDS-PAGE and DNA binding analysis of Pds5FL and Pds5T. (E) SDS-PAGE and DNA binding analysis of WaplC and WaplFL. For the gel showing WaplFL, the black vertical line indicates the position where the gel has been cropped. Concentration of the DNA duplex in the EMSA analysis was 1 μM. The DNA was incubated at different molar ratios, as indicated on top of each lane, with the indicated protein preparation. Native EMSA PAGE gels were stained with Sybr Safe Nucleic Acid Gel Stain. SDS-PAGE gels were stained with Coomassie.
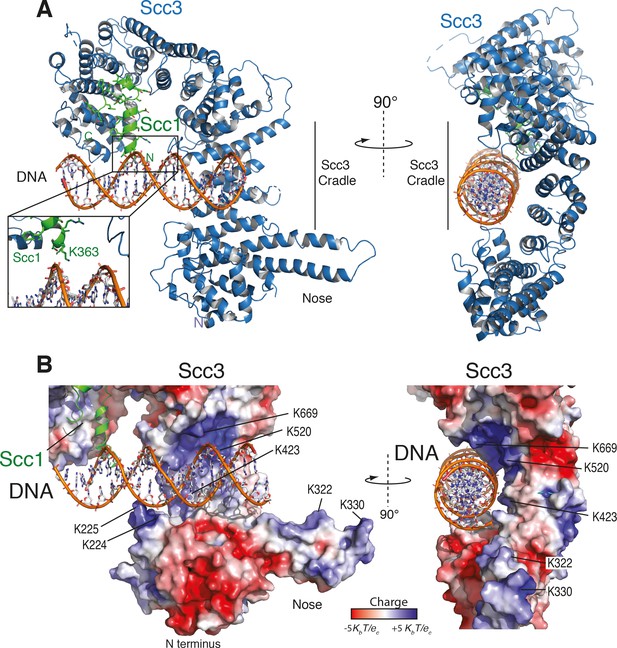
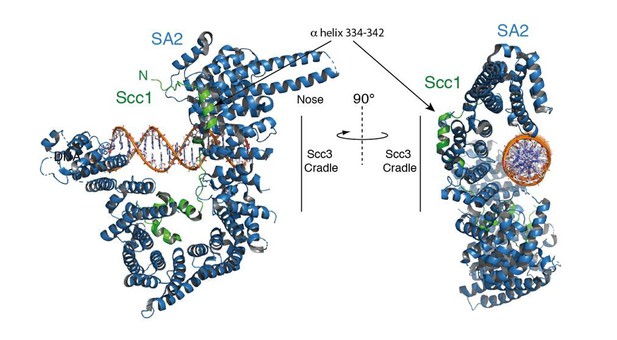
Structure of the Scc3-Scc1 subcomplex bound to DNA.
(A) Cartoon representation of the Scc3-Scc1 complex bound to a 19 bp dsDNA substrate. The N- and C- termini of Scc3 (violet) and Scc1 subunits (green) are shown. The inset shows a close-up view of the Scc1 amino acid K363. (B) Electrostatic surface potential representation of the Scc3-Scc1 subcomplex with bound dsDNA (calculated with APBS and displayed with Pymol).
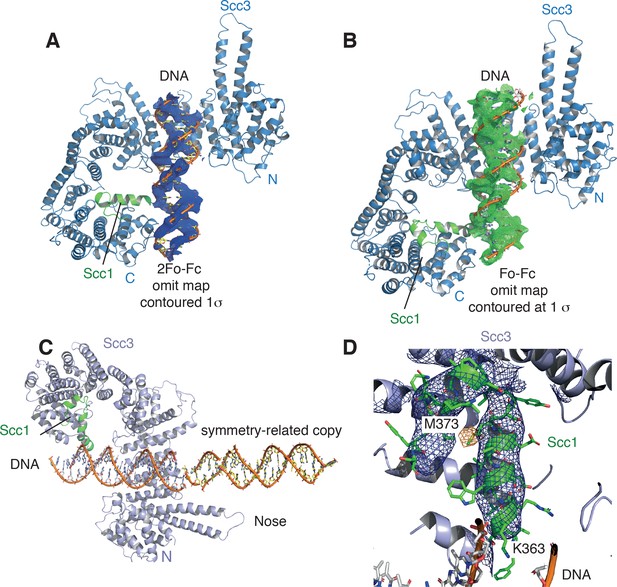
Electron density for the DNA molecule bound to the Scc3-Scc1 subcomplex.
(A) View of the DNA binding site of the Scc3-Scc1 subcomplex. DNA is shown as stick, along with its 2Fo-Fc electron density Fourier map (blue mesh) contoured at 1.0 σ. (B) Fo-Fc omit electron density Fourier map (grey mesh) contoured at 1.0 σ. (C) N-terminal ‘nose’ of Scc3 which interacts through a set of positively charged amino acid residues with the DNA of a neighbouring complex related by crystallographic symmetry. (D) Anomalous difference Fourier map (yellow mesh) contoured at 2.8 σ of a selenomethionine–substituted crystal, confirming the location of Scc1 residue M373. Blue mesh: 2Fo-Fc electron density Fourier map around Scc1 contoured at 1.5 s. K363 of Scc1 is located in close proximity to the DNA phosphate backbone.

A conserved DNA binding domain in the Scc3-Scc1 subcomplex is required for cohesin association with chromatin.
(A) Surface amino acid conservation of yeast Scc3. Residues in the DNA binding domain are well conserved. (B) DNA binding residues are located in three surface patches of Scc3. (C) DNA binding fluorescence polarization of 6-FAM labelled 32 bp dsDNA by variants of the Scc3-Scc1 subcomplex. Data points corresponding to the average of three independent experiments were fitted to a standard binding equation assuming a single binding site using Kaleidagraph. Standard deviations are depicted as vertical error bars. Apparent dissociation constants (KD) are noted below. (D) Tetrad analysis of diploid budding yeast strains expressing ectopic wild-type or mutant versions of Scc3 under control of the endogenous promoter in an SCC3/scc3Δ background (strains C5013, C5014, C5015, C5043, C5033). Images were recorded after three days at 30°C on rich media. Genetic marker analysis identified Scc3(mutant), scc3Δ cells (circles). (E) ChIP-qPCR analysis of binding to centromeric (cen), pericentromeric (pericen) or chromosome arm (arm) regions (chromosomes IV, V, and VI as indicated) of untagged (strain C3) or PK6-tagged wild-type or mutant versions of Scc3 expressed from an ectopic locus under its endogenous promoter (strains C5013, C5043, C5033). The fractions of immunoprecipitated DNA relative to input DNA are plotted as circles for two biological repeats with two technical repeats each (same colour pairs). Mean values of all four data points are shown as lines.
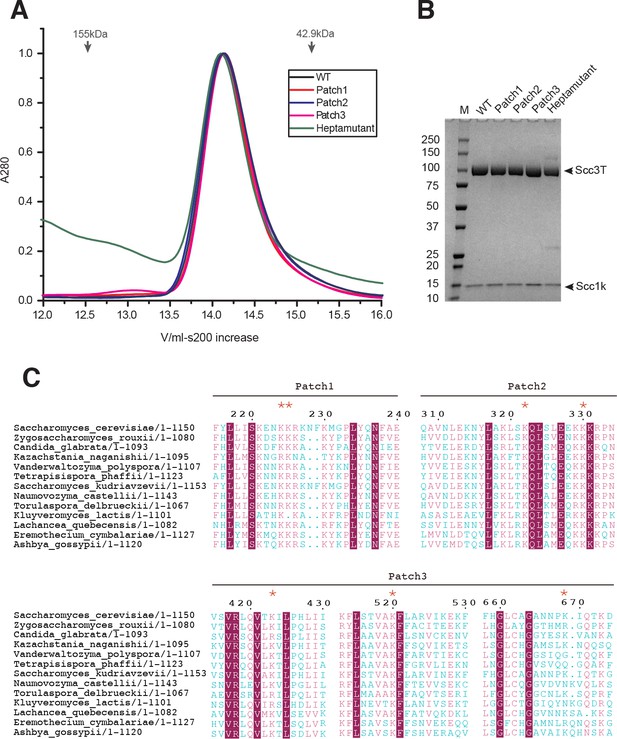
Biochemical analysis of Scc3-Scc1 subcomplexes and Scc3 conservation alignment.
(A) Size-exclusion chromatography of Scc1-Scc1 complexes. Arrows indicate the elution volume of molecular weight size markers. (B) SDS-PAGE analysis of Scc3-Scc1 complexes from peak fractions in panel (A), showing that the mutants do not impact Scc1 binding in vitro. (C) Alignment of Scc3 amino acid sequences. Residues that that were probed by mutagenesis are labeled with red stars (*). Magenta boxes indicate fully conserved amino acid residues while chemically conserved residues are colored magenta. Blue residues indicate divergent residues.
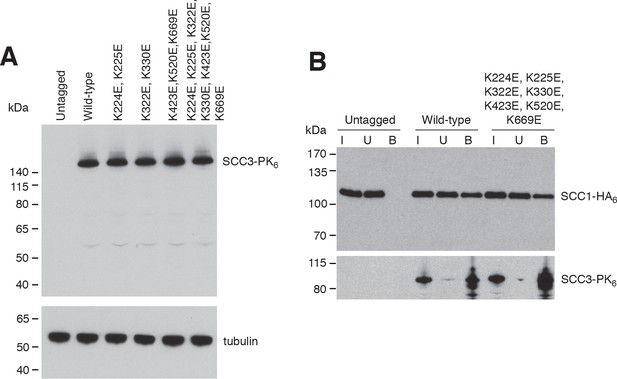
In vivo analysis of SCC3 mutants.
(A) Protein levels of ectopically expressed wild-type and mutant versions of Scc3 tagged with PK6 (strains C1073, C5013, C5014, C5015, C5043, C5033) measured by western blotting of whole cell lysates against the PK tag (top) and α-tubulin as loading control (bottom). (B) Co-immunoprecipitation of Scc1-HA6 with wild-type or mutant versions of PK6-tagged Scc3 expressed from an ectopic locus under the endogenous promoter (strains C501, C5165, C5166).
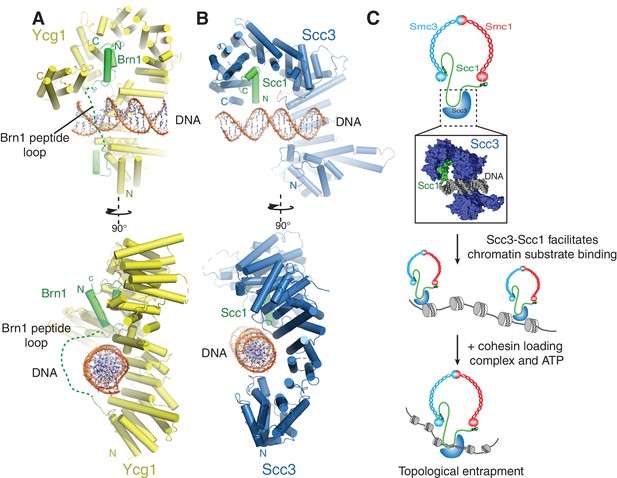
A conserved DNA binding interface in cohesin and condensin.
(A) Structure of the DNA-bound Ycg1–Brn1 subcomplex from condensin. (B) Structure of the Scc3-Scc1 complex. In the condensin structure, a peptide loop (green dashed line) of the Brn1 kleisin subunit encircles the bound DNA and prevents its dissociation. Alignments were generated by secondary structure matching using Cα atoms from the Scc3 HEAT-repeats and the structurally equivalent region of the condensin Ycg1 HEAT-repeat subunit. (C) Model for Scc3-mediated DNA binding by cohesin complexes. Scc3-Scc1 enables direct chromatin binding. Cohesin is loaded by Scc2-Scc4 in an ATP dependent fashion thus resulting in topological DNA entrapment.
Tables
Data collection and refinement statistics.
https://doi.org/10.7554/eLife.38356.005| Scc3T/Scc1K native | Scc3T/Scc1K SeMet | |
|---|---|---|
| Data collection | ||
| Space group | P21212 | P21212 |
| Cell dimensions | ||
| a, b, c (Å) | 109.9, 115.4, 295.6 | 109.9, 115.6, 296.2 |
| Wavelength (Å) | 1.282 | 1.282 |
| Resolution (Å) | 50–3.60 | 49.9–4.79 |
| No. reflections | 20963 | 10279 |
| Rmerge | 5.8 (122.6)* | 4.6 (112.3)* |
| I / σI | 11.9 (1.6)* | 10.6 2(.1) |
| CC 1/2 | 0.99 (0.56) | 0.99 (0.52) |
| Completeness (%) | 91.4 (63.5)* | 93.6 (71.2)* |
| Redundancy | 4.4 (6.0)* | 1.8 (1.8) |
| Refinement | ||
| Resolution (Å) | 50–3.60 | |
| Rwork/Rfree | 0.28/0.31 | |
| No. atoms | 16465 | |
| Protein | 14909 | |
| DNA | 1556 | |
| B-factors (mean) | ||
| Protein | 254.5 | |
| DNA | 266.4 | |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.002 | |
| Bond angles (°) | 0.5 |
-
*Values in parentheses are for highest-resolution shell.
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.38356.012
-
Supplementary file 1
DNA substrates
- https://doi.org/10.7554/eLife.38356.013
-
Supplementary file 2
Yeast genotypes.
All strains are derivatives of W303.
- https://doi.org/10.7554/eLife.38356.014
-
Supplementary file 3
ChIP-qPCR primer sequences (5’→3’).
- https://doi.org/10.7554/eLife.38356.015





